Identification and RNAi Profile of a Novel Iflavirus Infecting Senegalese Aedes vexans arabiensis Mosquitoes
Abstract
1. Introduction
2. Materials and Methods
2.1. Mosquito Samples and Collections
2.2. RNA Extraction and Small RNA Deep Sequencing
2.3. Bioinformatics: Virus Discovery and RNAi Profile Analysis
2.4. Virus Genome Annotation and Phylogenetic Analysis
2.5. Construction and Validation of an AvIFV RT-PCR Screen for Ae. vexans spp. Mosquitoes
3. Results
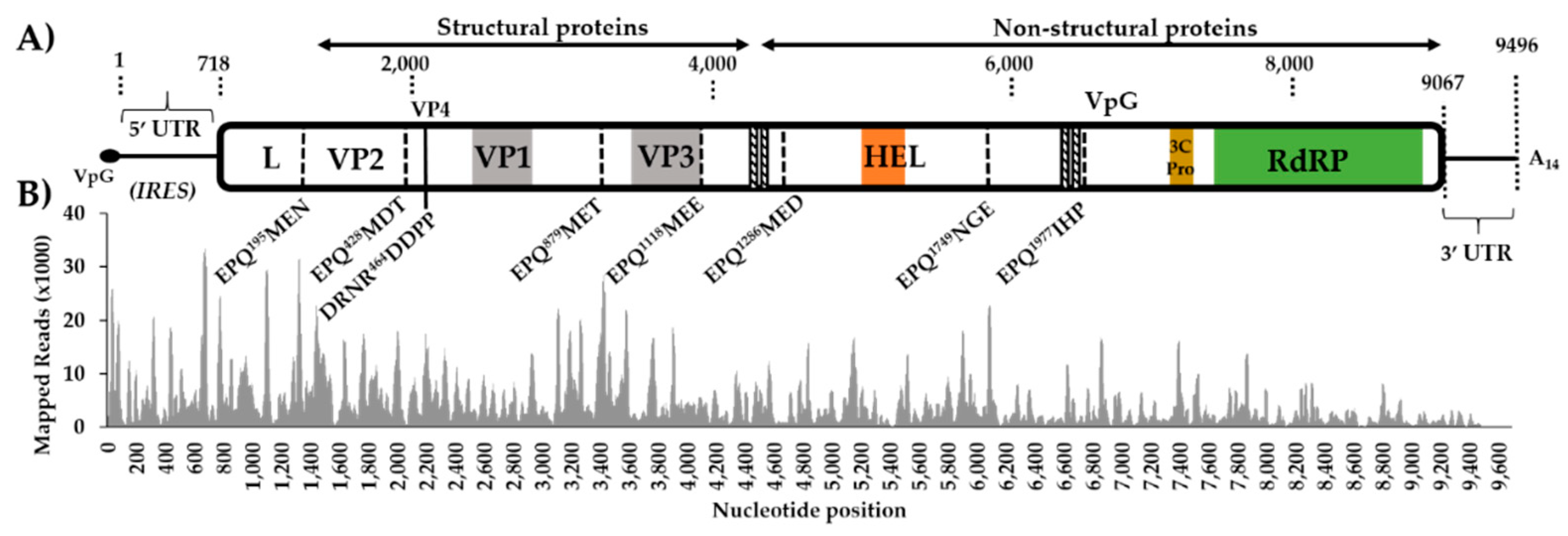
3.1. Aedes vexans arabiensis Mosquitoes from Barkédji Village, Senegal, Were infected with a Novel Iflavirus
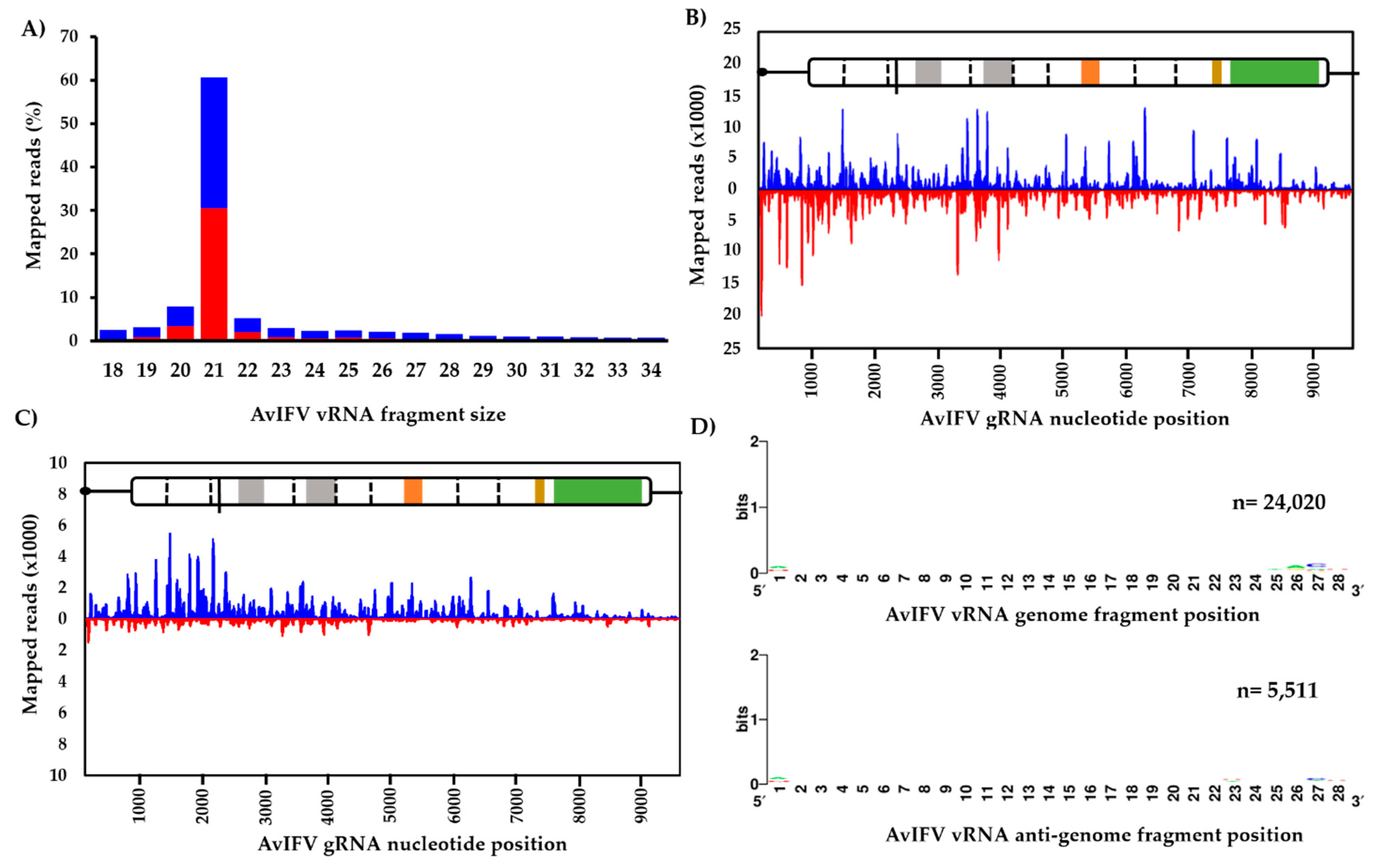
3.2. The Virus-Derived Small RNA Profile of Aedes Vexans Iflavirus
3.3. RT-PCR Screening of Ae. vexans spp. Samples from Senegal, Italy, the United Kingdom, Germany, and Sweden Suggests AvIFV is Exclusively Present in Ae. vexans Samples from Senegal
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reinert, J.F. Contributions to the mosquito fauna of Southeast Asia. XVI. Genus Aedes Meigen, subgenus Aedimorphus Theobald in Southeast Asia; Walter Reed Army Institute of Research: Washington, DC, USA, 1973. [Google Scholar]
- Johansen, C.A.; Lindsay, M.D.; Harrington, S.A.; Whelan, P.I.; Russell, R.C.; Broom, A.K. First record of Aedes (Aedimorphus) vexans vexans (Meigen) in Australia. J. Am. Mosq. Control Assoc. 2005, 21, 222–224. [Google Scholar] [CrossRef]
- White, G.B. Notes on a catalogue of Culicidae of the Ethiopian region. Mosq. Syst. 1975, 7, 303–344. [Google Scholar]
- Ndiaye el, H.; Fall, G.; Gaye, A.; Bob, N.S.; Talla, C.; Diagne, C.T.; Diallo, D.; B, A.Y.; Dia, I.; Kohl, A.; et al. Vector competence of Aedes vexans (Meigen), Culex poicilipes (Theobald) and Cx. quinquefasciatus Say from Senegal for West and East African lineages of Rift Valley fever virus. Parasites Vectors 2016, 9, 94. [Google Scholar] [CrossRef]
- Rodl, P.; Bardos, V.; Ryba, J. Experimental transmission of Tahyna virus (California group) to wild rabbits (Oryctolagus cuniculus) by mosquitoes. Folia Parasitol. 1979, 26, 61–64. [Google Scholar]
- Turell, M.J.; Dohm, D.J.; Sardelis, M.R.; Oguinn, M.L.; Andreadis, T.G.; Blow, J.A. An update on the potential of North American mosquitoes (Diptera: Culicidae) to transmit West Nile virus. J. Med. Entomol. 2005, 42, 57–62. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.L.; Bixby, M.A.; Morin, K.J.; Bradley, D.S.; Vaughan, J.A. Potential of a northern population of Aedes vexans (Diptera: Culicidae) to transmit Zika virus. J. Med. Entomol. 2017, 54, 1354–1359. [Google Scholar] [CrossRef] [PubMed]
- Takashima, I.; Hashimoto, N.; Arikawa, J.; Matsumoto, K. Getah virus in Aedes vexans nipponii and Culex tritaeniorhynchus: Vector susceptibility and ability to transmit. Arch. Virol. 1983, 76, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, R.; Edman, J.D.; Cooper, L.A.; Scott, T.W. Vector competence of mosquitoes (Diptera: Culicidae) from Massachusetts for a sympatric isolate of eastern equine encephalomyelitis virus. J. Med. Entomol. 1997, 34, 346–352. [Google Scholar] [CrossRef]
- Bockova, E.; Rudolf, I.; Kocisova, A.; Betasova, L.; Venclikova, K.; Mendel, J.; Hubalek, Z. Dirofilaria repens microfilariae in Aedes vexans mosquitoes in Slovakia. Parasitol. Res. 2013, 112, 3465–3470. [Google Scholar] [CrossRef]
- Shi, M.; Neville, P.; Nicholson, J.; Eden, J.S.; Imrie, A.; Holmes, E.C. High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J. Virol. 2017, 91, e00680-17. [Google Scholar] [CrossRef]
- Zakrzewski, M.; Rasic, G.; Darbro, J.; Krause, L.; Poo, Y.S.; Filipovic, I.; Parry, R.; Asgari, S.; Devine, G.; Suhrbier, A. Mapping the virome in wild-caught Aedes aegypti from Cairns and Bangkok. Sci. Rep. 2018, 8, 4690. [Google Scholar] [CrossRef] [PubMed]
- Parry, R.; Asgari, S. Aedes anphevirus: an insect-specific virus distributed worldwide in Aedes aegypti mosquitoes that has complex interplays with Wolbachia and dengue virus infection in cells. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Asad, S.; Khromykh, A.A.; Asgari, S. Cell fusing agent virus and dengue virus mutually interact in Aedes aegypti cell lines. Sci. Rep. 2017, 7, 6935. [Google Scholar] [CrossRef] [PubMed]
- Baidaliuk, A.; Miot, E.F.; Lequime, S.; Moltini-Conclois, I.; Delaigue, F.; Dabo, S.; Dickson, L.B.; Aubry, F.; Merkling, S.H.; Cao-Lormeau, V.-M.; et al. Cell-fusing agent virus reduces arbovirus dissemination in Aedes aegypti mosquitoes in vivo. J. Virol. 2019, 93, e00705–e00719. [Google Scholar] [CrossRef]
- Hall-Mendelin, S.; McLean, B.J.; Bielefeldt-Ohmann, H.; Hobson-Peters, J.; Hall, R.A.; van den Hurk, A.F. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasites Vectors 2016, 9, 414. [Google Scholar] [CrossRef]
- Sanborn, M.A.; Klein, T.A.; Kim, H.C.; Fung, C.K.; Figueroa, K.L.; Yang, Y.; Asafo-Adjei, E.A.; Jarman, R.G.; Hang, J. Metagenomic analysis reveals three novel and prevalent mosquito viruses from a single pool of Aedes vexans nipponii Collected in the Republic of Korea. Viruses 2019, 11, 222. [Google Scholar] [CrossRef]
- Lee, J.S.; Grubaugh, N.D.; Kondig, J.P.; Turell, M.J.; Kim, H.C.; Klein, T.A.; O’Guinn, M.L. Isolation and genomic characterization of Chaoyang virus strain ROK144 from Aedes vexans nipponii from the Republic of Korea. Virology 2013, 435, 220–224. [Google Scholar] [CrossRef]
- Asgari, S. microRNAs as regulators of insect host-pathogen interactions and immunity. Adv. Insect Physiol. 2018, 55, 19–45. [Google Scholar] [CrossRef]
- Bronkhorst, A.W.; van Rij, R.P. The long and short of antiviral defense: Small RNA-based immunity in insects. Curr. Opin. Virol. 2014, 7, 19–28. [Google Scholar] [CrossRef]
- Varjak, M.; Leggewie, M.; Schnettler, E. The antiviral piRNA response in mosquitoes? J. Gen. Virol. 2018, 99, 1551–1562. [Google Scholar] [CrossRef]
- Sabin, L.R.; Zheng, Q.; Thekkat, P.; Yang, J.; Hannon, G.J.; Gregory, B.D.; Tudor, M.; Cherry, S. Dicer-2 processes diverse viral RNA species. PLoS ONE 2013, 8, e55458. [Google Scholar] [CrossRef] [PubMed]
- Barnard, A.-C.; Nijhof, A.M.; Fick, W.; Stutzer, C.; Maritz-Olivier, C. RNAi in arthropods: insight into the machinery and applications for understanding the pathogen-vector interface. Genes 2012, 3, 702–741. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, E.R.; Olmo, R.P.; Paro, S.; Ferreira, F.V.; de Faria, I.J.; Todjro, Y.M.; Lobo, F.P.; Kroon, E.G.; Meignin, C.; Gatherer, D.; et al. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res. 2015, 43, 6191–6206. [Google Scholar] [CrossRef] [PubMed]
- van Oers, M.M. Genomics and biology of iflaviruses. In Insect Virology; Caister Academic Press: Norfolk, UK, 2010; pp. 231–250. [Google Scholar]
- Kobayashi, D.; Isawa, H.; Fujita, R.; Murota, K.; Itokawa, K.; Higa, Y.; Katayama, Y.; Sasaki, T.; Mizutani, T.; Iwanaga, S.; et al. Isolation and characterization of a new iflavirus from Armigeres spp. mosquitoes in the Philippines. J. Gen. Virol. 2017, 98, 2876–2881. [Google Scholar] [CrossRef]
- Dietrich, I.; Jansen, S.; Fall, G.; Lorenzen, S.; Rudolf, M.; Huber, K.; Heitmann, A.; Schicht, S.; Ndiaye, E.; Watson, M.; et al. RNA interference restricts Rift Valley Fever virus in multiple insect systems. Msphere 2017, 2, e00090. [Google Scholar] [CrossRef]
- Modlmaier, M.; Kuhn, R.; Kaaden, O.-R.; Pfeffer, M. Transmission studies of a European Sindbis virus in the floodwater mosquito Aedes vexans (Diptera: Culicidae). Int. J. Med. Microbiol. 2002, 291, 164–170. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Chilton, J.M.; Gruning, B.; Johnson, J.E.; Soranzo, N. NCBI BLAST plus integrated into Galaxy. Gigascience 2015, 4, 39. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.D.; Vasilakis, N.; Tian, J.H.; Li, C.X.; Chen, L.J.; Eastwood, G.; Diao, X.N.; Chen, M.H.; Chen, X.; et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 2016, 90, 659–669. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Kall, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Kolekar, P.; Pataskar, A.; Kulkarni-Kale, U.; Pal, J.; Kulkarni, A. IRESPred: Web server for prediction of cellular and viral Internal Ribosome Entry Site (IRES). Sci. Rep. 2016, 6, 27436. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchene, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kuhnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef]
- Staley, M.; Dorman, K.S.; Bartholomay, L.C.; Fernández-Salas, I.; Farfan-Ale, J.A.; Loroño-Pino, M.A.; Garcia-Rejon, J.E.; Ibarra-Juarez, L.; Blitvich, B.J. Universal primers for the amplification and sequence analysis of actin-1 from diverse mosquito species. J. Am. Mosq. Control Assoc. 2010, 26, 214–218. [Google Scholar] [CrossRef]
- Reineke, A.; Asgari, S. Presence of a novel small RNA-containing virus in a laboratory culture of the endoparasitic wasp Venturia canescens (Hymenoptera: Ichneumonidae). J. Insect Physiol. 2005, 51, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef] [PubMed]
- Ongus, J.R.; Peters, D.; Bonmatin, J.M.; Bengsch, E.; Vlak, J.M.; van Oers, M.M. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 2004, 85, 3747–3755. [Google Scholar] [CrossRef] [PubMed]
- Isawa, H.; Asano, S.; Sahara, K.; Iizuka, T.; Bando, H. Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: Evidence for evolutionary relationships among insect, mammalian and plant picorna(-like) viruses. Arch. Virol. 1998, 143, 127–143. [Google Scholar] [CrossRef]
- Ye, S.; Xia, H.; Dong, C.; Cheng, Z.; Xia, X.; Zhang, J.; Zhou, X.; Hu, Y. Identification and characterization of Iflavirus 3C-like protease processing activities. Virology 2012, 428, 136–145. [Google Scholar] [CrossRef]
- Luke, G.A.; de Felipe, P.; Lukashev, A.; Kallioinen, S.E.; Bruno, E.A.; Ryan, M.D. Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J. Gen. Virol. 2008, 89, 1036–1042. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef]
- Chejanovsky, N.; Ophir, R.; Schwager, M.S.; Slabezki, Y.; Grossman, S.; Cox-Foster, D. Characterization of viral siRNA populations in honey bee colony collapse disorder. Virology 2014, 454–455, 176–183. [Google Scholar] [CrossRef]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Jakubowska, A.K.; D’Angiolo, M.; Gonzalez-Martinez, R.M.; Millan-Leiva, A.; Carballo, A.; Murillo, R.; Caballero, P.; Herrero, S. Simultaneous occurrence of covert infections with small RNA viruses in the lepidopteran Spodoptera exigua. J. Invertebr. Pathol. 2014, 121, 56–63. [Google Scholar] [CrossRef]
- Geng, P.; Li, W.; de Miranda, J.R.; Qian, Z.; An, L.; Terenius, O. Studies on the transmission and tissue distribution of Antheraea pernyi iflavirus in the Chinese oak silkmoth Antheraea pernyi. Virology 2017, 502, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Brettell, L.E.; Pachori, P.; Villalobos, E.M.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Moku virus; a new Iflavirus found in wasps, honey bees and Varroa. Sci. Rep. 2016, 6, 34983. [Google Scholar] [CrossRef] [PubMed]
- Cholleti, H.; Hayer, J.; Fafetine, J.; Berg, M.; Blomstrom, A.L. Genetic characterization of a novel picorna-like virus in Culex spp. mosquitoes from Mozambique. Virol. J. 2018, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Virto, C.; Navarro, D.; Tellez, M.M.; Herrero, S.; Williams, T.; Murillo, R.; Caballero, P. Natural populations of Spodoptera exigua are infected by multiple viruses that are transmitted to their offspring. J. Invertebr. Pathol. 2014, 122, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef]
- Bolling, B.G.; Olea-Popelka, F.J.; Eisen, L.; Moore, C.G.; Blair, C.D. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 2012, 427, 90–97. [Google Scholar] [CrossRef]
- Hobson-Peters, J.; Yam, A.W.; Lu, J.W.; Setoh, Y.X.; May, F.J.; Kurucz, N.; Walsh, S.; Prow, N.A.; Davis, S.S.; Weir, R.; et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE 2013, 8, e56534. [Google Scholar] [CrossRef]
- Kent, R.J.; Crabtree, M.B.; Miller, B.R. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex Flavivirus Izabal. PLoS Negl. Trop. Dis. 2010, 4, e671. [Google Scholar] [CrossRef]
- Talavera, S.; Birnberg, L.; Nunez, A.I.; Munoz-Munoz, F.; Vazquez, A.; Busquets, N. Culex flavivirus infection in a Culex pipiens mosquito colony and its effects on vector competence for Rift Valley fever phlebovirus. Parasites Vectors 2018, 11, 310. [Google Scholar] [CrossRef]
- Jakubowska, A.K.; Murillo, R.; Carballo, A.; Williams, T.; van Lent, J.W.M.; Caballero, P.; Herrero, S. Iflavirus increases its infectivity and physical stability in association with baculovirus. PeerJ 2016, 4, e1687. [Google Scholar] [CrossRef]
- Carballo, A.; Murillo, R.; Jakubowska, A.; Herrero, S.; Williams, T.; Caballero, P. Co-infection with iflaviruses influences the insecticidal properties of Spodoptera exigua multiple nucleopolyhedrovirus occlusion bodies: Implications for the production and biosecurity of baculovirus insecticides. PLoS ONE 2017, 12, e0177301. [Google Scholar] [CrossRef] [PubMed]
- van Cleef, K.W.; van Mierlo, J.T.; Miesen, P.; Overheul, G.J.; Fros, J.J.; Schuster, S.; Marklewitz, M.; Pijlman, G.P.; Junglen, S.; van Rij, R.P. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 2014, 42, 8732–8744. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Wagner, V.; Rasmussen, S.B.; Hartmann, R.; Paludan, S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006, 80, 5059. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, I.; Shi, X.H.; McFarlane, M.; Watson, M.; Blomstrom, A.L.; Skelton, J.K.; Kohl, A.; Elliott, R.M.; Schnettler, E. The antiviral RNAi response in vector and non-vector cells against Orthobunyaviruses. PLoS Negl. Trop. Dis. 2017, 11. [Google Scholar] [CrossRef]
- Ruckert, C.; Prasad, A.N.; Garcia-Luna, S.M.; Robison, A.; Grubaugh, N.D.; Weger-Lucarelli, J.; Ebel, G.D. Small RNA responses of Culex mosquitoes and cell lines during acute and persistent virus infection. Insect Biochem. Mol. Biol. 2019, 109, 13–23. [Google Scholar] [CrossRef]
- Parry, R.; Bishop, C.; De Hayr, L.; Asgari, S. Density-dependent enhanced replication of a densovirus in Wolbachia-infected Aedes cells is associated with production of piRNAs and higher virus-derived siRNAs. Virology 2019, 528, 89–100. [Google Scholar] [CrossRef]
- Hermanns, K.; Marklewitz, M.; Zirkel, F.; Overheul, G.J.; Page, R.A.; Loaiza, J.R.; Drosten, C.; van Rij, R.P.; Junglen, S. Agua Salud alphavirus defines a novel lineage of insect-specific alphaviruses discovered in the New World. J. Gen. Virol. 2020, 101, 96–104. [Google Scholar] [CrossRef]
- Franzke, K.; Leggewie, M.; Sreenu, V.B.; Jansen, S.; Heitmann, A.; Welch, S.R.; Brennan, B.; Elliott, R.M.; Tannich, E.; Becker, S.C.; et al. Detection, infection dynamics and small RNA response against Culex Y virus in mosquito-derived cells. J. Gen. Virol. 2018, 99, 1739–1745. [Google Scholar] [CrossRef]

| Country | Location | Laboratory or Field | Coordinates | Date | Pools Screened | RT-PCR Result |
|---|---|---|---|---|---|---|
| SEN | Barkédji village | Laboratory | 15°17′ N, 14°53′ W | MAR 2020 | 10 | + |
| SEN | Barkédji village | Laboratory | 15°17′ N, 14°53′ W | 2013 | 10 | + |
| SEN | Barkédji village | Laboratory | 15°17′ N, 14°53′ W | 2013 | 10 | + |
| SEN | Barkédji village | Field | 15°17′ N, 14°53′ W | 2011 | 10 | + |
| GER | Großheide | Field | 53°36′ N, 7°21′ E | JUL 2017 | 1 | − |
| GER | Varel | Field | 53°24 N, 8°8′ E | JUL 2017 | 1 | − |
| GER | Rastede | Field | 53°14′ N, 8°12′ E | JUL 2017 | 1 | − |
| GER | Oldenburg | Field | 53°6′ N, 8°15′ E | JUL 2017 | 1 | − |
| GER | Bremen | Field | 53°5′ N, 8°51′ E | AUG 2017 | 1 | − |
| GER | Hamburg | Field | 53°28′ N, 9°49′ E | AUG 2017 | 12 | − |
| GER | Frankfurt a.M. | Field | 50°6′ N, 8°40′ E | AUG 2017 | 3 | − |
| GER | Frankfurt a.M. | Field | 50°6′ N, 8°40′ E | SEP 2017 | 14 | − |
| UK | Nottinghamshire | Field | 53°7′ N, 0°54′ W | JUL 2018 | 13 | − |
| ITA | Guarda Veneta | Field | 44°58′ N, 11°47′ E | JUN 2019 | 1 | − |
| ITA | Porto Tolle | Field | 44°54N, 12°27′ E | JUN 2019 | 1 | − |
| ITA | Papozze | Field | 44°59′ N, 12°2′ E | JUN 2019 | 1 | − |
| ITA | Ceggia | Field | 45°40′ N, 12°39′ E | JUN 2019 | 1 | − |
| ITA | Riese Pio X | Field | 45°43′ N, 11°54′ E | JUN 2019 | 1 | − |
| SWE | Forshaga | Field | 59°31′ N, 13°29′ E | JUN 2019 | 2 | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parry, R.; Naccache, F.; Ndiaye, E.H.; Fall, G.; Castelli, I.; Lühken, R.; Medlock, J.; Cull, B.; Hesson, J.C.; Montarsi, F.; et al. Identification and RNAi Profile of a Novel Iflavirus Infecting Senegalese Aedes vexans arabiensis Mosquitoes. Viruses 2020, 12, 440. https://doi.org/10.3390/v12040440
Parry R, Naccache F, Ndiaye EH, Fall G, Castelli I, Lühken R, Medlock J, Cull B, Hesson JC, Montarsi F, et al. Identification and RNAi Profile of a Novel Iflavirus Infecting Senegalese Aedes vexans arabiensis Mosquitoes. Viruses. 2020; 12(4):440. https://doi.org/10.3390/v12040440
Chicago/Turabian StyleParry, Rhys, Fanny Naccache, El Hadji Ndiaye, Gamou Fall, Ilaria Castelli, Renke Lühken, Jolyon Medlock, Benjamin Cull, Jenny C. Hesson, Fabrizio Montarsi, and et al. 2020. "Identification and RNAi Profile of a Novel Iflavirus Infecting Senegalese Aedes vexans arabiensis Mosquitoes" Viruses 12, no. 4: 440. https://doi.org/10.3390/v12040440
APA StyleParry, R., Naccache, F., Ndiaye, E. H., Fall, G., Castelli, I., Lühken, R., Medlock, J., Cull, B., Hesson, J. C., Montarsi, F., Failloux, A.-B., Kohl, A., Schnettler, E., Diallo, M., Asgari, S., Dietrich, I., & Becker, S. C. (2020). Identification and RNAi Profile of a Novel Iflavirus Infecting Senegalese Aedes vexans arabiensis Mosquitoes. Viruses, 12(4), 440. https://doi.org/10.3390/v12040440







