Successive Passage In Vitro Led to Lower Virulence and Higher Titer of A Variant Porcine Epidemic Diarrhea Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Antibody
2.2. Clinical Samples Collection
2.3. Virus Isolation and Propagation
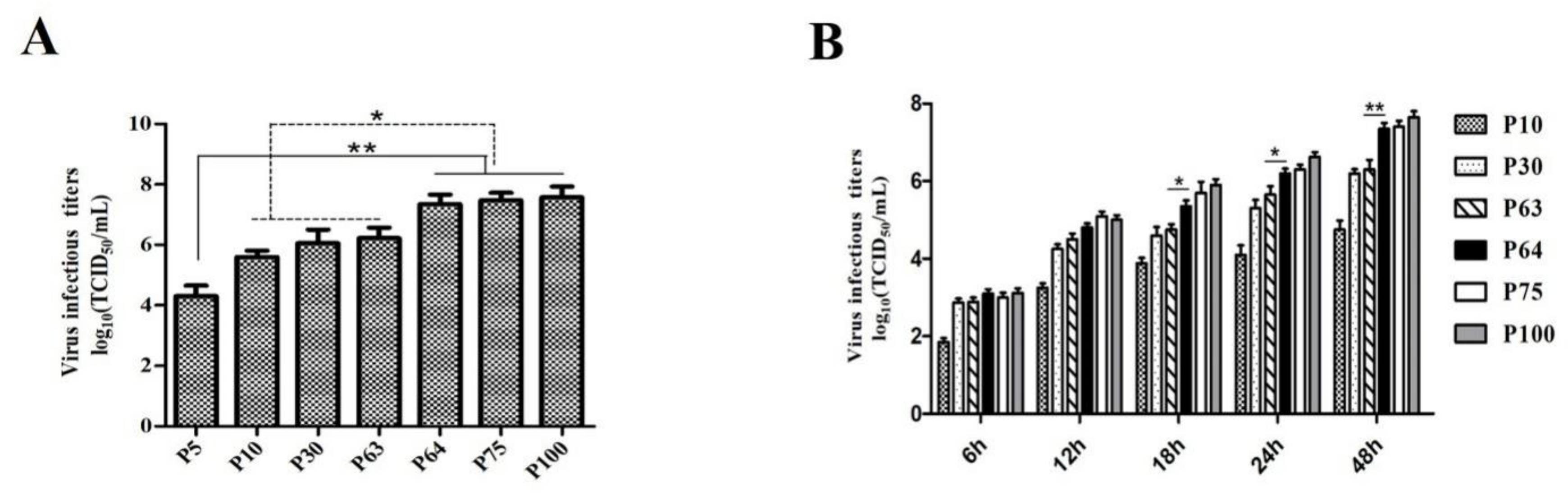
2.4. Growth Kinetics and Virus Titration
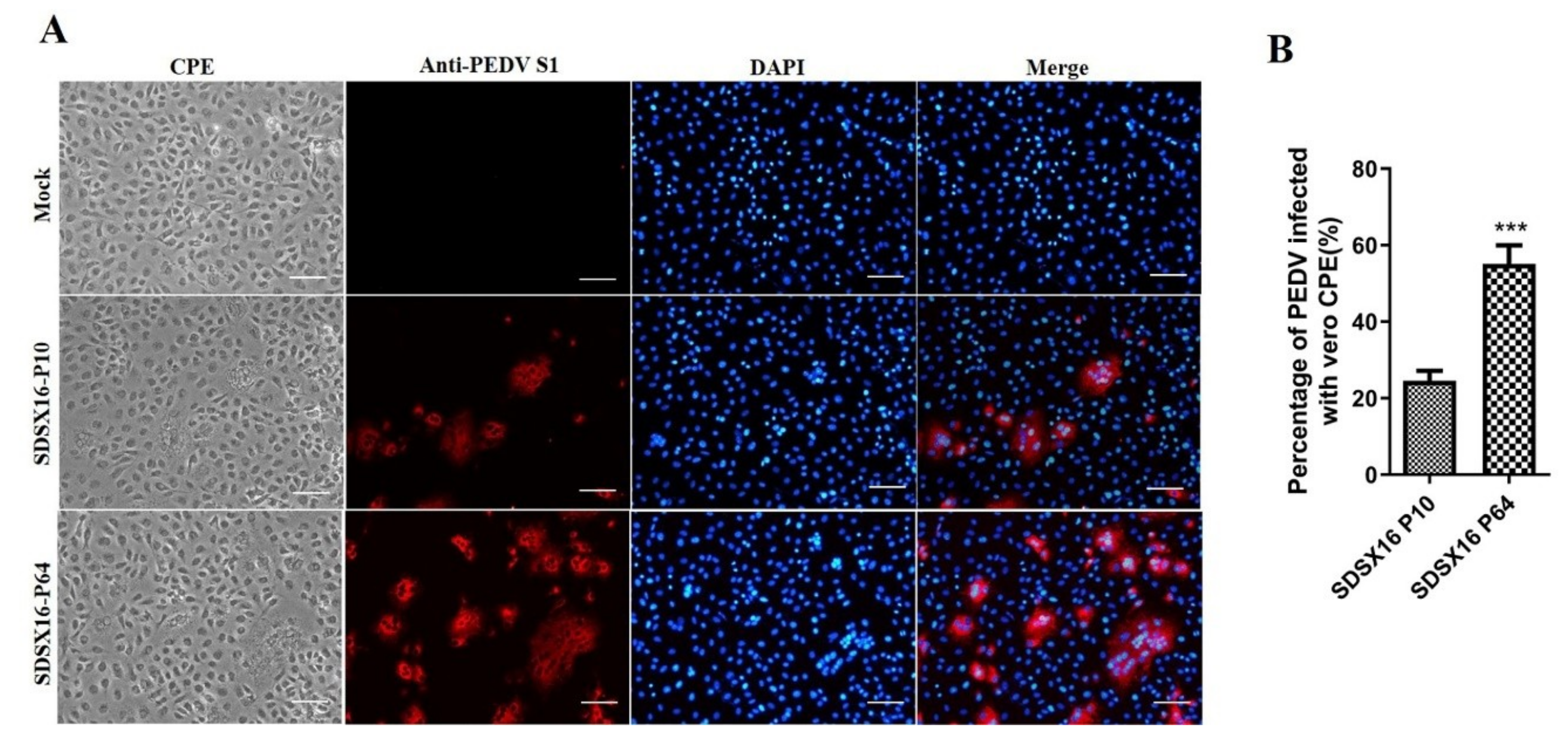
2.5. Immunofluorescence Assay (IFA)
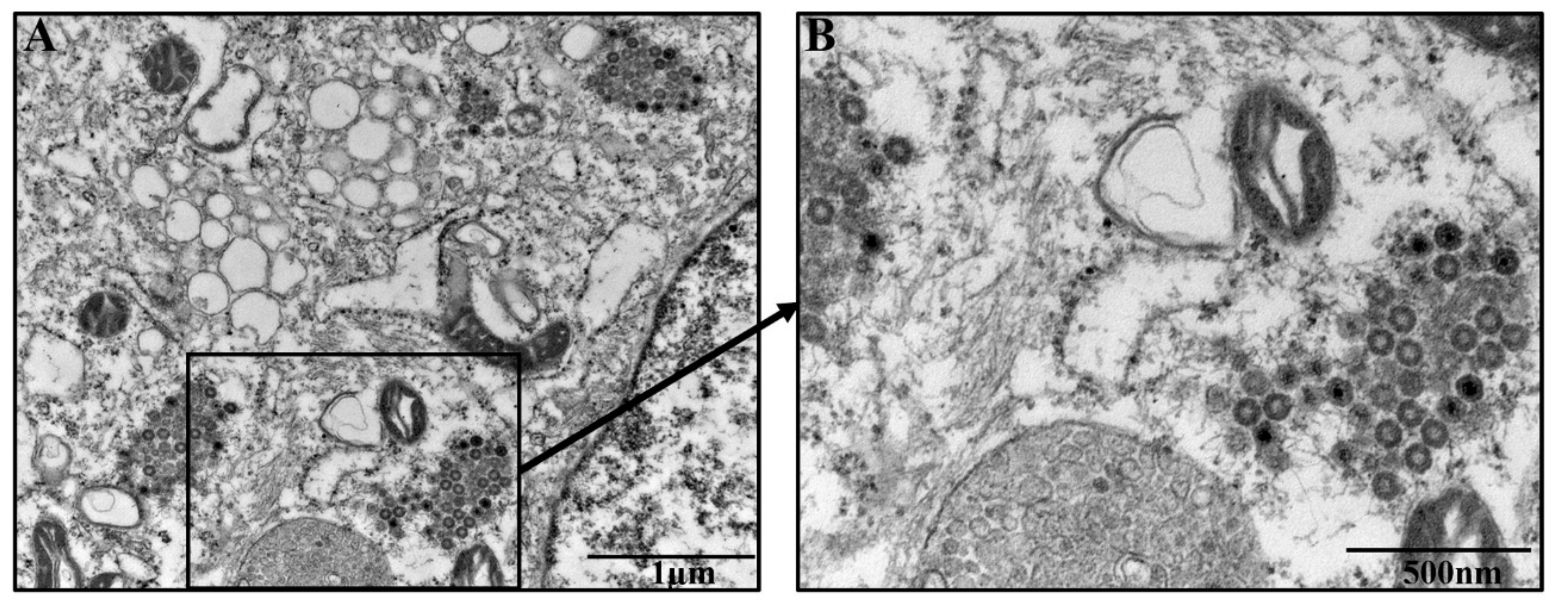
2.6. Morphologic Observation of Virus by TEM
2.7. RNA Extraction, RT-PCR and Sequence Analysis
2.8. Pig Infection Experiment
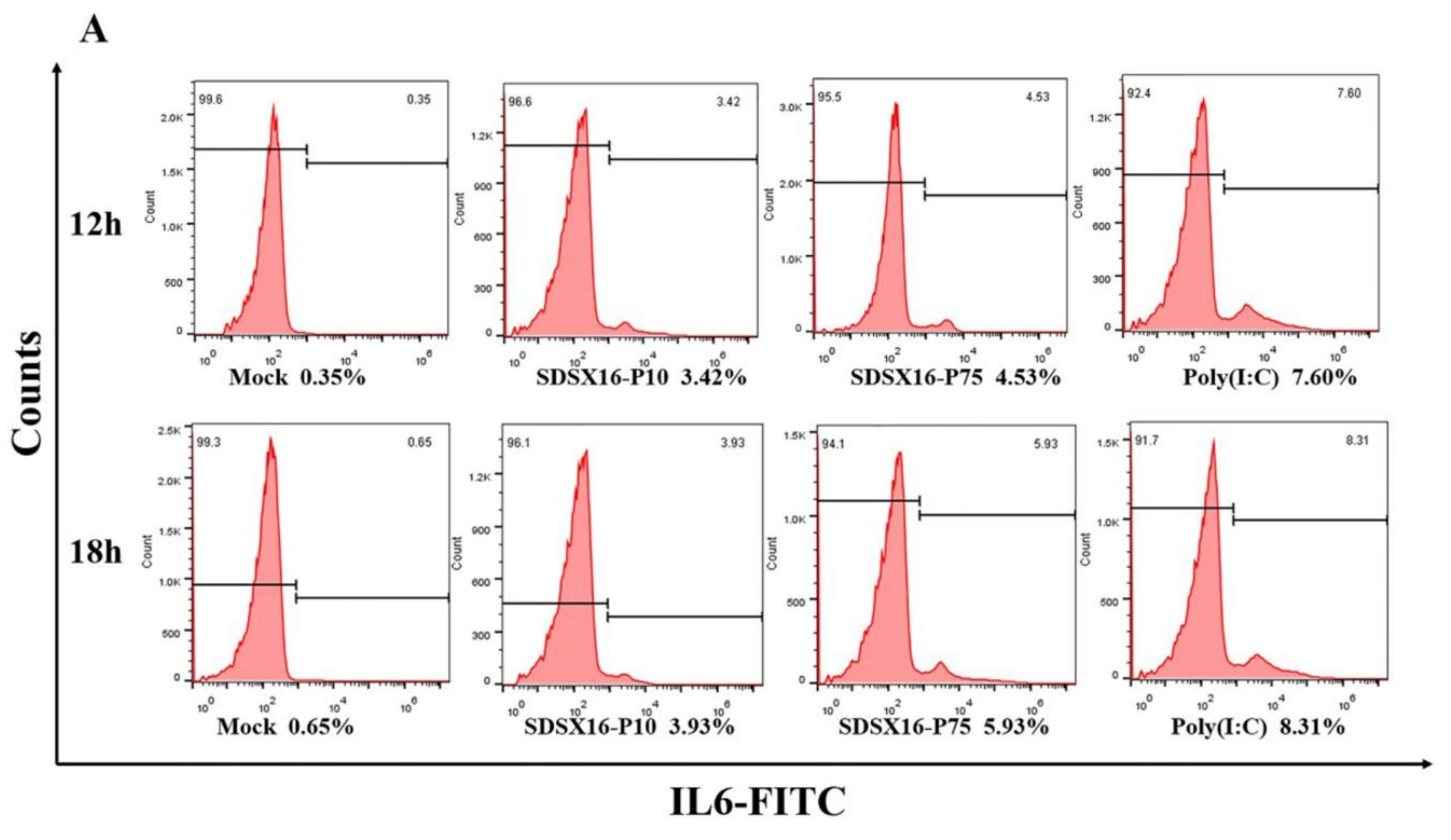
2.9. Detection of Cytokines
2.10. Statistical Analysis
3. Results
3.1. Virus Isolation and In Vitro Characterization
3.2. Morphologic Characterization of Virus
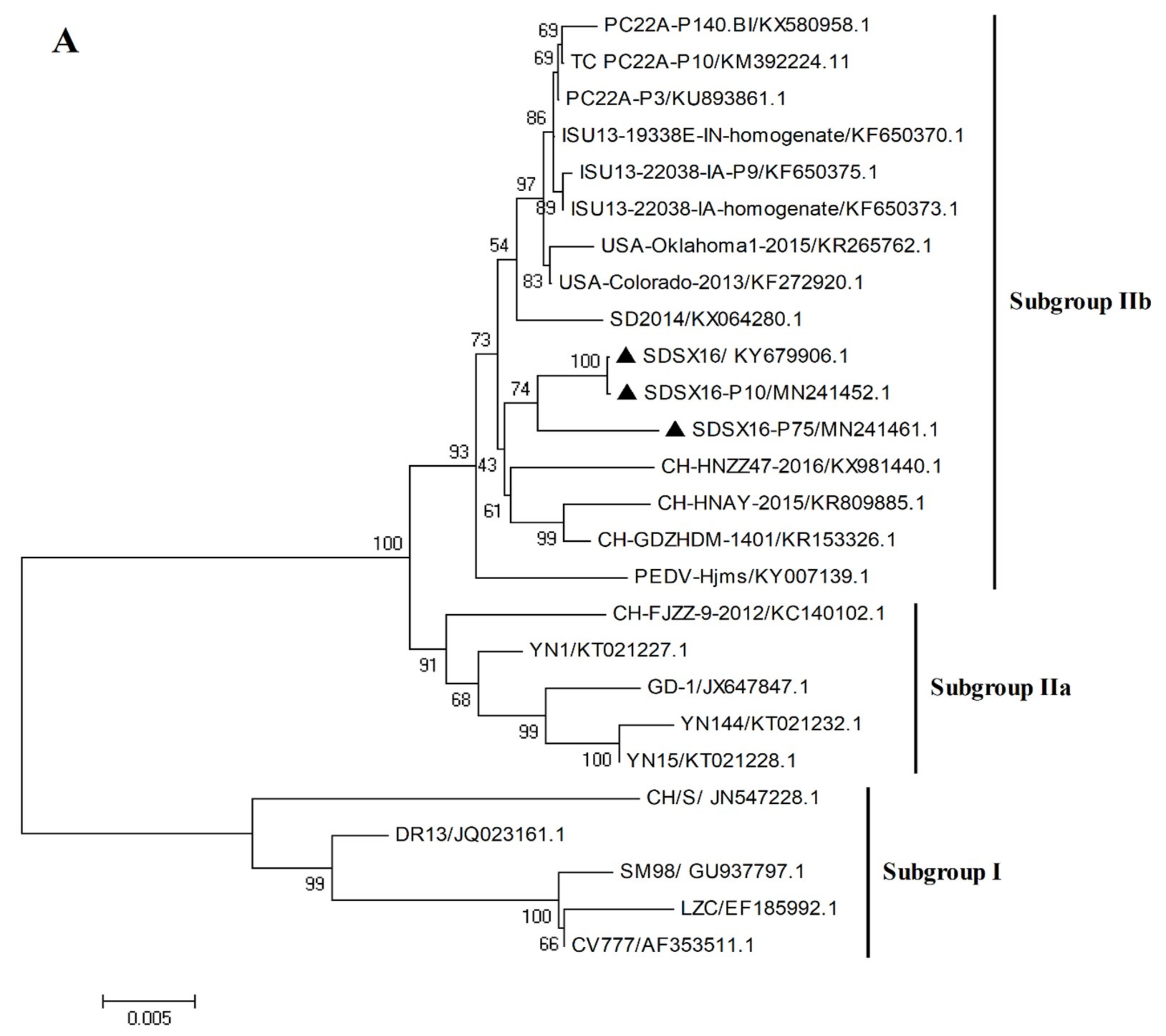
3.3. Sequence Analysis
3.4. Pathogenicity of SDSX16
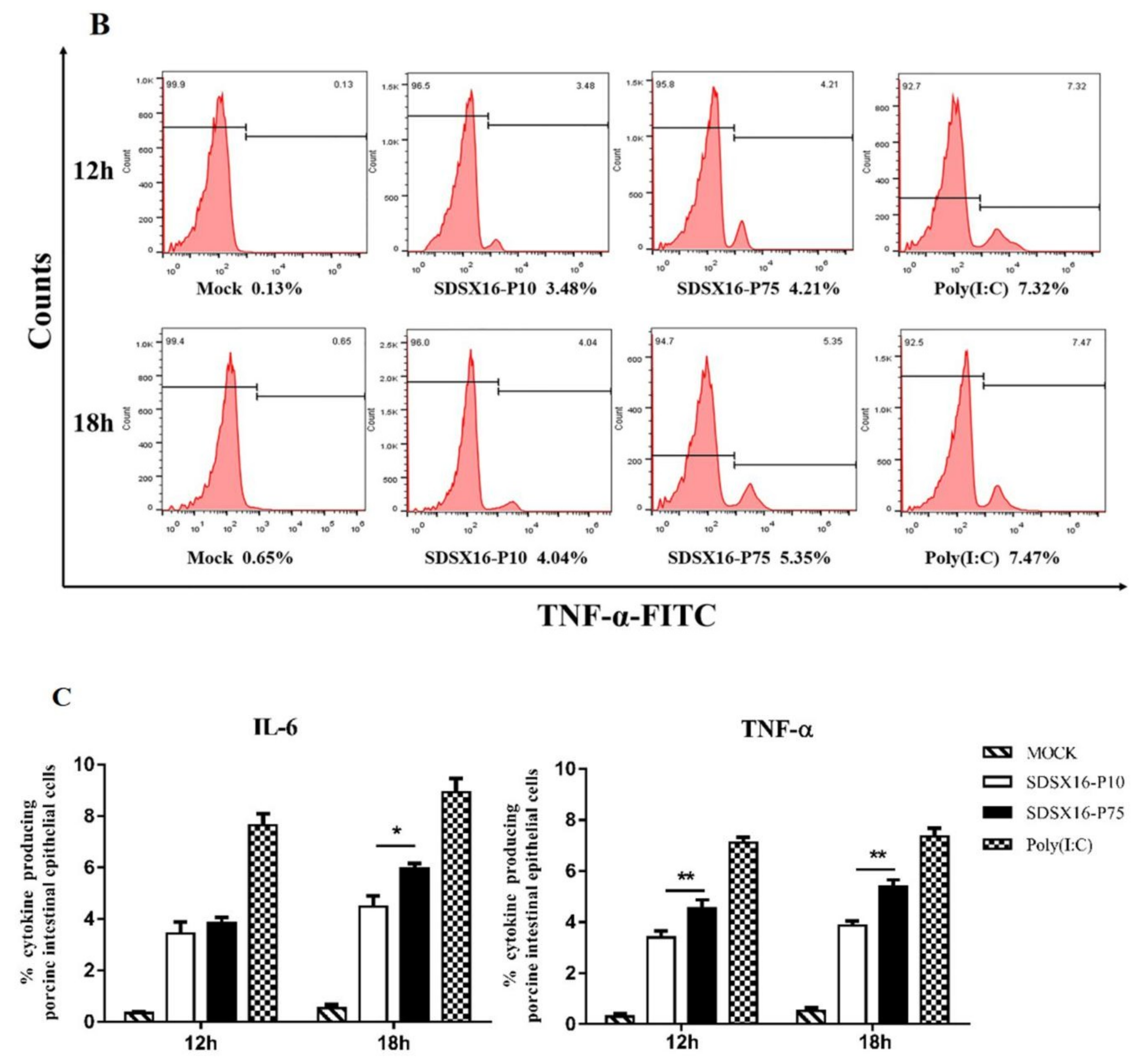
3.5. Analysis of Cytokine Expression by SDSX16 Infected Porcine Intestinal Epithelial Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gao, Y.; Kou, Q.; Ge, X.; Zhou, L.; Guo, X.; Yang, H. Phylogenetic analysis of porcine epidemic diarrhea virus field strains prevailing recently in China. Arch. Virol. 2013, 158, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, P.; Guo, L.; Liu, Y.; Du, Y.; Ren, S.; Li, J.; Zhang, Y.; Fan, Y.; Huang, B.; et al. Porcine epidemic diarrhea virus variants with high pathogenicity, China. Emerg. Infect. Dis. 2013, 19, 2048–2049. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Chen, L.; Gao, L.; Yuan, X.; Ma, Z.; Fan, H. Epidemic strain YC2014 of porcine epidemic diarrhea virus could provide piglets against homologous challenge. Virol. J. 2016, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Puranaveja, S.; Poolperm, P.; Lertwatcharasarakul, P.; Kesdaengsakonwut, S.; Boonsoongnern, A.; Urairong, K.; Kitikoon, P.; Choojai, P.; Kedkovid, R.; Teankum, K.; et al. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg. Infect. Dis. 2009, 15, 1112–1115. [Google Scholar] [CrossRef]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef]
- Dastjerdi, A.; Carr, J.; Ellis, R.J.; Steinbach, F.; Williamson, S. Porcine Epidemic Diarrhea Virus among Farmed Pigs, Ukraine. Emerg. Infect. Dis. 2015, 21, 2235–2237. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Kabemura, M.; Masatani, T.; Matsuu, A.; Ozawa, M. Isolation and molecular characterization of porcine epidemic diarrhea viruses collected in Japan in 2014. Arch. Virol. 2016, 161, 2189–2195. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Dickerman, A.W.; Pineyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 2013, 4, e00737-13. [Google Scholar] [CrossRef] [PubMed]
- Ojkic, D.; Hazlett, M.; Fairles, J.; Marom, A.; Slavic, D.; Maxie, G.; Alexandersen, S.; Pasick, J.; Alsop, J.; Burlatschenko, S. The first case of porcine epidemic diarrhea in Canada. Can. Vet. J. 2015, 56, 149–152. [Google Scholar] [PubMed]
- Vlasova, A.N.; Marthaler, D.; Wang, Q.; Culhane, M.R.; Rossow, K.D.; Rovira, A.; Collins, J.; Saif, L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014, 20, 1620–1628. [Google Scholar] [CrossRef]
- Stevenson, G.W.; Hoang, H.; Schwartz, K.J.; Burrough, E.R.; Sun, D.; Madson, D.; Cooper, V.L.; Pillatzki, A.; Gauger, P.; Schmitt, B.J.; et al. Emergence of Porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn Invest. 2013, 25, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Marthaler, D.; Jiang, Y.; Otterson, T.; Goyal, S.; Rossow, K.; Collins, J. Complete Genome Sequence of Porcine Epidemic Diarrhea Virus Strain USA/Colorado/2013 from the United States. Genome Announc. 2013, 1, e00555-13. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Park, B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 2012, 44, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, X.; Shi, D.; Shi, H.; Zhang, X.; Li, C.; Chi, Y.; Feng, L. Detection and molecular diversity of spike gene of porcine epidemic diarrhea virus in China. Viruses 2013, 5, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Lee, K.W.; Choi, H.W.; Lee, C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch. Virol. 2014, 159, 2977–2987. [Google Scholar] [CrossRef]
- Hain, K.S.; Joshi, L.R.; Okda, F.; Nelson, J.; Singrey, A.; Lawson, S.; Martins, M.; Pillatzki, A.; Kutish, G.F.; Nelson, E.A.; et al. Immunogenicity of a recombinant parapoxvirus expressing the spike protein of Porcine epidemic diarrhea virus. J. Gen. Virol. 2016, 97, 2719–2731. [Google Scholar] [CrossRef]
- Gerber, P.F.; Gong, Q.; Huang, Y.W.; Wang, C.; Holtkamp, D.; Opriessnig, T. Detection of antibodies against porcine epidemic diarrhea virus in serum and colostrum by indirect ELISA. Vet. J. 2014, 202, 33–36. [Google Scholar] [CrossRef]
- Chen, J.F.; Sun, D.B.; Wang, C.B.; Shi, H.Y.; Cui, X.C.; Liu, S.W.; Qiu, H.J.; Feng, L. Molecular characterization and phylogenetic analysis of membrane protein genes of porcine epidemic diarrhea virus isolates in China. Virus Genes 2008, 36, 355–364. [Google Scholar] [CrossRef]
- Tian, Y.; Yu, Z.; Cheng, K.; Liu, Y.; Huang, J.; Xin, Y.; Li, Y.; Fan, S.; Wang, T.; Huang, G.; et al. Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses 2013, 5, 1991–2004. [Google Scholar] [CrossRef]
- Suzuki, T.; Murakami, S.; Takahashi, O.; Kodera, A.; Masuda, T.; Itoh, S.; Miyazaki, A.; Ohashi, S.; Tsutsui, T. Molecular characterization of pig epidemic diarrhoea viruses isolated in Japan from 2013 to 2014. Infect. Genet. Evol. 2015, 36, 363–368. [Google Scholar] [CrossRef]
- Wong, G.H.; Goeddel, D.V. Tumour necrosis factors α and β inhibit virus replication and synergize with interferons. Nature 1986, 323, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Sergerie, Y.; Rivest, S.; Boivin, G. Tumor necrosis factor-alpha and interleukin-1 beta play a critical role in the resistance against lethal herpes simplex virus encephalitis. J. Infect. Dis. 2007, 196, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, K.; Yoo, D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 2016, 489, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, J.; Yoo, D. Inhibition of NF-kappaB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 2017, 510, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Wyler, R. Quantitation, biological and physicochemical properties of cell culture-adapted porcine epidemic diarrhea coronavirus (PEDV). Vet. Microbiol. 1989, 20, 131–142. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.; Lee, C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015, 208, 215–224. [Google Scholar] [CrossRef]
- Madson, D.M.; Magstadt, D.R.; Arruda, P.H.; Hoang, H.; Sun, D.; Bower, L.P.; Bhandari, M.; Burrough, E.R.; Gauger, P.C.; Pillatzki, A.E.; et al. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet. Microbiol. 2014, 174, 60–68. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Chen, Q.; Li, G.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.J.; et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Wang, Q.; Scheuer, K.A.; Lu, Z.; Zhang, Y.; Saif, L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014, 20, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.M.; Hou, Y.; Marthaler, D.G.; Gao, X.; Liu, X.; Zheng, L.; Saif, L.J.; Wang, Q. Attenuation of an original US porcine epidemic diarrhea virus strain PC22A via serial cell culture passage. Vet. Microbiol. 2017, 201, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.H.; Bae, J.L.; Kang, T.J.; Kim, J.; Chung, G.H.; Lim, C.W.; Laude, H.; Yang, M.S.; Jang, Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells 2002, 14, 295–299. [Google Scholar] [PubMed]
- Cruz, D.J.; Kim, C.J.; Shin, H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008, 132, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Li, C.; Qi, S.; Yang, D.; Jiang, N.; Yin, B.; Guo, D.; Kong, F.; Yuan, D.; Feng, L.; et al. A molecular epidemiological investigation of PEDV in China: Characterization of co-infection and genetic diversity of S1-based genes. Transbound Emerg. Dis. 2019. [Google Scholar] [CrossRef]
- Chen, P.; Wang, K.; Hou, Y.; Li, H.; Li, X.; Yu, L.; Jiang, Y.; Gao, F.; Tong, W.; Yu, H.; et al. Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011-2017. Infect. Genet. Evol. 2019, 69, 153–165. [Google Scholar] [CrossRef]
- Sun, R.Q.; Cai, R.J.; Chen, Y.Q.; Liang, P.S.; Chen, D.K.; Song, C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; Zhang, Q.; Zhou, P.; Fang, Y.; Zhao, D.; Feng, J.; Li, W.; Zhang, Y.; Wang, Y. Evaluation and comparison of immunogenicity and cross-protective efficacy of two inactivated cell culture-derived GIIa- and GIIb-genotype porcine epidemic diarrhea virus vaccines in suckling piglets. Vet. Microbiol. 2019, 230, 278–282. [Google Scholar] [CrossRef]
- De Haan, C.A.; Masters, P.S.; Shen, X.; Weiss, S.; Rottier, P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 2002, 296, 177–189. [Google Scholar] [CrossRef]
- Haijema, B.J.; Volders, H.; Rottier, P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol 2004, 78, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Takeyama, N.; Katsumata, A.; Tuchiya, K.; Kodama, T.; Kusanagi, K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes 2011, 43, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kim, H.K.; Song, D.S.; An, D.J.; Park, B.K. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J. Virol. 2012, 86, 5964. [Google Scholar] [CrossRef] [PubMed]
- Muth, D.; Corman, V.M.; Roth, H.; Binger, T.; Dijkman, R.; Gottula, L.T.; Gloza-Rausch, F.; Balboni, A.; Battilani, M.; Rihtaric, D.; et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018, 8, 15177. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Ea, C.K.; Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Ren, Y.; Suo, S.; Sun, X.; Li, X.; Li, P.; Yang, W.; Li, G.; Li, L.; Schwegmann-Wessels, C.; et al. Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing spike genes from porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. PLoS ONE 2013, 8, e57468. [Google Scholar] [CrossRef]
- Park, J.-E.; Shin, H.-J. Resistance of field-isolated porcine epidemic diarrhea virus to interferon and neutralizing antibody. J. Immunol. 2018, 200, 59.32. [Google Scholar]
- Gerdts, V.; Zakhartchouk, A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017, 206, 45–51. [Google Scholar] [CrossRef]
- Won, H.; Lee, D.U.; Jang, G.; Noh, Y.H.; Lee, S.C.; Choi, H.W.; Yoon, I.J.; Yoo, H.S.; Lee, C. Generation and protective efficacy of a cold-adapted attenuated genotype 2b porcine epidemic diarrhea virus. J. Vet. Sci 2019, 20, e32. [Google Scholar] [CrossRef]
- Koonpaew, S.; Teeravechyan, S.; Frantz, P.N.; Chailangkarn, T.; Jongkaewwattana, A. PEDV and PDCoV Pathogenesis: The Interplay Between Host Innate Immune Responses and Porcine Enteric Coronaviruses. Front. Vet. Sci. 2019, 6, 34. [Google Scholar] [CrossRef]
- Cao, L.; Ge, X.; Gao, Y.; Herrler, G.; Ren, Y.; Ren, X.; Li, G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Fang, L.; Jing, H.; Zeng, S.; Wang, D.; Liu, L.; Zhang, H.; Luo, R.; Chen, H.; Xiao, S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014, 88, 8936–8945. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Sequence 5′–3′ | Size (bp) |
|---|---|---|
| PEDV-F1 | CGCAAATGGGCGGTAGGCGTG | 1169 |
| PEDV-R1 | GCATGAAGCCACAAGGAC | |
| PEDV-F2 | GTTGTCTTGTCTGAGCCA | 1119 |
| PEDV-R2 | CAGGATGTTCAATGACAA | |
| PEDV-F3 | CAATGTGAATCTCGTCGT | 1259 |
| PEDV-R3 | GATCCTGTGAGATCGTGTC | |
| PEDV-F4 | CTCCCTTGAATTTGAGTTCG | 1326 |
| PEDV-R4 | TGACACCACGTCCAACTTTA | |
| PEDV-F5 | CAGACGTTGAGCCTGTATT | 1157 |
| PEDV-R5 | GTTGTCTGTGACAACGGT | |
| PEDV-F6 | ATGTGGAGCGTTTCTACGCA | 1284 |
| PEDV-R6 | AACATCATCACAAACAGGGC | |
| PEDV-F7 | GCCTTTCGTAAGAGGGAT | 1460 |
| PEDV-R7 | AGGGCAATGAAATCACGTA | |
| PEDV-F8 | TTGCTGAGGCTCATCGTTA | 1493 |
| PEDV-R8 | AGAGCCTACGAACTTGTCA | |
| PEDV-F9 | TCGGTGATATGTCTGTTGGC | 1346 |
| PEDV-R9 | TGCAGAGACTGGATTGAGGC | |
| PEDV-F10 | TGGTACCGTTGAGTTTTGCT | 1330 |
| PEDV-R10 | TTCATGCTAGAGAGACAGCC | |
| PEDV-F11 | GCTTCTACGGTATTCTCTACT | 1249 |
| PEDV-R11 | TGCACCATTAGGAGAATCCA | |
| PEDV-F12 | CCTGGTAAGCTGAAGCAGCG | 1462 |
| PEDV-R12 | AAGCACTCACTAGCAAGGCA | |
| PEDV-F13 | ATTCATCGTGTCTATGCATTG | 1367 |
| PEDV-R13 | TGCTCTGTCGCACTTTGGG | |
| PEDV-F14 | CCATGACTACTCGGCAGTAT | 1232 |
| PEDV-R14 | CACACATATGGAGTGATGGC | |
| PEDV-F15 | AATCTGCAGGGCTTTGTGTT | 1318 |
| PEDV-R15 | GACAGCGATAACACTTGTGC | |
| PEDV-F16 | TGGTGGATGAGGTCTCTATG | 1342 |
| PEDV-R16 | GGGCATGTTTGAAACAACAG | |
| PEDV-F17 | GGTTGCGTTGTAACTGAGTC | 1358 |
| PEDV-R17 | GTCCTACCTTACGTTTGGCA | |
| PEDV-F18 | TCGGCGGGCTTTACTATTTG | 1306 |
| PEDV-R18 | CACCAAGATGTAGCACACG | |
| PEDV-F19 | CTCCAGACATTTTATCCGCA | 1294 |
| PEDV-R19 | CGCGCAGTAGCATTAGTGTT | |
| PEDV-F20 | GGCGGTTATCTACCTATTGG | 1345 |
| PEDV-R20 | CTGTTCATGACTCAGAAGG | |
| PEDV-F21 | TGTTGGACGCTGTCACAAT | 1518 |
| PEDV-R21 | GACCATTAGAACAGCGCTTA | |
| PEDV-F22 | TCACCCAGTACACTGCAGC | 1412 |
| PEDV-R22 | GAGAGACTCTGAACGCTGC | |
| PEDV-F23 | GCCTTGACTCTACGTGAGCC | 1423 |
| PEDV-R23 | CGCTATTACACAACCGGTGA | |
| PEDV-F24 | GACACTTTCTTTCCTCAATG | 1475 |
| PEDV-R24 | AGTCCTATAGCGGAGGTCGG | |
| PEDV-F25 | GGACGTGTTGGTCGTTCAGT | 1242 |
| PEDV-R25 | CATTTGGATCAGACTTTGGC | |
| PEDV-F26 | AGATGCGGAATTTGTCGAA | 1121 |
| PEDV-R26 | GACTTCAAGTCACGTGAA | |
| 5′UTR1 | GCATCGCTCCATTATCC | 623 |
| 5′UTR2 | TTACCACCACGACGACC | |
| 3′UTR1 | GGCATAAGCAACAGCAG | 690 |
| 3′UTR2 | ATCGCCAGTTTAGCACCA |
| Gene | Sequence 5′–3′ | GenBank NO. |
|---|---|---|
| IL-1β-F | GAAAGCCATACCCAGAGGTC | NM_214055.1 |
| IL-1β-R | GCACTAATCTAGGGAAGACAGC | |
| IL-6-F | TGGGTTCAATCAGGAGACCT | AF518322.1 |
| IL-6-R | CAGCCTCGACATTTCCCTTA | |
| TNF-α-F | CCCCCAGAAGGAAGAGTTTC | JF831365.1 |
| TNF-α-R | CGGGCTTATCTGAGGTTTGA | |
| IFN-β-F | CTCCAAATCGCTCTCCTGAT | GQ415073.1 |
| IFN-β-R | GGGACCTCAAAGTTCATCCT | |
| β-actin-F | TCTGGCACCACACCTTCT | AY550069.1 |
| β-actin-R | GATCTGGGTCATCTTCTCAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, P.; Wang, S.; Chen, Z.; Yu, J.; Tang, R.; Qiu, W.; Zhao, L.; Liu, Y.; Guo, X.; He, H.; et al. Successive Passage In Vitro Led to Lower Virulence and Higher Titer of A Variant Porcine Epidemic Diarrhea Virus. Viruses 2020, 12, 391. https://doi.org/10.3390/v12040391
Zhao P, Wang S, Chen Z, Yu J, Tang R, Qiu W, Zhao L, Liu Y, Guo X, He H, et al. Successive Passage In Vitro Led to Lower Virulence and Higher Titer of A Variant Porcine Epidemic Diarrhea Virus. Viruses. 2020; 12(4):391. https://doi.org/10.3390/v12040391
Chicago/Turabian StyleZhao, Pengwei, Song Wang, Zhi Chen, Jiang Yu, Rongzhi Tang, Wenbin Qiu, Lu Zhao, Yueyue Liu, Xiaozhen Guo, Hongbin He, and et al. 2020. "Successive Passage In Vitro Led to Lower Virulence and Higher Titer of A Variant Porcine Epidemic Diarrhea Virus" Viruses 12, no. 4: 391. https://doi.org/10.3390/v12040391
APA StyleZhao, P., Wang, S., Chen, Z., Yu, J., Tang, R., Qiu, W., Zhao, L., Liu, Y., Guo, X., He, H., Xu, G., Li, J., & Wu, J. (2020). Successive Passage In Vitro Led to Lower Virulence and Higher Titer of A Variant Porcine Epidemic Diarrhea Virus. Viruses, 12(4), 391. https://doi.org/10.3390/v12040391





