A Heterologous Viral Protein Scaffold for Chimeric Antigen Design: An Example PCV2 Virus Vaccine Candidate
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid, Gene Cloning, and Strain
2.2. Design of the Recombinant Chimeric Antigen (Qm1)
2.2.1. Protein Sequence Similarity Analyses
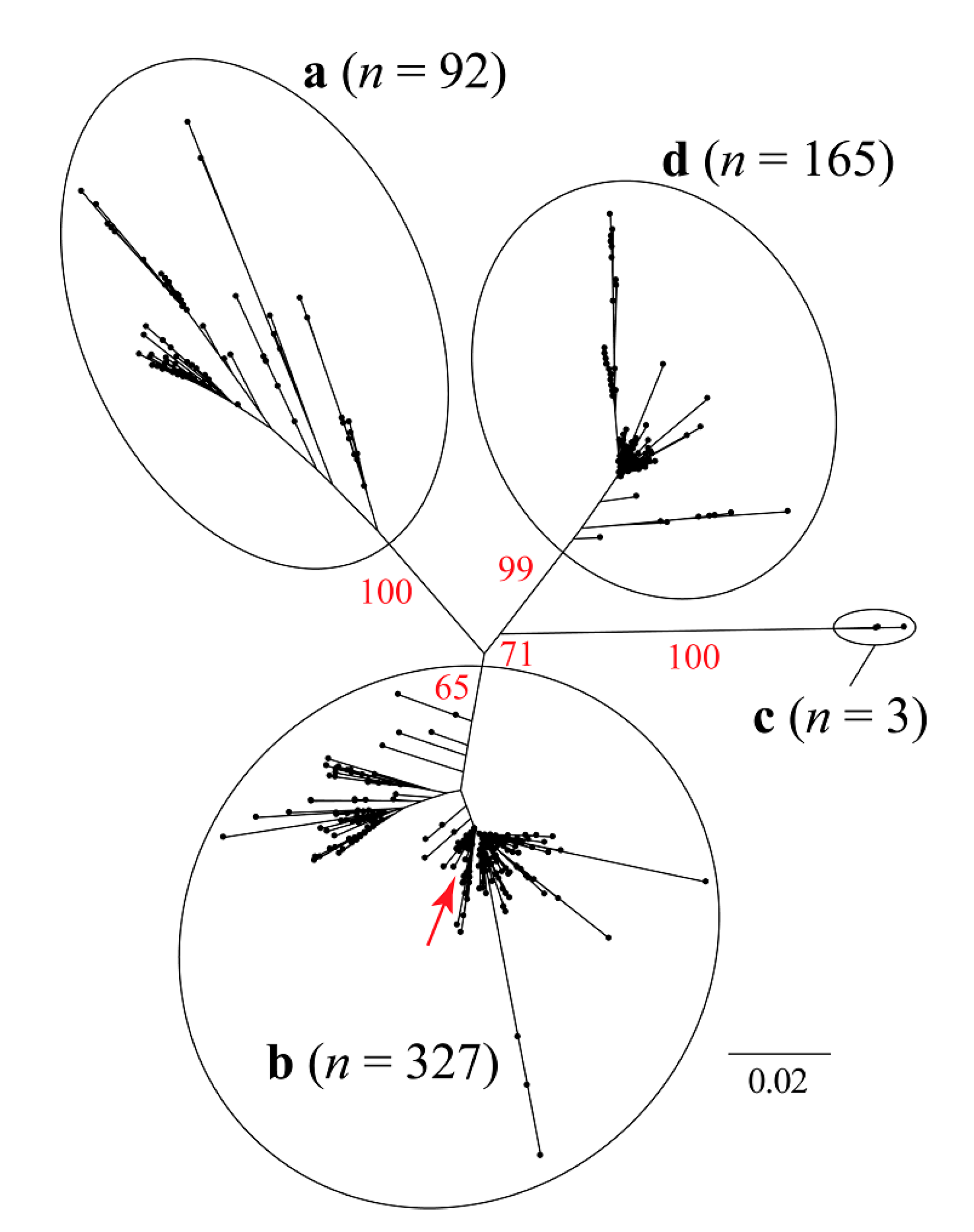
2.2.2. Phylogenetic Analysis
2.2.3. Structural Homologs for PCV2 Capsid Protein
2.2.4. Recombinant Chimeric Antigen Design
2.2.5. Protein Modeling of Chimeras
2.3. Bacterial Transformation and Small-Scale Expression Analysis
2.4. SDS-PAGE and Western Blotting
2.5. Release of the Polyclonal Antibody against Proteins of the SHuffle ® T7 E. coli Strain
2.6. Batch Fermentation Conditions
2.7. Biomass Disruption and Protein Detection
2.8. Qm1 Purification by Immobilized Metal Affinity Chromatography (IMAC)
2.9. Formulation and Dose Distribution
2.10. Animal Trials
2.10.1. Safety Evaluation Test
2.10.2. Immunization Assay
2.10.3. Challenge Test in Immunized Pigs with Recombinant Chimeric Antigens
2.10.4. Immune Enzymatic Assay for Immune Response Evaluation
2.10.5. Real-Time PCR for Viral Titer Analysis
3. Results
3.1. Similarity Analysis of Sequences of PCV2 Isolates Worldwide
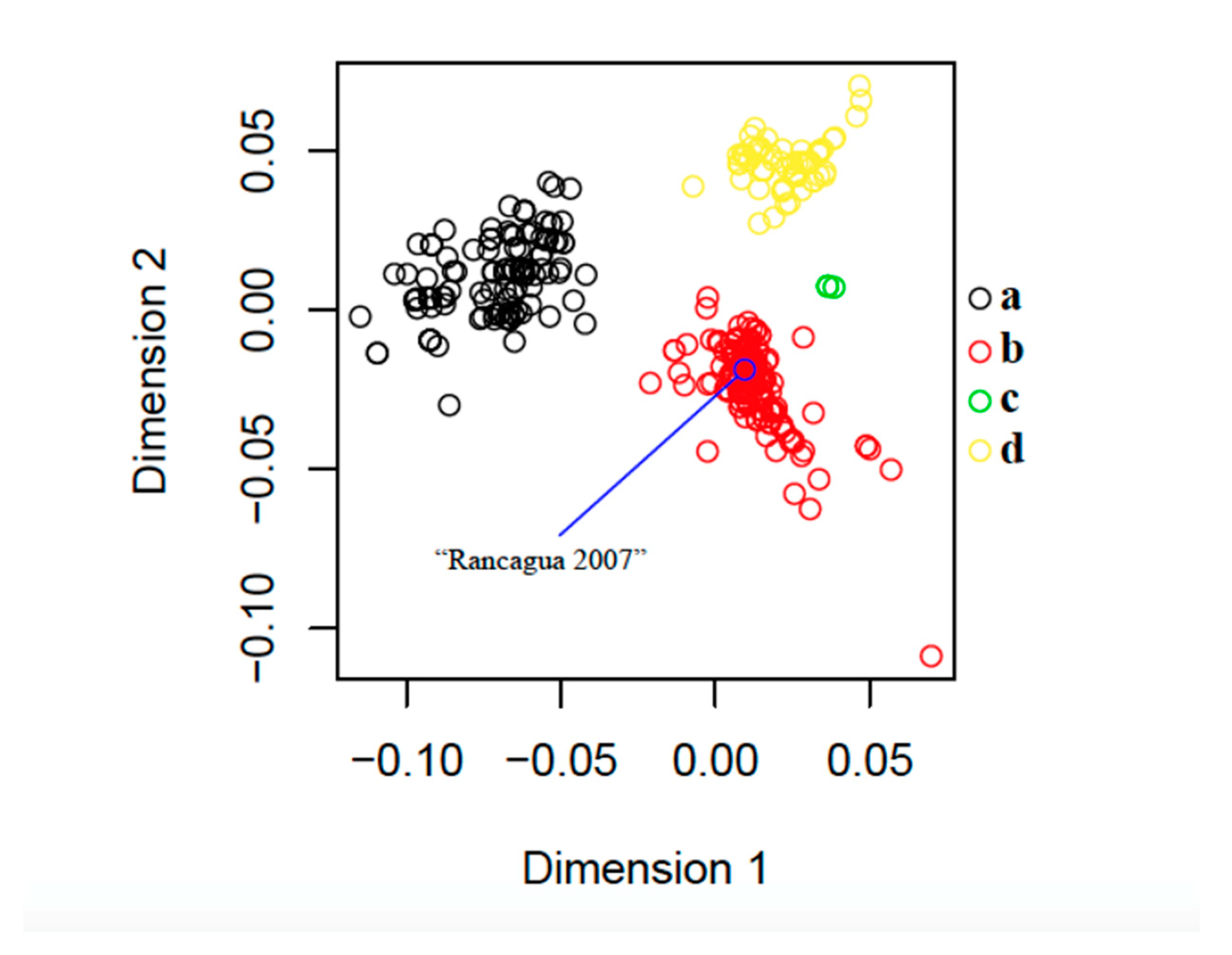
3.2. Phylogenetic and Classical Multidimensional Scaling Analysis
3.3. Structural Homology for Cap Protein
3.4. Modeling of the Antigenic Variant Qm1 vs. Cap
3.5. pET22b-Qm1 Expression Vector Construction
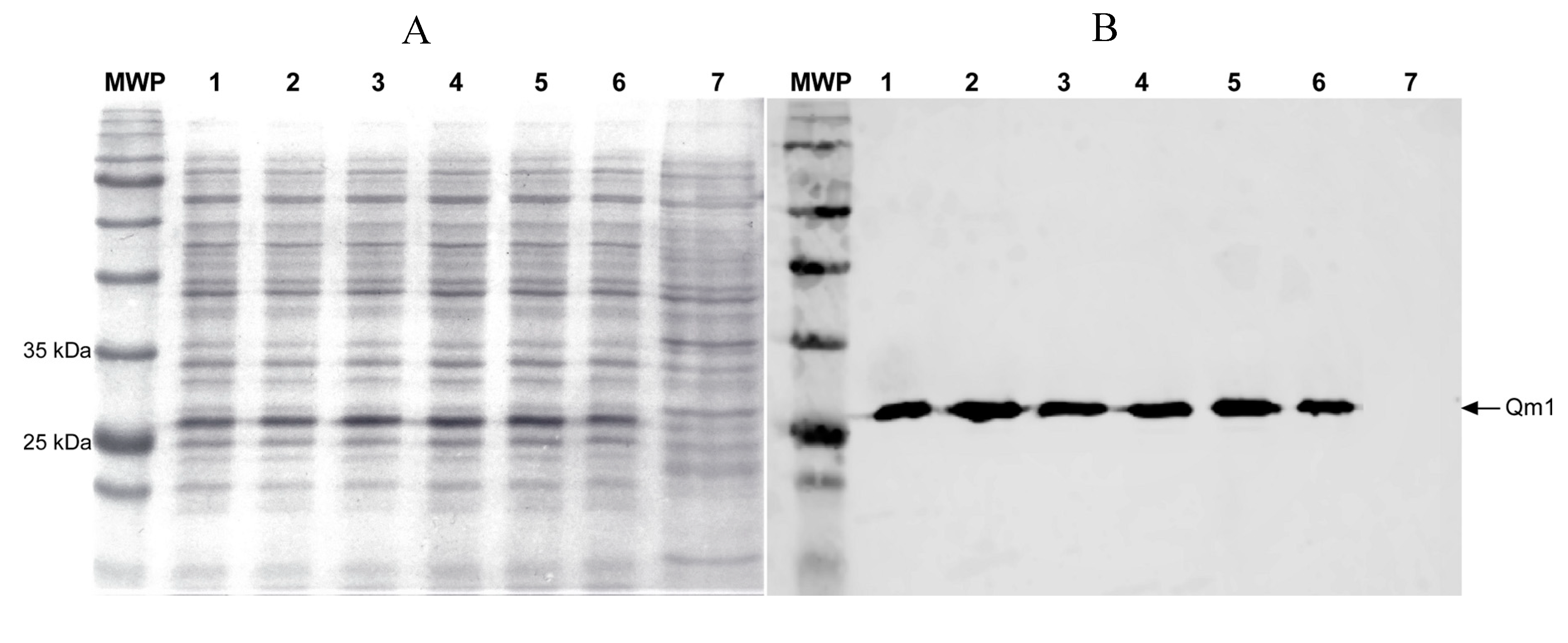
3.6. Qm1 Expression in SHuffle® T7
3.7. Qm1 Specific Antibody Detection
3.8. Qm1 Batch Fermentation
3.9. Qm1 Protects Pigs against PCV2 Infection
4. Discussion
4.1. Design of Qm1 Chimeric Antigen
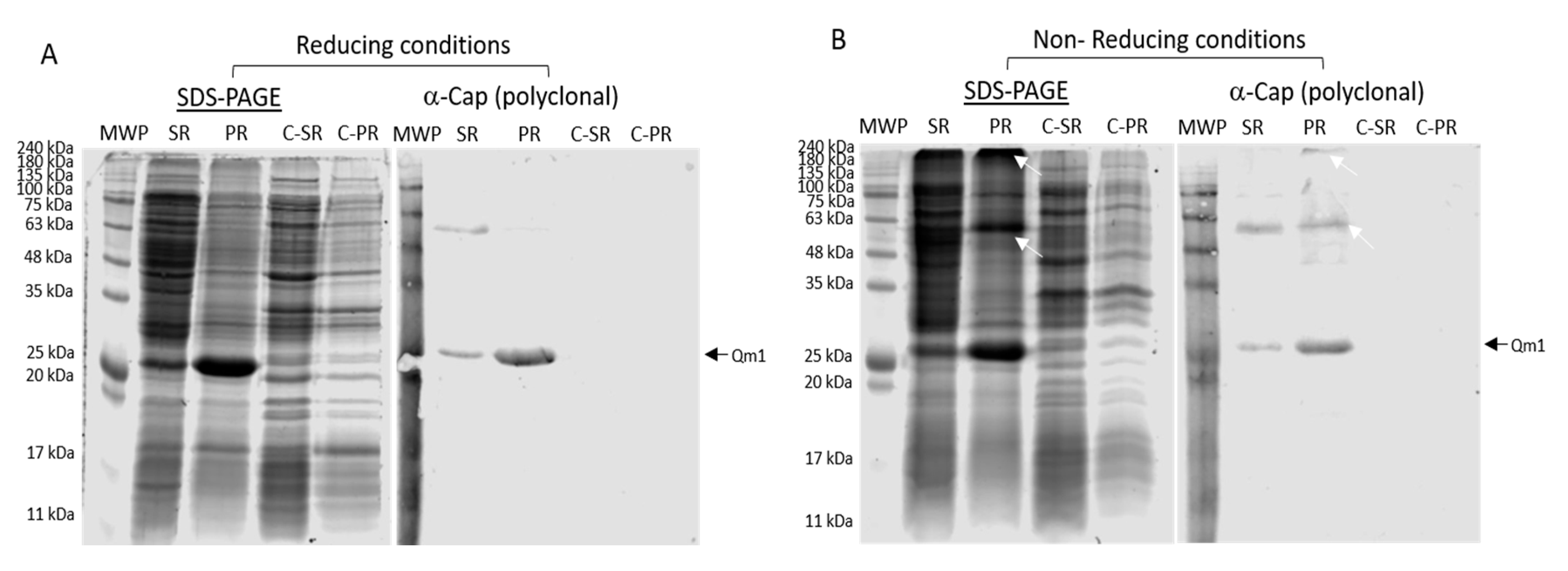
4.2. Anti-Cap Antibody Recognition
4.3. Qm1 Production
4.4. Protection against PCV2 Induced by the Qm1 Antigen
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Oliveira, C.; Domingues, L. Guidelines to reach high-quality purified recombinant proteins. Appl. Microbiol. Biotechnol. 2018, 102, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Jozala, A.F.; Geraldes, D.C.; Tundisi, L.L.; Feitosa, V.A.; Breyer, C.A.; Cardoso, S.L.; Mazzola, P.G.; Oliveira-Nascimento, L.; Rangel-Yagui, C.O.; Magalhaes, P.O.; et al. Biopharmaceuticals from microorganisms: From production to purification. Braz. J. Microbiol. 2016, 47 (Suppl. 1), 51–63. [Google Scholar] [CrossRef] [PubMed]
- Owczarek, B.; Gerszberg, A.; Hnatuszko-Konka, K. A brief reminder of systems of production and chromatography-based recovery of recombinant protein biopharmaceuticals. Biomed. Res. Int. 2019, 2019, 4216060. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Garcia, L.; Martin, L.; Mangues, R.; Ferrer-Miralles, N.; Vazquez, E.; Villaverde, A. Recombinant pharmaceuticals from microbial cells: A 2015 update. Microb. Cell Fact. 2016, 15, 33. [Google Scholar] [CrossRef] [PubMed]
- Karch, C.P.; Burkhard, P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochem. Pharm. 2016, 120, 1–14. [Google Scholar] [CrossRef]
- Ada, G. Overview of vaccines and vaccination. Mol. Biotechnol. 2005, 29, 255–272. [Google Scholar] [CrossRef]
- Cruz, T.F.; Magro, A.J.; de Castro, A.; Pedraza-Ordonez, F.J.; Tsunemi, M.H.; Perahia, D.; Araujo, J.P., Jr. In vitro and in silico studies reveal capsid-mutant porcine circovirus 2b with novel cytopathogenic and structural characteristics. Virus Res. 2018, 251, 22–33. [Google Scholar] [CrossRef]
- Chae, C. Commercial porcine circovirus type 2 vaccines: Efficacy and clinical application. Vet. J. 2012, 194, 151–157. [Google Scholar] [CrossRef]
- Afghah, Z.; Webb, B.; Meng, X.J.; Ramamoorthy, S. Ten years of pcv2 vaccines and vaccination: Is eradication a possibility? Vet. Microbiol. 2017, 206, 21–28. [Google Scholar] [CrossRef]
- Meng, X.J. Porcine circovirus type 2 (pcv2): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 2013, 1, 43–64. [Google Scholar] [CrossRef]
- Chae, C. Porcine respiratory disease complex: Interaction of vaccination and porcine circovirus type 2, porcine reproductive and respiratory syndrome virus, and mycoplasma hyopneumoniae. Vet. J. 2016, 212, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, A.; Marois-Crehan, C.; Helie, P.; Gagnon, C.A.; Gottschalk, M.; Archambault, M. Genetic diversity of mycoplasma hyopneumoniae isolates of abattoir pigs. Vet. Microbiol. 2014, 168, 348–356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Segales, J. Porcine circovirus type 2 (pcv2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 2012, 164, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Ssemadaali, M.A.; Ilha, M.; Ramamoorthy, S. Genetic diversity of porcine circovirus type 2 and implications for detection and control. Res. Vet. Sci. 2015, 103, 179–186. [Google Scholar] [CrossRef]
- Hayashi, S.; Ohshima, Y.; Furuya, Y.; Nagao, A.; Oroku, K.; Tsutsumi, N.; Sasakawa, C.; Sato, T. First detection of porcine circovirus type 3 in japan. J. Vet. Med. Sci. 2018, 80, 1468–1472. [Google Scholar] [CrossRef]
- Van Renne, N.; Wei, R.; Pochet, N.; Nauwynck, H.J. Dissecting clinical outcome of porcine circovirus type 2 with in vivo derived transcriptomic signatures of host tissue responses. BMC Genom. 2018, 19, 831. [Google Scholar] [CrossRef]
- Vincent, I.E.; Carrasco, C.P.; Guzylack-Piriou, L.; Herrmann, B.; McNeilly, F.; Allan, G.M.; Summerfield, A.; McCullough, K.C. Subset-dependent modulation of dendritic cell activity by circovirus type 2. Immunology 2005, 115, 388–398. [Google Scholar] [CrossRef]
- Wei, R.; Trus, I.; Yang, B.; Huang, L.; Nauwynck, H.J. Breed differences in pcv2 uptake and disintegration in porcine monocytes. Viruses 2018, 10, 562. [Google Scholar] [CrossRef]
- Vincent, I.E.; Balmelli, C.; Meehan, B.; Allan, G.; Summerfield, A.; McCullough, K.C. Silencing of natural interferon producing cell activation by porcine circovirus type 2 DNA. Immunology 2007, 120, 47–56. [Google Scholar] [CrossRef]
- Kekarainen, T.; Montoya, M.; Dominguez, J.; Mateu, E.; Segales, J. Porcine circovirus type 2 (pcv2) viral components immunomodulate recall antigen responses. Vet. Immunol. Immunopathol. 2008, 124, 41–49. [Google Scholar] [CrossRef]
- Wikstrom, F.H.; Meehan, B.M.; Berg, M.; Timmusk, S.; Elving, J.; Fuxler, L.; Magnusson, M.; Allan, G.M.; McNeilly, F.; Fossum, C. Structure-dependent modulation of alpha interferon production by porcine circovirus 2 oligodeoxyribonucleotide and cpg dnas in porcine peripheral blood mononuclear cells. J. Virol. 2007, 81, 4919–4927. [Google Scholar] [CrossRef] [PubMed]
- Ober, R.A.; Thissen, J.B.; Jaing, C.J.; Cino-Ozuna, A.G.; Rowland, R.R.R.; Niederwerder, M.C. Increased microbiome diversity at the time of infection is associated with improved growth rates of pigs after co-infection with porcine reproductive and respiratory syndrome virus (prrsv) and porcine circovirus type 2 (pcv2). Vet. Microbiol. 2017, 208, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Crowther, R.A.; Berriman, J.A.; Curran, W.L.; Allan, G.M.; Todd, D. Comparison of the structures of three circoviruses: Chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J. Virol. 2003, 77, 13036–13041. [Google Scholar] [CrossRef] [PubMed]
- Khayat, R.; Brunn, N.; Speir, J.A.; Hardham, J.M.; Ankenbauer, R.G.; Schneemann, A.; Johnson, J.E. The 2.3-angstrom structure of porcine circovirus 2. J. Virol. 2011, 85, 7856–7862. [Google Scholar] [CrossRef]
- Karuppannan, A.K.; Opriessnig, T. Porcine circovirus type 2 (pcv2) vaccines in the context of current molecular epidemiology. Viruses 2017, 9, 99. [Google Scholar] [CrossRef]
- Duan, J.; Yang, D.; Chen, L.; Yu, Y.; Zhou, J.; Lu, H. Efficient production of porcine circovirus virus-like particles using the nonconventional yeast kluyveromyces marxianus. Appl. Microbiol. Biotechnol. 2019, 103, 833–842. [Google Scholar] [CrossRef]
- Wu, P.C.; Chen, T.Y.; Chi, J.N.; Chien, M.S.; Huang, C. Efficient expression and purification of porcine circovirus type 2 virus-like particles in Escherichia coli. J. Biotechnol. 2016, 220, 78–85. [Google Scholar] [CrossRef]
- Afolabi, K.O.; Iweriebor, B.C.; Okoh, A.I.; Obi, L.C. Global status of porcine circovirus type 2 and its associated diseases in sub-saharan africa. Adv. Virol. 2017, 2017, 6807964. [Google Scholar] [CrossRef]
- Kekarainen, T.; Segales, J. Porcine circovirus 2 immunology and viral evolution. Porc. Health Manag. 2015, 1, 17. [Google Scholar] [CrossRef]
- Li, X.; Meng, X.; Wang, S.; Li, Z.; Yang, L.; Tu, L.; Diao, W.; Yu, C.; Yu, Y.; Yan, C.; et al. Virus-like particles of recombinant pcv2b carrying fmdv-vp1 epitopes induce both anti-pcv and anti-fmdv antibody responses. Appl. Microbiol. Biotechnol. 2018, 102, 10541–10550. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, S.; Zou, Y.; Yu, W.; Jiang, Y.; Zhan, Y.; Wang, N.; Dong, Y.; Yang, Y. Structure-based design of porcine circovirus type 2 chimeric vlps (cvlps) displays foreign peptides on the capsid surface. Front. Cell Infect. Microbiol. 2018, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, N.; Yu, W.; Wang, Z.; Zou, Y.; Zhang, Y.; Wang, A.; Deng, Z.; Yang, Y. Generation and immunogenicity of porcine circovirus type 2 chimeric virus-like particles displaying porcine reproductive and respiratory syndrome virus gp5 epitope b. Vaccine 2016, 34, 1896–1903. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Phylip—phylogeny inference package (version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.6; Department of Genome Sciences, University of Washington: Seattle, WA, USA, 2005. [Google Scholar]
- Kosiol, C.; Goldman, N. Different versions of the dayhoff rate matrix. Mol. Biol. Evol. 2005, 22, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Franzo, G.; Cortey, M.; Olvera, A.; Novosel, D.; Castro, A.M.; Biagini, P.; Segales, J.; Drigo, M. Revisiting the taxonomical classification of porcine circovirus type 2 (pcv2): Still a real challenge. Virol. J. 2015, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids. Symp. 1999, 41, 95–98. [Google Scholar]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. Partitionfinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Stamatakis, A. Raxml-vi-hpc: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree Version 1.4.0. 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 31 March 2020).
- Shang, S.B.; Jin, Y.L.; Jiang, X.T.; Zhou, J.Y.; Zhang, X.; Xing, G.; He, J.L.; Yan, Y. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol. Immunol. 2009, 46, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Rajalingam, D.; Loftis, C.; Xu, J.J.; Kumar, T.K. Trichloroacetic acid-induced protein precipitation involves the reversible association of a stable partially structured intermediate. Protein Sci. 2009, 18, 980–993. [Google Scholar] [CrossRef]
- Franzo, G.; Cortey, M.; Segales, J.; Hughes, J.; Drigo, M. Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol. Phylogenet Evol. 2016, 100, 269–280. [Google Scholar] [CrossRef]
- Neira, V.; Ramos, N.; Tapia, R.; Arbiza, J.; Neira-Carrillo, A.; Quezada, M.; Ruiz, A.; Bucarey, S.A. Genetic analysis of porcine circovirus type 2 from pigs affected with pmws in chile reveals intergenotypic recombination. Virol. J. 2017, 14, 191. [Google Scholar] [CrossRef] [PubMed]
- Dei Giudici, S.; Lo Presti, A.; Bonelli, P.; Angioi, P.P.; Sanna, G.; Zinellu, S.; Balzano, F.; Salis, F.; Ciccozzi, M.; Oggiano, A. Phylogenetic analysis of porcine circovirus type 2 in sardinia, italy, shows genotype 2d circulation among domestic pigs and wild boars. Infect. Genet. Evol. 2019, 71, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, Q.; Chen, H.; Luo, X.; Wu, Q.; Tan, C.; Pan, Q.; Chen, J.L. Evolution and genetic diversity of porcine circovirus 3 in china. Viruses 2019, 11, 786. [Google Scholar] [CrossRef]
- Segales, J. Best practice and future challenges for vaccination against porcine circovirus type 2. Expert Rev. Vaccines 2015, 14, 473–487. [Google Scholar] [CrossRef]
- Huan, C.; Fan, M.; Cheng, Q.; Wang, X.; Gao, Q.; Wang, W.; Gao, S.; Liu, X. Evaluation of the efficacy and cross-protective immunity of live-attenuated chimeric pcv1-2b vaccine against pcv2b and pcv2d subtype challenge in pigs. Front. Microbiol. 2018, 9, 455. [Google Scholar] [CrossRef]
- Cox, M.M. Recombinant protein vaccines produced in insect cells. Vaccine 2012, 30, 1759–1766. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Gomez, A.; Sanchez-Miron, A.; Garcia-Camacho, F.; Molina-Grima, E.; Chisti, Y. Protein production using the baculovirus-insect cell expression system. Biotechnol. Prog. 2014, 30, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Sun, S.; Yang, S.; Shang, Y.; Cai, X.; Liu, X. Self-assembly of virus-like particles of porcine circovirus type 2 capsid protein expressed from Escherichia coli. Virol. J. 2010, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Mo, X.; Xiao, Y.; Yin, B.; Lv, C.; Wang, Y.; Sun, Z.; Yang, Q.; Yao, Y.; Xuan, Y.; et al. Production of Escherichia coli-based virus-like particle vaccine against porcine circovirus type 2 challenge in piglets: Structure characterization and protective efficacy validation. J. Biotechnol. 2016, 223, 8–12. [Google Scholar] [CrossRef]
- Peternel, S.; Komel, R. Isolation of biologically active nanomaterial (inclusion bodies) from bacterial cells. Microb. Cell Fact. 2010, 9, 66. [Google Scholar] [CrossRef]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 2015, 14, 41. [Google Scholar] [CrossRef]
- Jurgen, B.; Breitenstein, A.; Urlacher, V.; Buttner, K.; Lin, H.; Hecker, M.; Schweder, T.; Neubauer, P. Quality control of inclusion bodies in Escherichia coli. Microb. Cell Fact. 2010, 9, 41. [Google Scholar] [CrossRef]
- Invernizzi, G.; Aprile, F.A.; Natalello, A.; Ghisleni, A.; Penco, A.; Relini, A.; Doglia, S.M.; Tortora, P.; Regonesi, M.E. The relationship between aggregation and toxicity of polyglutamine-containing ataxin-3 in the intracellular environment of Escherichia coli. PLoS ONE 2012, 7, e51890. [Google Scholar] [CrossRef]
- Wedrychowicz, H.; Kesik, M.; Kaliniak, M.; Kozak-Cieszczyk, M.; Jedlina-Panasiuk, L.; Jaros, S.; Plucienniczak, A. Vaccine potential of inclusion bodies containing cysteine proteinase of fasciola hepatica in calves and lambs experimentally challenged with metacercariae of the fluke. Vet. Parasitol. 2007, 147, 77–88. [Google Scholar] [CrossRef]
- Myers, J.A.; Curtis, B.S.; Curtis, W.R. Improving accuracy of cell and chromophore concentration measurements using optical density. BMC Biophys. 2013, 6, 4. [Google Scholar] [CrossRef]
- Sorensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.; Ramadan, H.A.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of biopharmaceuticals in e. Coli: Current scenario and future perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [CrossRef] [PubMed]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. Shuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012, 11, 56. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Soto, M.E.; Seibel, J. Expression of functional human sialyltransferases st3gal1 and st6gal1 in Escherichia coli. PLoS ONE 2016, 11, e0155410. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Ke, N.; Berkmen, M. Use of the shuffle strains in production of proteins. Curr. Protoc. Protein Sci. 2016, 85, 5–26. [Google Scholar] [CrossRef]
- Nguyen, J.T.; Fong, J.; Fong, D.; Fong, T.; Lucero, R.M.; Gallimore, J.M.; Burata, O.E.; Parungao, K.; Rascon, A.A., Jr. Soluble expression of recombinant midgut zymogen (native propeptide) proteases from the aedes aegypti mosquito utilizing e. Coli as a host. BMC Biochem. 2018, 19, 12. [Google Scholar] [CrossRef]
- Fathi-Roudsari, M.; Maghsoudi, N.; Maghsoudi, A.; Niazi, S.; Soleiman, M. Auto-induction for high level production of biologically active reteplase in Escherichia coli. Protein Expr. Purif. 2018, 151, 18–22. [Google Scholar] [CrossRef]
- Chen, G.L.; Fu, P.F.; Wang, L.Q.; Li, X.S.; Chen, H.Y. Immune responses of piglets immunized by a recombinant plasmid containing porcine circovirus type 2 and porcine interleukin-18 genes. Viral. Immunol. 2014, 27, 521–528. [Google Scholar] [CrossRef]
- Canelli, E.; Borghetti, P.; Ferrari, L.; De Angelis, E.; Ferrarini, G.; Catella, A.; Ogno, G.; Martelli, P. Immune response to pcv2 vaccination in prrsv viraemic piglets. Vet. Rec. 2016, 178, 193. [Google Scholar] [CrossRef]
- Ouyang, T.; Zhang, X.; Liu, X.; Ren, L. Co-infection of swine with porcine circovirus type 2 and other swine viruses. Viruses 2019, 11, 185. [Google Scholar] [CrossRef]
- Zhang, H.; Qian, P.; Peng, B.; Shi, L.; Chen, H.; Li, X. A novel subunit vaccine co-expressing gm-csf and pcv2b cap protein enhances protective immunity against porcine circovirus type 2 in piglets. Vaccine 2015, 33, 2449–2456. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Lyoo, K.S.; Kim, K.; Do, V.T.; Lee, K.W.; Hahn, T.W. A pilot comparative study of recombinant protein and whole-virus inactivated vaccines against porcine circovirus type 2 in conventionally reared pigs. Res. Vet. Sci. 2019, 123, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Enders, J.F.; Katz, S.L.; Milovanovic, M.V.; Holloway, A. Studies on an attenuated measles-virus vaccine. I. Development and preparations of the vaccine: Technics for assay of effects of vaccination. N. Engl. J. Med. 1960, 263, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Okuno, Y.; Otsuka, T.; Osame, J.; Takamizawa, A. Development of a live attenuated varicella vaccine. Biken. J. 1975, 18, 25–33. [Google Scholar] [PubMed]
- Mishra, R.P.N.; Yadav, R.S.P.; Jones, C.; Nocadello, S.; Minasov, G.; Shuvalova, L.A.; Anderson, W.F.; Goel, A. Structural and immunological characterization of e. Coli derived recombinant crm197 protein used as carrier in conjugate vaccines. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Gonzalez-Silvera, D.; Guardiola, F.A.; Espinosa, C.; Chaves-Pozo, E.; Esteban, M.A.; Cuesta, A. Recombinant nodavirus vaccine produced in bacteria and administered without purification elicits humoral immunity and protects european sea bass against infection. Fish. Shellfish Immunol. 2019, 88, 458–463. [Google Scholar] [CrossRef]
- Kay, E.; Cuccui, J.; Wren, B.W. Recent advances in the production of recombinant glycoconjugate vaccines. NPJ Vaccines 2019, 4, 16. [Google Scholar] [CrossRef]
- Jia, B.; Jeon, C.O. High-throughput recombinant protein expression in Escherichia coli: Current status and future perspectives. Open Biol. 2016, 6. [Google Scholar] [CrossRef]
- Ozturk, S.; Ergun, B.G.; Calik, P. Double promoter expression systems for recombinant protein production by industrial microorganisms. Appl. Microbiol. Biotechnol. 2017, 101, 7459–7475. [Google Scholar] [CrossRef]
- Zepeda-Cervantes, J.; Cruz-Resendiz, A.; Sampieri, A.; Carreon-Napoles, R.; Sanchez-Betancourt, J.I.; Vaca, L. Incorporation of orf2 from porcine circovirus type 2(pcv2) into genetically encoded nanoparticles as a novel vaccine using a self-aggregating peptide. Vaccine 2019, 37, 1928–1937. [Google Scholar] [CrossRef]
- Montesino, R.; Toledo, J.R.; Sanchez, O.; Zamora, Y.; Barrera, M.; Royle, L.; Rudd, P.M.; Dwek, R.A.; Harvey, D.J.; Cremata, J.A. N-glycosylation pattern of e2 glycoprotein from classical swine fever virus. J. Proteome Res. 2009, 8, 546–555. [Google Scholar] [CrossRef] [PubMed]

| Fermentation | Total Biomass (g in 5 L) | Total Protein (mg/g Of Disrupted Biomass) | Qm1 (mg/g Of Disrupted Biomass) | % Qm1 (Of Protein) | Qm1 Total (mg) | Qm1 Productivity (mg/L/h) |
|---|---|---|---|---|---|---|
| F1 | 10.6 | 32.0 | 2.83 | 8.8 | 107.6 | 2.7 |
| F2 | 6.4 | 42.7 | 3.10 | 7.3 | 71.3 | 1.8 |
| F3 | 8.1 | 52.9 | 5.24 | 9.9 | 152.0 | 3.8 |
| Average | 8.4 ± 1.7 | 42.5 ± 8.5 | 3.7 ± 1.1 | 8.7 ± 1.1 | 110.3 ± 33 | 2.8 ± 0.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamazares, E.; Gutiérrez, F.; Hidalgo, A.; Gutiérrez, N.A.; Espinoza, F.I.; Sánchez, O.; Cortez-San Martín, M.; Mascayano, C.; González, J.; Saavedra, J.; et al. A Heterologous Viral Protein Scaffold for Chimeric Antigen Design: An Example PCV2 Virus Vaccine Candidate. Viruses 2020, 12, 385. https://doi.org/10.3390/v12040385
Lamazares E, Gutiérrez F, Hidalgo A, Gutiérrez NA, Espinoza FI, Sánchez O, Cortez-San Martín M, Mascayano C, González J, Saavedra J, et al. A Heterologous Viral Protein Scaffold for Chimeric Antigen Design: An Example PCV2 Virus Vaccine Candidate. Viruses. 2020; 12(4):385. https://doi.org/10.3390/v12040385
Chicago/Turabian StyleLamazares, Emilio, Fernando Gutiérrez, Angela Hidalgo, Nicolas A. Gutiérrez, Felipe I. Espinoza, Oliberto Sánchez, Marcelo Cortez-San Martín, Carolina Mascayano, Javier González, José Saavedra, and et al. 2020. "A Heterologous Viral Protein Scaffold for Chimeric Antigen Design: An Example PCV2 Virus Vaccine Candidate" Viruses 12, no. 4: 385. https://doi.org/10.3390/v12040385
APA StyleLamazares, E., Gutiérrez, F., Hidalgo, A., Gutiérrez, N. A., Espinoza, F. I., Sánchez, O., Cortez-San Martín, M., Mascayano, C., González, J., Saavedra, J., Altamirano, C., Mansur, M., Ruiz, Á., & Toledo, J. R. (2020). A Heterologous Viral Protein Scaffold for Chimeric Antigen Design: An Example PCV2 Virus Vaccine Candidate. Viruses, 12(4), 385. https://doi.org/10.3390/v12040385





