Abstract
Hepatitis B virus (HBV) infects the liver resulting in end stage liver disease, cirrhosis, and hepatocellular carcinoma. Despite an effective vaccine, HBV poses a serious health problem globally, accounting for 257 million chronic carriers. Unique features of HBV, including its narrow virus–host range and its hepatocyte tropism, have led to major challenges in the development of suitable in vivo and in vitro model systems to recapitulate the HBV replication cycle and to test various antiviral strategies. Moreover, HBV is classified into at least nine genotypes and 35 sub-genotypes with distinct geographical distributions and prevalence, which have different natural histories of infection, clinical manifestation, and response to current antiviral agents. Here, we review various in vitro systems used to study the molecular biology of the different (sub)genotypes of HBV and their response to antiviral agents, and we discuss their strengths and limitations. Despite the advances made, no system is ideal for pan-genotypic HBV research or drug development and therefore further improvement is required. It is necessary to establish a centralized repository of HBV-related generated materials, which are readily accessible to HBV researchers, with international collaboration toward advancement and development of in vitro model systems for testing new HBV antivirals to ensure their pan-genotypic and/or customized activity.
1. Introduction
Hepatitis B is an inflammatory disease of the liver caused by a partially double-stranded enveloped hepatitis B virus (HBV). The virus persistently infects the liver resulting in end stage liver disease, cirrhosis, and hepatocellular carcinoma. Since its discovery in the 1960s [1,2,3,4,5], HBV continues to pose a serious health problem worldwide, accounting for 257 million chronically infected cases in 2015 [6], despite the existence of an effective vaccine. Approved HBV treatment regimens are restricted to interferon and nucleos(t)ide analogues (NAs), but these drugs can only efficiently suppress viral replication, without eliminating the virus [7,8,9]. Interferons, which act as immunomodulators and interfere indirectly with HBV replication, are administered by injection and require long-term use, which can be associated with significant side effects [10]. On the other hand, NAs suppress HBV replication primarily by hindering the process of reverse transcription, a characteristic and crucial step in the HBV replication cycle. A number of the earlier NAs, such as lamivudine, give rise to drug resistance HBV strains, which can result in adverse long-term clinical effects. Therefore, to achieve better treatment outcomes and the ultimate elimination of the virus, there is a need for more potent inhibitors and in vitro systems in which to test them and biomarkers to measure their effect [11].
The discovery of the sodium–taurocholate co-transporting polypeptide (NTCP) as the receptor for HBV entry into hepatocytes has stimulated new efforts for the development of novel antiviral strategies and newer systems to test them [12]. HBV is classified into at least nine genotypes and 35 sub-genotypes, with distinct geographical distributions and prevalence [13,14,15]. These (sub)genotypes can have different natural histories of infection, clinical manifestation, and response to antiviral agents [16]. Therefore it is important that the systems used to monitor antiviral efficacy of various agents are able to test the response of the various (sub)genotypes in order to ensure that the antiviral modalities are pan-genotypic (or customized if a pan-genotypic effect is not possible) and can be used in all regions of the world, especially where HBV is endemic, such as in Africa and Asia. Here we review the various in vitro systems that have been used to study the molecular biology of the different (sub)genotypes of HBV and their response to antiviral agents, and we discuss their strengths and limitations.
2. Molecular Biology of Hepatitis B Virus
HBV, the prototype member of the genus Orthohepadnavirus, family Hepadnaviridae, is the smallest DNA virus infecting humans. It has a partially double stranded 3200 base pair (bp) genome, with four open reading frames encoding seven proteins. The four open reading frames, which are completely or partly overlapping include: The precore/core (preC/C) for HBeAg and HBcAg (capsid protein); P for polymerase (including reverse transcriptase); PreS1/PreS2/S for three envelope proteins (large Hepatitis B surface: LHBs, middle Hepatitis B surface: MHBs, and small Hepatitis B surface: SHBs (HBsAg)); and X for a transcriptional trans-activator protein, x [17].
HBV binds to the NTCP, which is the viral receptor on the hepatocytes [12]. Once in the cytoplasm, the genome is uncoated and imported into the nucleus, where the partially double stranded genome is repaired to give rise to covalently closed circular DNA (cccDNA), which is the template for HBV transcription by the cellular RNA polymerase II [18]. In the cytoplasm, the HBV transcripts are translated into the structural and non-structural proteins. The pre-genomic RNA (pgRNA) together with the viral polymerase are encapsidated into the capsid, where the RNA intermediate is reverse transcribed into the negative strand DNA from which the plus strand is synthesized. The final stage in the viral assembly is the envelopment of the nucleocapsid, with the viral envelope proteins, by budding into the endoplasmic reticulum to form the mature virions, which are then released from the cells. Alternatively, the nucleocapsid can be recycled back into the nucleus, where the relaxed circular DNA is repaired, to maintain the reservoir of cccDNA [18]. In addition to the complete virions, HBV expresses non-infectious filamentous and spherical sub-viral particles composed mainly of HBsAg, the extraparticulate HBeAg, and x protein (Figure 1).
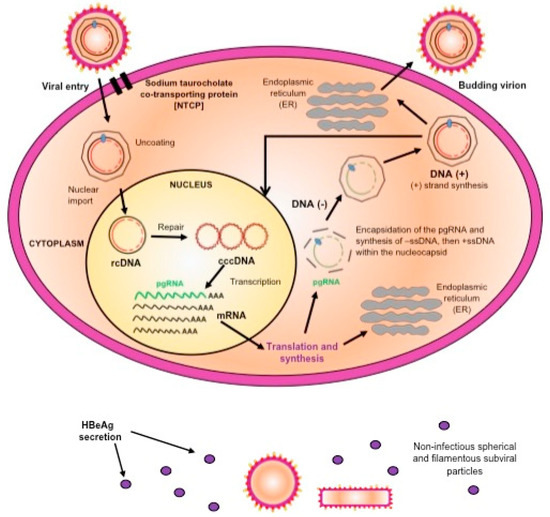
Figure 1.
A schematic representation of the replication cycle of hepatitis B virus (HBV).
3. Genotypes/Sub-Genotypes of HBV
Sequence heterogeneity is characteristic of HBV because the viral-encoded polymerase, a reverse transriptase, lacks proof-reading ability. HBV has been classified phylogenetically into nine genotypes, A to I [13,14,19,20], based on an intergroup divergence of greater than 7.5% across the complete genome, with a putative 10th genotype, “J”, isolated from a single individual [21], which is a recombinant of genotype C and gibbon HBV [22] and clustering with sub-genotype C4 [23] in the S region. Genotypes A–D, F, H, and I are classified further into at least 35 sub-genotypes, having between a ~4% and 8% intergroup nucleotide difference across the complete genome and good bootstrap support [13]. The genotypes differ in genome length, the size of ORFs and the proteins translated [14], as well as the development of various mutations [24]. HBV has been classified into nine serological subtypes, ayw1, ayw2, ayw3, ayw4, ayr, adw2, adw4, adwq, adr, and adrq based on HBsAg heterogeneity [14]. A broad, highly statistically significant relationship exists between serological subtypes and genotypes: adw is associated with genotypes A, B, F, G, and H; adr with C; and ayw with D and E [25], but there are exceptions. The (sub)genotype of HBV can influence the outcome of HBV infection because it can affect the frequency of HBeAg-positivity, the age at which HBeAg loss occurs and the mode of transmission [26]. Therefore, the natural history of HBV infection can differ in different geographical regions [12,13,24,27,28] and references cited therein. Furthermore, the (sub)genotype can affect the response to antiviral therapy [24,27] and possibly vaccination [29,30].
4. In Vitro Systems for the Study of HBV
Significant features of HBV including its narrow virus–host range and its strong tropism for hepatocytes [31], have led to major challenges in the development of suitable in vivo and in vitro model systems to recapitulate the in vivo human hepatocyte HBV replication cycle [32,33]. The chimpanzee (Pan troglodytes) and the macaque (Macaca fascicularis) are the only non-human primate models for HBV infection, whereas the tree shrew (Tupaia belangeri) is susceptible to HBV infection [33,34]. However, these in vivo animal models have several limitations, including ethical restrictions, high costs and large size in the case of chimpanzees [35], lack of reproducibility in the case of macaques [34,36], and poor infection efficiency and mild and transient infection in the case of the tree shrew [37]. Although a Mauritian macaque colony has been found to be naturally infected with HBV closely related to genotype D [34], this infection could not be recapitulated in vitro or in vivo by others, unless the macaque hepatocytes were transduced by NTCP [36]. A lack of a robust and reproducible in vitro cell culture system that is capable of supporting all the steps of the HBV replication cycle, including infection and formation of cccDNA, has also led to the hindrance of the study of the virus in terms of the mechanisms of the early stages of virus–cell interactions, and the development of anti-HBV drugs [38,39,40,41]. Therefore, to date, most of our understanding on how the HBV functions has come from in vitro and in vivo studies using the duck HBV (DHBV) [42], woodchuck HBV (WHBV) [43], and ground squirrel HBV (GSHBV) [44] models. Over the years, various in vitro cell culture systems have been developed that have enabled the study of the molecular and genetic characteristics of HBV, and the history of their development is depicted in Figure 2.
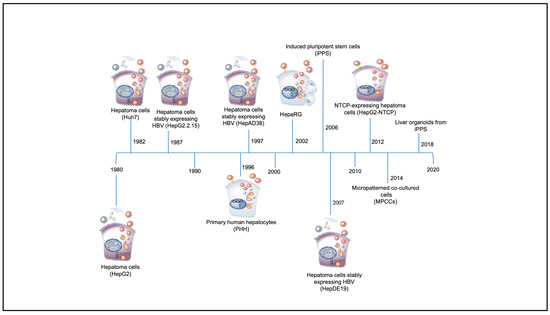
Figure 2.
Timeline of discovery of in vitro hepatitis B virus (HBV) systems (adapted from [45]).
The various in vitro model systems for the study of HBV have been extensively reviewed elsewhere [32,33,40,41,45,46] and Table 1 provides a brief summary of their strengths and limitations.

Table 1.
Summary of the in vitro model systems suitable for studying hepatitis B virus (HBV).
4.1. In Vitro Model Systems Based on Hepatoma Cells (HepG2, Huh7, HepG2.2.15, and HepAD38)
Using recombinant HBV DNA constructs carrying over-length HBV genomes, well-differentiated human hepatic cell lines (such as HepG2, Huh6, and Huh7 derived from HCCs) have been transfected to study mechanisms of HBV replication and morphogenesis [59,80,91]. These neoplastic immortal cells are easier to culture and have stable enzyme concentrations when compared to primary hepatocytes. However, they have absent or low expression levels of drug metabolizing enzymes, thus restricting their application [92,93]. Although HBV cannot infect these cell lines most likely as a result of the loss of cell surface receptors during de-differentiation of the hepatocyte [94], they have been widely used and have been invaluable in the study of the various aspects of the life cycle following either: (i) transient transfection or transduction of recombinant HBV DNA using baculoviral, adenoviral, and lentiviral vectors for delivery; or (ii) transfection with recombinant cccDNA (rcccDNA), generated by a minicircle-based technique. This transfection can recapitulate the expression of HBV RNAs and proteins in Huh-7 cells [95] and (iii) the generation of stably transfected cell lines containing integrated HBV DNA genomes [59,91,96]; or (iv) the development of stable rcccDNA-producing cell lines termed HepG2-HBV/loxP [97]. However, these cell culture systems are unsuitable for studying HBV-host cell infection mechanisms such as viral attachment, penetration, and uncoating of the virus and to follow the development of hepatitis and hepatocellular carcinoma (HCC) [98,99]. The rcccDNA systems are excellent for studying the molecular biology of cccDNA and for screening of antiviral agents, which can silence or eliminate HBV cccDNA. Surrogate in vivo mouse models of chronic hepatitis have been established by delivering rcccDNA to mouse hepatocytes using adenoviral vectors [100].
In 1987, the widely used HepG2.2.15 cell line generated by Sell et al. [91], which contains multiple copies of the HBV genotype D genome, was shown to stably express HBV viral gene products. These HepG2.2.15 cells were later used to study the occurrence of spontaneous HBV integrations in the host genome, showing that DNA damage increases the frequency of integration. However, because HBV viral particles produced are generated from chromosomally integrated DNA, the HepAD38 cell line, established by Ladner and colleagues expressing HBV with Tet-OFF and Tet-ON regulatory systems was generated [74]. This was possible following co-transfection of HepG2 cells with plasmids ptetHBV and pUHD15-1neo under the influence of a tetracycline responsive promoter. The successful establishment of this system has permitted for an improved and more strongly controlled platform to study HBV, as well as resulting in a more robust production of viral particles with increased accumulation of cccDNA in the cells.
Another cell line the HepDE19, was generated containing a 1.1 mer HBV transgene mutated in its 5′ pre-core ATG leaving the 3′ pre-core ATG unchanged. Using this strategy, the expression of HBV e-antigen (HBeAg) was now from the episomal DNA and not from the integrated DNA, thus providing a platform for screening cccDNA-targeting drugs on a large-scale [72,101]. A “second-generation” cccDNA reporter cell line, termed HepBHAe82 was developed where an in-frame haemagglutinin (HA) epitope tag was introduced into the precore domain of HBeAg open reading frame in the transgene of HepBHAe82 cells without disrupting any cis-element critical for HBV replication and HBeAg secretion [102]. These developments have allowed for HBV production in vitro, however, their use is limited when it comes to studying the regulation of HBV replication. This is because some carry greater-than genome length HBV and a neomycin resistance gene. So, the quest continues to establish stable HBV-expressing cell lines that will enable the study of the relationship between HBV and host genes.
4.2. In Vitro Model Systems Based on Primary Human Hepatocytes (PHH)
The discovery of PHHs in 1996 as the only cells to be infected by the authentic HBV in vitro in their fully differentiated form has long enabled in vitro studies of HBV infection in a system with an intact host defense system [53,103]. However, in addition to their limited availability, infection of PHHs with HBV is inefficient because these cells are only viable for a few days after culturing even upon supplementation with dimethyl sulphoxide (DMSO) [54,103]. Also, indefinite maintenance in culture results in loss of liver-specific functions and de-differentiation into fibroblasts, resulting in the impairment of HBV replication, which relies on hepatocyte nuclear factors for transcription within these cells [41,92,94,104]. Further, heterogeneity in the quality of PHHs and the variation in susceptibility to HBV infection results in maintenance difficulty and generation of experiments of poor reproducibility, with great inter-experimental variation that is difficult to control [53,105].
4.3. In Vitro Model Systems Based on Differentiated Hepatoma Cell Lines (HepaRG)
In 2002, HepaRG cells, a human hepatoma cell line derived from HCC from a female with chronic hepatitis C infection [106], was shown to be permissive to HBV infection under certain conditions and extended culture time. This cell line contains hepatic progenitor cells that make them susceptible to HBV/HDV infection after differentiation by the addition of DMSO and hydrocortisone [72,73,77,106]. Although HepRG can support HBV infection, cccDNA formation, and secretion of infectious viral particles into the culture medium [72,73], it is an unsuitable system to study the complete HBV life cycle and to evaluate antiviral compounds because of several disadvantages, which include: (i) a complex and time consuming induction of the differentiation process prior to infection with HBV, which requires the addition of DMSO [72]; (ii) cells exhibit heterogeneity in albumin expression and chromosomal abnormalities [41,46]; and (iii) the activity of the number of enzymes involved in drug metabolism varies when compared to PHHs [107,108]. These cells also allow for low to minimal cell-to-cell spread of HBV.
4.4. In Vitro Model Systems Based on NTCP Expressing Cell Lines
The middle HBsAg of HBV initially attaches to heparan sulfate proteoglycans (HSP) on the hepatocyte [109]. This is followed by binding of the large HbsAg to the NTCP, which is the essential receptor for HBV infection [12]. NTCP is a sodium-bile acid pump, coded by the SLC10A1 gene, largely expressed in liver cells and is restricted to the sinusoidal plasma membrane. The only established susceptible cell line expressing NTCP is HepaRG. However, its heterogeneity in albumin expression and chromosomal abnormalities do not allow the study the complete HBV life cycle and the evaluation of antiviral compounds. Thus, DMSO-induced differentiation is required [71].
The exogenous expression of NTCP in hepatoma cell lines can render these cell lines susceptible to HBV infection. The establishment of HepG2 and Huh7-based cell lines in which NTCP is over expressed provides a much-needed and easily accessible platform for studying HBV. HepG2-NTCP cells could also be used to identify chemicals targeting key steps of the virus life cycle including HBV cccDNA, and enable the development of novel antivirals against the infection. However, although improved techniques, such as spinoculation, during HBV inoculation and the addition of DMSO in culture media has greatly enhanced infection efficiency of NTCP expressing cells, the system still fails to recap the full HBV life cycle [85]. The reasons being that in contrast to in vivo, the system requires very high multiplicity of infection (MOI), the infection is short-lived, does not result in substantial viral spreading, and only a modest amount of cccDNA is detected. As previously reported [103,110,111,112], HBV infection in cell culture systems including adult primary human hepatocytes, HepaRG hepatoma cells, and more recently HepG2-NTCP cells, is enhanced after the addition of polyethylene glycol 8000 (PEG), which promotes the binding of HBV to HSP. Furthermore, a study by Michailidis and coworkers, showed that maintaining PEG in cell culture medium increases infection by at least one order of magnitude, as a result of improved viral spread [113].
4.5. In Vitro Model Systems Based on Inducible Pluripotent Stem Cell (iPSCs)
iPSCs are pluripotent reprogrammed cells derived from either adult skin or blood cells and were first discovered by Japanese researchers following the introduction of genes necessary for the expression of a set of transcription factors in specialized adult cells [114]. The iPSCs are capable of self-renewal and differentiation into different body cell types, except for extra-embryonic tissue cells, like the placenta, making them a promising cell source for regenerative therapy in several disease states. To limit donor variability biases and in an attempt to increase hepatocyte availability, in 2006, hepatocyte-like cells (iHeps/HLCs) were differentiated from iPSCs [89,115], and in 2014 Shlomai and colleagues, first established that HBV could infect iPSC-derived HLCs [89]. They showed that HBsAg was efficiently produced in the supernatant after infection; however, the HBsAg levels gradually dropped to background levels. In their study, Sakurai and coworkers observed an increase in HBsAg secretion in culture supernatant up to 17 days following HBV infection in iPS-HLCs [87]. This indicated that iPS-HLCs could support long-term HBV infection. Xia and coworkers supported this finding when they utilized human iPS-derived HLCs as a robust and convenient in vitro model to study HBV [86]. Thus iPS-HLCs provide a promising in vitro HBV infection model and pave the way to dissect the underlying mechanisms of HBV infection and the development of novel anti-HBV drugs.
4.6. In Vitro Model Systems Based on Micropatterned Co-Cultured Cells (MPPCs)
Using a combination of microtechnology and tissue engineering techniques, Khetani and Bhatia [116] established a miniature-like multi-well culture system for human liver cells termed micropatterned co-cultured (MPCC) system. The MPCC system was shown to preserve hepatocyte functions for a prolonged period following their plating, thereby serving as a platform for drug toxicity and drug interaction studies. In their study, Shlomai et al. [89] showed that MPCCs support productive HBV infection and, by blocking elements of the hepatocyte innate immune response related to the initiation of IFN-stimulated genes, HBV infectivity can be enhanced. The ability to sustain lengthy and productive HBV infection, makes MPCCs a facile platform for studying virus–host interactions and developing antiviral medications. Despite its advantage in HBV research in providing useful information with respect to the activation of the innate immune response following HBV infection, the MPCC system does not fully support the spread of infection with only minimal infection efficiency of less than 50%. In addition, the system does not provide sustained cccDNA, pre-genomic RNA, HBeAg as well HBsAg production, and re-infection of naïve cells with medium collected from infected cells is not possible, suggesting that the MPCC system is not robust enough to yield highly infectious viral particles [116]. Furthermore, it should be noted that this culture system is time consuming and technically challenging to establish [116,117].
4.7. In Vitro Model Systems Based on Liver Organoids
Over the years, the use of human hepatoma cell lines and humanized mouse models in HBV research has increased, but these systems remain poor in recapitulating the complex biology of hepatocytes. Their high cost, difficulty to obtain, and impracticality in drug screening on a large scale has led to the continued search for better cell culture models [118]. Organoid cultures have arisen as a substitute in vitro system to mimic tissues and join the gap between 2D cultures and in vivo mouse/human models. These liver organoids have been established for multiple species derived from induced pluripotent stem cells, embryonic stem cells, hepatoblasts, and adult tissue-derived cells. To some degree, liver organoid cultures recapitulate the complexity and design of the liver and may offer new insights into host-to-organism interactions. Differentiated liver organoids maintain innate immune responses and retain cell polarity of hepatocytes, mimicking the natural entry of HBV, thus permitting their cell-to-cell transmission [119]. Following the establishment of a functional human induced pluripotent stem cell liver organoid (hiPPSC-LO), recent studies show that these are a suitable in vitro culture system to study and model HBV infections [90]. This system is an advancement in the models for generating fundamental knowledge of HBV biology and providing a promising platform toward screening potential new therapies and the development of customized hepatitis treatment.
Although we have come a long way, the development of the ideal in vitro system continues as none of the systems currently available are without limitations. The ultimate in vitro model for the study of HBV infection and its response to various antiviral agents should ideally:
- Express NTCP
- Maintain hepatocyte function and susceptibility to HBV infection indefinitely
- Not require the addition of DMSO to maintain hepatocyte function or PEG to promote infection
- Be capable of being infected with high efficiency with multiple HBV genotypes/sub-genotypes and variants.
- Express the host factors necessary to support HBV infection
- Have high longevity to support the complete viral life cycle
- Have an intact innate immune response
- Have functional pathways
- Be genetically homogeneous
- Recapitulate HBV infection seen in patients or in vivo systems
- Be renewable
- Be of unlimited supply
- Allow for the testing of a wide range of antiviral and immunomodulatory agents
- Be low cost
- Allow for miniaturization
- Be ethically acceptable
5. The Use of In Vitro Systems to Study Genotypes/Sub-Genotypes of HBV
Although a number of in vitro model systems have been used to study HBV, very few have compared the different (sub)genotypes. Moreover, a panel of strains representative of each (sub)genotype has not been established, making comparisons across studies difficult because of the range of constructs, methods of transfection/infection/transduction, and cell lines used. It is important to note that many earlier studies did not classify the genotype of the virus used [47,53,54,56,57]. Table 2 provides a summary of studies using either viral particles for infection and replication competent HBV DNA, in various delivery vectors, to follow the replication of different serotypes and/or (sub)genotypes in the different in vitro model systems.

Table 2.
Studies conducted for the various genotypes/sub-genotypes of hepatitis B virus (HBV) in different in vitro model systems.
As is evident from the above table, the majority of studies to functionally characterize the different (sub)genotypes of HBV in vitro have been carried out on genotype D, with a single study for each of genotypes I [19] and J [130]. The only stably transfected cell line expressing HBV and currently widely available is Hep2.2.15 expressing genotype D and this is the most frequent source of cell-line derived inoculum. It will be important to have cell lines expressing other (sub)genotypes generated and a number of these are currently under development. Although genotype D is a cosmopolitan and diverse genotype with serological subtype ayw, it differs from other genotypes by having a 33-nucleotide deletion at the amino terminus of the pre-S1. Therefore, it is important that systems are developed that express HBV without this deletion and with serological subtype adw (genotypes A, B, F and H) and adr (genotype C) [13].
The genotypes and in some cases sub-genotypes of HBV have distinct geographical distributions [13,14]. In the two regions of the world where HBV is endemic different genotypes prevail: in Asia, genotype B and C; whereas in sub-Saharan Africa, genotypes A, D, and E. Moreover, for the sub-genotype of A, A1 circulating in Africa is different from A2, which is prevailing outside Africa, and they differ in molecular characteristics and natural history [137,138,139,140]. The studies carried out in one region cannot necessarily be extrapolated to other regions. The natural history and response to various antiviral agents can be influenced by the genetic heterogeneity of the (sub)genotypes and therefore this should be taken into account when designing and testing various antiviral modalities. It is important to be aware that even though in some cases the efficacy of antiviral agents may not be affected by sequence heterogeneity, in other cases it is of utmost importance. As all the (sub)genotypes use NTCP to enter to the hepatocyte, the efficacy of entry-inhibitors will not differ between the different (sub)genotypes. On the other hand, sequence heterogeneity of HBV may be challenging when designing guide RNA (gRNA) for the different CRISPR/Cas9 systems and avoiding off-target effects [135]. Thus, in some cases antiviral agents may be pangenotypic, whereas in others the agent will have to be customized for the (sub)genotype prevailing in a particular geographic region and/or population.
6. Knowledge Gaps and Future Prospective
Systems to transfect cells with HBV DNA and study its replication have been available for over three decades enabling the study of processes of chronic HBV infection and the identification of many agents that target the later stages of the HBV life cycle. These include HBV DNA transcription, RNA stability, capsid assembly, RNaseH digestion, virion secretion, HBsAg secretion, and reverse transcription inhibitors. Studies of HepG2-NTCP cells and other cell types that express the NTCP transgene have led to the identification of Myrcludex B, an agent that interrupts the entry phase of the HBV life cycle as well as other HBV entry inhibitors. Systems targeting cccDNA could facilitate efforts to find a cure for chronic HBV infection [11]. Despite the advances made, no system is ideal for pan-genotypic HBV research or drug development and therefore their robustness and reliability require further improvement. With the WHO’s vision to end HBV infection by 2030, more work is needed to develop better models that will facilitate the search for a cure for chronic HBV infection. This can be achieved through the establishment of cell culture systems that most strictly resemble human hepatocytes but are more convenient, less costly, limitless in supply or more readily available, and allow more efficient amplification of infection and spread. Improved efforts to stably maintain primary human hepatocyte culture conditions and optimization of iHep cells are of interest. This emphasizes the need to establish a centralized repository of all HBV-related generated materials [141] and protocols [142], which are readily accessible to HBV researchers, with international collaboration toward the advancement and development of in vitro model systems for testing new HBV antivirals with either pan-genotypic or (sub)genotypic activity, depending on the requirement.
Author Contributions
All the authors contributed equally to the work reported. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the South African National Research Foundation (NRF) with a Postdoctoral Fellowship (WOSE016 to CNWK)”, the Poliomyelitis Research Foundation South Africa (grant number 16/29 to CNWK), University of the Witwatersrand URC Postdoctoral Fellowship (to CNWK), Cancer Association of South Africa (CANSA to AK), and Deutsche Forschungsgemeinschaft. (DFG; German Research Foundation; DFGR000 to AK).
Acknowledgments
Figure 1 was prepared by Hillary Vos. We thank her for allowing us to use the figure.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Prince, A.M. Relation of Australia and SH antigens. Lancet 1968, 2, 462–463. [Google Scholar] [CrossRef]
- Prince, A.M. An antigen detected in the blood during the incubation period of serum hepatitis. Proc. Natl. Acad. Sci. USA 1968, 60, 814–821. [Google Scholar] [CrossRef]
- Blumberg, B.S.; Alter, H.J.; Visnich, S. A “New” Antigen in Leukemia Sera. JAMA 1965, 191, 541–546. [Google Scholar] [CrossRef]
- Blumberg, B.S.; Gerstley, B.J.; Hungerford, D.A.; London, W.T.; Sutnick, A.I. A serum antigen (Australia antigen) in Down’s syndrome, leukemia, and hepatitis. Ann. Intern. Med. 1967, 66, 924–931. [Google Scholar] [CrossRef]
- Alter, H.J.; Blumberg, B.S. Further studies on a “new” human isoprecipitin system (Australia antigen). Blood 1966, 27, 297–309. [Google Scholar] [CrossRef]
- World Health Organization. Global Hepatitis Report, 2017; WHO: Geneva, Switzerland, 2017; ISBN 978-92-4-156545-5. [Google Scholar]
- Flink, H.J.; van Zonneveld, M.; Hansen, B.E.; de Man, R.A.; Schalm, S.W.; Janssen, H.L.A.; Grp, H.-S. Treatment with Peg-interferon alpha-2b for HBeAg-positive chronic hepatitis B: HBsAg loss is associated with HBV genotype. Am. J. Gastroenterol. 2006, 101, 297–303. [Google Scholar] [CrossRef]
- Sonneveld, M.J.; Rijckborst, V.; Boucher, C.A.B.; Hansen, B.E.; Janssen, H.L.A. Prediction of Sustained Response to Peginterferon Alfa-2b for Hepatitis B e Antigen Positive Chronic Hepatitis B Using On-Treatment Hepatitis B Surface Antigen Decline. Hepatology 2010, 52, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.C.; Yu, M.L.; Liu, C.J.; Lin, C.L.; Huang, Y.W.; Hsu, C.S.; Liu, C.H.; Kuo, S.F.T.; Pan, C.J.H.; Yang, S.S.; et al. Effect of host and viral factors on hepatitis B e antigen-positive chronic hepatitis B patients receiving pegylated interferon-alpha-2a therapy. Antivir. Ther. 2011, 16, 629–637. [Google Scholar] [CrossRef]
- Arends, P.; Rijckborst, V.; Zondervan, P.E.; Buster, E.; Cakaloglu, Y.; Ferenci, P.; Tabak, F.; Akarca, U.S.; Simon, K.; Sonneveld, M.J.; et al. Loss of intrahepatic HBsAg expression predicts sustained response to peginterferon and is reflected by pronounced serum HBsAg decline. J. Viral Hepat. 2014, 21, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Revill, P.A.; Chisari, F.V.; Block, J.M.; Dandri, M.; Gehring, A.J.; Guo, H.; Hu, J.; Kramvis, A.; Lampertico, P.; Janssen, H.L.A.; et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 2019, 4, 545–558. [Google Scholar] [CrossRef]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 2012, 1, e00049. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Kew, M.; Francois, G. Hepatitis B virus genotypes. Vaccine 2005, 23, 2409–2423. [Google Scholar] [CrossRef] [PubMed]
- Velkov, S.; Ott, J.J.; Protzer, U.; Michler, T. The Global Hepatitis B Virus Genotype Distribution Approximated from Available Genotyping Data. Genes 2018, 9, 495. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Kew, M.C. Epidemiology of hepatitis B virus in Africa, its genotypes and clinical associations of genotypes. Hepatol. Res. 2007, 37, S9–S19. [Google Scholar] [CrossRef] [PubMed]
- Tiollais, P.; Pourcel, C.; Dejean, A. The hepatitis B virus. Nature 1985, 317, 489–495. [Google Scholar] [CrossRef]
- Beck, J.; Nassal, M. Hepatitis B virus replication. World J. Gastroenterol. 2007, 13, 48–64. [Google Scholar] [CrossRef]
- Yu, H.; Yuan, Q.; Ge, S.X.; Wang, H.Y.; Zhang, Y.L.; Chen, Q.R.; Zhang, J.; Chen, P.J.; Xia, N.S. Molecular and phylogenetic analyses suggest an additional hepatitis B virus genotype “I”. PLoS ONE 2010, 5, e9297. [Google Scholar] [CrossRef]
- Norder, H.; Courouce, A.M.; Coursaget, P.; Echevarria, J.M.; Lee, S.D.; Mushahwar, I.K.; Robertson, B.H.; Locarnini, S.; Magnius, L.O. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology 2004, 47, 289–309. [Google Scholar] [CrossRef]
- Tatematsu, K.; Tanaka, Y.; Kurbanov, F.; Sugauchi, F.; Mano, S.; Maeshiro, T.; Nakayoshi, T.; Wakuta, M.; Miyakawa, Y.; Mizokami, M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J. Virol. 2009, 83, 10538–10547. [Google Scholar] [CrossRef]
- Locarnini, S.; Littlejohn, M.; Aziz, M.N.; Yuen, L. Possible origins and evolution of the hepatitis B virus (HBV). Semin. Cancer Biol. 2013, 23, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Littlejohn, M.; Davies, J.; Yuen, L.; Edwards, R.; Sozzi, T.; Jackson, K.; Cowie, B.; Tong, S.; Davis, J.; Locarnini, S. Molecular virology of hepatitis B virus, sub-genotype C4 in northern Australian Indigenous populations. J. Med. Virol. 2014, 86, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Kew, M.C. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J. Viral Hepat. 2005, 12, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Arakawa, K.; Yu, M.C.; Nogueira, R.; Stram, D.O.; Kew, M.C. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J. Med. Virol. 2008, 80, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Kostaki, E.G.; Hatzakis, A.; Paraskevis, D. Immunomodulatory Function of HBeAg Related to Short-Sighted Evolution, Transmissibility, and Clinical Manifestation of Hepatitis B Virus. Front. Microbiol. 2018, 9, 2521. [Google Scholar] [CrossRef]
- Kao, J.H. Hepatitis B viral genotypes: Clinical relevance and molecular characteristics. J. Gastroenterol. Hepatol. 2002, 17, 643–650. [Google Scholar] [CrossRef]
- McMahon, B.J. The influence of hepatitis B virus genotype and subgenotype on the natural history of chronic hepatitis B. Hepatol. Int. 2009, 3, 334–342. [Google Scholar] [CrossRef]
- Gerlich, W.H. Prophylactic vaccination against hepatitis B: Achievements, challenges and perspectives. Med. Microbiol. Immunol. 2015, 204, 39–55. [Google Scholar] [CrossRef]
- Gerlich, W.H. Do we need better hepatitis B vaccines? Indian J. Med. Res. 2017, 145, 414–419. [Google Scholar] [CrossRef]
- De Meyer, S.; Gong, Z.J.; Suwandhi, W.; van Pelt, J.; Soumillion, A.; Yap, S.H. Organ and species specificity of hepatitis B virus (HBV) infection: A review of literature with a special reference to preferential attachment of HBV to human hepatocytes. J. Viral Hepat. 1997, 4, 145–153. [Google Scholar] [CrossRef]
- Chen, J.; Wu, M.; Liu, K.; Zhang, W.; Li, Y.; Zhou, X.; Bai, L.; Yuan, Z. New insights into hepatitis B virus biology and implications for novel antiviral strategies. Natl. Sci. Rev. 2015, 2, 296–313. [Google Scholar] [CrossRef]
- Lamontagne, J.; Mell, J.C.; Bouchard, M.J. Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression. PLoS Pathog. 2016, 12, e1005438. [Google Scholar] [CrossRef] [PubMed]
- Dupinay, T.; Gheit, T.; Roques, P.; Cova, L.; Chevallier-Queyron, P.; Tasahsu, S.I.; Le Grand, R.; Simon, F.; Cordier, G.; Wakrim, L.; et al. Discovery of naturally occurring transmissible chronic hepatitis B virus infection among Macaca fascicularis from Mauritius Island. Hepatology 2013, 58, 1610–1620. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.H.; London, W.T.; McAuliffe, V.J.; Palmer, A.E.; Kaplan, P.M.; Gerin, J.L.; Wagner, J.; Popper, H.; Lvovsky, E.; Wong, D.C.; et al. Modification of chronic hepatitis-B virus infection in chimpanzees by administration of an interferon inducer. Lancet 1976, 2, 757–761. [Google Scholar] [CrossRef]
- Burwitz, B.J.; Wettengel, J.M.; Muck-Hausl, M.A.; Ringelhan, M.; Ko, C.; Festag, M.M.; Hammond, K.B.; Northrup, M.; Bimber, B.N.; Jacob, T.; et al. Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat. Commun. 2017, 8, 2146. [Google Scholar] [CrossRef] [PubMed]
- von Weizsacker, F.; Kock, J.; MacNelly, S.; Ren, S.; Blum, H.E.; Nassal, M. The tupaia model for the study of hepatitis B virus: Direct infection and HBV genome transduction of primary tupaia hepatocytes. Methods Mol. Med. 2004, 96, 153–161. [Google Scholar] [CrossRef]
- Guha, C.; Mohan, S.; Roy-Chowdhury, N.; Roy-Chowdhury, J. Cell culture and animal models of viral hepatitis. Part I: Hepatitis B. Lab. Anim. 2004, 33, 37–46. [Google Scholar] [CrossRef]
- Lampertico, P.; Malter, J.S.; Gerber, M.A. Development and application of an in vitro model for screening anti-hepatitis B virus therapeutics. Hepatology 1991, 13, 422–426. [Google Scholar]
- Witt-Kehati, D.; Bitton Alaluf, M.; Shlomai, A. Advances and Challenges in Studying Hepatitis B Virus In Vitro. Viruses 2016, 8, 21. [Google Scholar] [CrossRef]
- Hayes, C.N.; Chayama, K. HBV culture and infectious systems. Hepatol. Int. 2016, 10, 559–566. [Google Scholar] [CrossRef]
- Tuttleman, J.S.; Pugh, J.C.; Summers, J.W. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J. Virol. 1986, 58, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Tyler, G.V.; Snyder, R.L.; Summers, J. Experimental infection of the woodchuck (Marmota monax monax) with woodchuck hepatitis virus. Lab. Investig. 1986, 55, 51–55. [Google Scholar] [PubMed]
- Marion, P.L.; Oshiro, L.S.; Regnery, D.C.; Scullard, G.H.; Robinson, W.S. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc. Natl. Acad. Sci. USA 1980, 77, 2941–2945. [Google Scholar] [CrossRef] [PubMed]
- Allweiss, L.; Dandri, M. Experimental in vitro and in vivo models for the study of human hepatitis B virus infection. J. Hepatol. 2016, 64, S17–S31. [Google Scholar] [CrossRef]
- Sandhu, P.; Haque, M.; Humphries-Bickley, T.; Ravi, S.; Song, J. Hepatitis B Virus Immunopathology, Model Systems, and Current Therapies. Front. Immunol. 2017, 8, 436. [Google Scholar] [CrossRef]
- Kock, J.; Nassal, M.; MacNelly, S.; Baumert, T.F.; Blum, H.E.; von Weizsacker, F. Efficient infection of primary tupaia hepatocytes with purified human and woolly monkey hepatitis B virus. J. Virol. 2001, 75, 5084–5089. [Google Scholar] [CrossRef]
- Meier, A.; Mehrle, S.; Weiss, T.S.; Mier, W.; Urban, S. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology 2013, 58, 31–42. [Google Scholar] [CrossRef]
- Bouchard, M.J.; Navas-Martin, S. Hepatitis B and C virus hepatocarcinogenesis: Lessons learned and future challenges. Cancer Lett. 2011, 305, 123–143. [Google Scholar] [CrossRef]
- Gearhart, T.L.; Bouchard, M.J. The hepatitis B virus HBx protein modulates cell cycle regulatory proteins in cultured primary human hepatocytes. Virus Res. 2011, 155, 363–367. [Google Scholar] [CrossRef]
- Rawat, S.; Bouchard, M.J. The hepatitis B virus (HBV) HBx protein activates AKT to simultaneously regulate HBV replication and hepatocyte survival. J. Virol. 2015, 89, 999–1012. [Google Scholar] [CrossRef]
- Seeger, M.C.; Zoulim, F.; Mason, W.S. Hepadnaviruses. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, PA, USA, 2013; pp. 3376–3436. [Google Scholar]
- Galle, P.R.; Hagelstein, J.; Kommerell, B.; Volkmann, M.; Schranz, P.; Zentgraf, H. In vitro experimental infection of primary human hepatocytes with hepatitis B virus. Gastroenterology 1994, 106, 664–673. [Google Scholar] [CrossRef]
- Gripon, P.; Diot, C.; Theze, N.; Fourel, I.; Loreal, O.; Brechot, C.; Guguen-Guillouzo, C. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J. Virol. 1988, 62, 4136–4143. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Chen, Q.; Yang, L.Y.; Li, W.Y.; Cao, X.B.; Wu, J.R.; Peng, Y.P.; Chen, M.R. Hepatitis B virus infection and replication in primarily cultured human fetal hepatocytes. World J. Gastroenterol. 2007, 13, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ochiya, T.; Tsurimoto, T.; Ueda, K.; Okubo, K.; Shiozawa, M.; Matsubara, K. An in vitro system for infection with hepatitis B virus that uses primary human fetal hepatocytes. Proc. Natl. Acad. Sci. USA 1989, 86, 1875–1879. [Google Scholar] [CrossRef]
- Tyakht, A.V.; Ilina, E.N.; Alexeev, D.G.; Ischenko, D.S.; Gorbachev, A.Y.; Semashko, T.A.; Larin, A.K.; Selezneva, O.V.; Kostryukova, E.S.; Karalkin, P.A.; et al. RNA-Seq gene expression profiling of HepG2 cells: The influence of experimental factors and comparison with liver tissue. BMC Genom. 2014, 15, 1108. [Google Scholar] [CrossRef]
- Belloni, L.; Allweiss, L.; Guerrieri, F.; Pediconi, N.; Volz, T.; Pollicino, T.; Petersen, J.; Raimondo, G.; Dandri, M.; Levrero, M. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Investig. 2012, 122, 529–537. [Google Scholar] [CrossRef]
- Chang, C.M.; Jeng, K.S.; Hu, C.P.; Lo, S.J.; Su, T.S.; Ting, L.P.; Chou, C.K.; Han, S.H.; Pfaff, E.; Salfeld, J.; et al. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987, 6, 675–680. [Google Scholar] [CrossRef]
- Chen, R.Y.; Edwards, R.; Shaw, T.; Colledge, D.; Delaney, W.E.t.; Isom, H.; Bowden, S.; Desmond, P.; Locarnini, S.A. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology 2003, 37, 27–35. [Google Scholar] [CrossRef]
- Delaney, W.E.t.; Miller, T.G.; Isom, H.C. Use of the hepatitis B virus recombinant baculovirus-HepG2 system to study the effects of (-)-beta-2’,3’-dideoxy-3’-thiacytidine on replication of hepatitis B virus and accumulation of covalently closed circular DNA. Antimicrob. Agents Chemother. 1999, 43, 2017–2026. [Google Scholar] [CrossRef]
- Knowles, B.B.; Howe, C.C.; Aden, D.P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980, 209, 497–499. [Google Scholar] [CrossRef]
- Levrero, M.; Pollicino, T.; Petersen, J.; Belloni, L.; Raimondo, G.; Dandri, M. Control of cccDNA function in hepatitis B virus infection. J. Hepatol. 2009, 51, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, H.; Taketa, K.; Miyano, K.; Yamane, T.; Sato, J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982, 42, 3858–3863. [Google Scholar] [PubMed]
- Pollicino, T.; Belloni, L.; Raffa, G.; Pediconi, N.; Squadrito, G.; Raimondo, G.; Levrero, M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 2006, 130, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Seeger, C.; Mason, W.S. Sodium-dependent taurocholic cotransporting polypeptide: A candidate receptor for human hepatitis B virus. Gut 2013, 62, 1093–1095. [Google Scholar] [CrossRef]
- Sureau, C.; Eichberg, J.W.; Hubbard, G.B.; Romet-Lemonne, J.L.; Essex, M. A molecularly cloned hepatitis B virus produced in vitro is infectious in a chimpanzee. J. Virol. 1988, 62, 3064–3067. [Google Scholar] [CrossRef]
- Yaginuma, K.; Shirakata, Y.; Kobayashi, M.; Koike, K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc. Natl. Acad. Sci. USA 1987, 84, 2678–2682. [Google Scholar] [CrossRef]
- Ding, X.R.; Yang, J.; Sun, D.C.; Lou, S.K.; Wang, S.Q. Whole genome expression profiling of hepatitis B virus-transfected cell line reveals the potential targets of anti-HBV drugs. Pharm. J. 2008, 8, 61–70. [Google Scholar] [CrossRef][Green Version]
- Gripon, P.; Cannie, I.; Urban, S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 2005, 79, 1613–1622. [Google Scholar] [CrossRef]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef]
- Guo, H.; Jiang, D.; Zhou, T.; Cuconati, A.; Block, T.M.; Guo, J.T. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: An intermediate of covalently closed circular DNA formation. J. Virol. 2007, 81, 12472–12484. [Google Scholar] [CrossRef]
- Hantz, O.; Parent, R.; Durantel, D.; Gripon, P.; Guguen-Guillouzo, C.; Zoulim, F. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J. Gen. Virol. 2009, 90, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Ladner, S.K.; Otto, M.J.; Barker, C.S.; Zaifert, K.; Wang, G.H.; Guo, J.T.; Seeger, C.; King, R.W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: A novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 1997, 41, 1715–1720. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Q.; Xu, W.Z.; Wang, J.X.; Deng, W.W.; Li, D.; Gu, H.X. Combination of small interfering RNA and lamivudine on inhibition of human B virus replication in HepG2.2.15 cells. World J. Gastroenterol. 2007, 13, 2324–2327. [Google Scholar] [CrossRef] [PubMed]
- Luangsay, S.; Gruffaz, M.; Isorce, N.; Testoni, B.; Michelet, M.; Faure-Dupuy, S.; Maadadi, S.; Ait-Goughoulte, M.; Parent, R.; Rivoire, M.; et al. Early inhibition of hepatocyte innate responses by hepatitis B virus. J. Hepatol. 2015, 63, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Lucifora, J.; Durantel, D.; Testoni, B.; Hantz, O.; Levrero, M.; Zoulim, F. Control of hepatitis B virus replication by innate response of HepaRG cells. Hepatology 2010, 51, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppensteiner, H.; Makowska, Z.; Volz, T.; et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014, 343, 1221–1228. [Google Scholar] [CrossRef]
- Sun, D.; Nassal, M. Stable HepG2- and Huh7-based human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus. J. Hepatol. 2006, 45, 636–645. [Google Scholar] [CrossRef]
- Tsurimoto, T.; Fujiyama, A.; Matsubara, K. Stable expression and replication of hepatitis B virus genome in an integrated state in a human hepatoma cell line transfected with the cloned viral DNA. Proc. Natl. Acad. Sci. USA 1987, 84, 444–448. [Google Scholar] [CrossRef]
- Bouezzedine, F.; Fardel, O.; Gripon, P. Interleukin 6 inhibits HBV entry through NTCP down regulation. Virology 2015, 481, 34–42. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Chen, J.; Li, Y.; Wang, W.; Du, X.; Song, W.; Zhang, W.; Lin, L.; Yuan, Z. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J. Virol. 2015, 89, 2287–2300. [Google Scholar] [CrossRef]
- Hoh, A.; Heeg, M.; Ni, Y.; Schuch, A.; Binder, B.; Hennecke, N.; Blum, H.E.; Nassal, M.; Protzer, U.; Hofmann, M.; et al. Hepatitis B Virus-Infected HepG2hNTCP Cells Serve as a Novel Immunological Tool To Analyze the Antiviral Efficacy of CD8+ T Cells In Vitro. J. Virol. 2015, 89, 7433–7438. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Cai, D.; Liu, Y.; Cuconati, A.; Guo, H. Spinoculation Enhances HBV Infection in NTCP-Reconstituted Hepatocytes. PLoS ONE 2015, 10, e0129889. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.; Liang, T.J. Experimental models of hepatitis B and C—New insights and progress. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Carpentier, A.; Cheng, X.; Block, P.D.; Zhao, Y.; Zhang, Z.; Protzer, U.; Liang, T.J. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J. Hepatol. 2017, 66, 494–503. [Google Scholar] [CrossRef]
- Sakurai, F.; Mitani, S.; Yamamoto, T.; Takayama, K.; Tachibana, M.; Watashi, K.; Wakita, T.; Iijima, S.; Tanaka, Y.; Mizuguchi, H. Human induced-pluripotent stem cell-derived hepatocyte-like cells as an in vitro model of human hepatitis B virus infection. Sci. Rep. 2017, 7, 45698. [Google Scholar] [CrossRef]
- Schwartz, R.E.; Fleming, H.E.; Khetani, S.R.; Bhatia, S.N. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol. Adv. 2014, 32, 504–513. [Google Scholar] [CrossRef]
- Shlomai, A.; Schwartz, R.E.; Ramanan, V.; Bhatta, A.; de Jong, Y.P.; Bhatia, S.N.; Rice, C.M. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc. Natl. Acad. Sci. USA 2014, 111, 12193–12198. [Google Scholar] [CrossRef]
- Nie, Y.Z.; Zheng, Y.W.; Miyakawa, K.; Murata, S.; Zhang, R.R.; Sekine, K.; Ueno, Y.; Takebe, T.; Wakita, T.; Ryo, A.; et al. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine 2018, 35, 114–123. [Google Scholar] [CrossRef]
- Sells, M.A.; Chen, M.L.; Acs, G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 1987, 84, 1005–1009. [Google Scholar] [CrossRef]
- Brandon, E.F.; Raap, C.D.; Meijerman, I.; Beijnen, J.H.; Schellens, J.H. An update on in vitro test methods in human hepatic drug biotransformation research: Pros and cons. Toxicol. Appl. Pharmacol. 2003, 189, 233–246. [Google Scholar] [CrossRef]
- Choi, S.; Sainz, B., Jr.; Corcoran, P.; Uprichard, S.; Jeong, H. Characterization of increased drug metabolism activity in dimethyl sulfoxide (DMSO)-treated Huh7 hepatoma cells. Xenobiotica 2009, 39, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Clayton, R.F.; Rinaldi, A.; Kandyba, E.E.; Edward, M.; Willberg, C.; Klenerman, P.; Patel, A.H. Liver cell lines for the study of hepatocyte functions and immunological response. Liver Int. 2005, 25, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, P.; Hou, X.; Xu, W.; Wang, D.; Wang, T.Y.; Zhang, L.; Zheng, G.; Gao, Z.L.; He, C.Y.; et al. The recombined cccDNA produced using minicircle technology mimicked HBV genome in structure and function closely. Sci. Rep. 2016, 6, 25552. [Google Scholar] [CrossRef] [PubMed]
- Delaney, W.E.t.; Isom, H.C. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology 1998, 28, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, J.; Yue, L.; Bai, L.; Li, Y.; Chen, J.; Zhang, X.; Yuan, Z. Establishment of Cre-mediated HBV recombinant cccDNA (rcccDNA) cell line for cccDNA biology and antiviral screening assays. Antivir. Res. 2018, 152, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Miyazaki, J.; Hino, O.; Tomita, N.; Chisaka, O.; Matsubara, K.; Yamamura, K. Expression and replication of hepatitis B virus genome in transgenic mice. Proc. Natl. Acad. Sci. USA 1989, 86, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wang, A.H.; Shao, L.H.; Wang, G.; Yao, Y.Y.; Sai, L.T.; Chen, F.Z.; Zheng, F.; Li, Y.; Ma, L.X. A new cell culture system for infection with hepatitis B virus that fuses HepG2 cells with primary human hepatocytes. J. Int. Med. Res. 2009, 37, 650–661. [Google Scholar] [CrossRef]
- Li, G.; Zhu, Y.; Shao, D.; Chang, H.; Zhang, X.; Zhou, D.; Gao, Y.; Lan, K.; Deng, Q. Recombinant covalently closed circular DNA of hepatitis B virus induces long-term viral persistence with chronic hepatitis in a mouse model. Hepatology 2018, 67, 56–70. [Google Scholar] [CrossRef]
- Cai, D.; Mills, C.; Yu, W.; Yan, R.; Aldrich, C.E.; Saputelli, J.R.; Mason, W.S.; Xu, X.; Guo, J.T.; Block, T.M.; et al. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob. Agents Chemother. 2012, 56, 4277–4288. [Google Scholar] [CrossRef]
- Cai, D.; Wang, X.; Yan, R.; Mao, R.; Liu, Y.; Ji, C.; Cuconati, A.; Guo, H. Establishment of an inducible HBV stable cell line that expresses cccDNA-dependent epitope-tagged HBeAg for screening of cccDNA modulators. Antivir. Res. 2016, 132, 26–37. [Google Scholar] [CrossRef]
- Gripon, P.; Diot, C.; Guguen-Guillouzo, C. Reproducible high level infection of cultured adult human hepatocytes by hepatitis B virus: Effect of polyethylene glycol on adsorption and penetration. Virology 1993, 192, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Runge, D.; Runge, D.M.; Jager, D.; Lubecki, K.A.; Beer Stolz, D.; Karathanasis, S.; Kietzmann, T.; Strom, S.C.; Jungermann, K.; Fleig, W.E.; et al. Serum-free, long-term cultures of human hepatocytes: Maintenance of cell morphology, transcription factors, and liver-specific functions. Biochem. Biophys. Res. Commun. 2000, 269, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Runge, D.M.; Runge, D.; Dorko, K.; Pisarov, L.A.; Leckel, K.; Kostrubsky, V.E.; Thomas, D.; Strom, S.C.; Michalopoulos, G.K. Epidermal growth factor- and hepatocyte growth factor-receptor activity in serum-free cultures of human hepatocytes. J. Hepatol. 1999, 30, 265–274. [Google Scholar] [CrossRef]
- Guillouzo, A.; Corlu, A.; Aninat, C.; Glaise, D.; Morel, F.; Guguen-Guillouzo, C. The human hepatoma HepaRG cells: A highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem. Biol. Interact. 2007, 168, 66–73. [Google Scholar] [CrossRef]
- Kanebratt, K.P.; Andersson, T.B. Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab. Dispos. 2008, 36, 1444–1452. [Google Scholar] [CrossRef]
- Kanebratt, K.P.; Andersson, T.B. HepaRG cells as an in vitro model for evaluation of cytochrome P450 induction in humans. Drug Metab. Dispos. 2008, 36, 137–145. [Google Scholar] [CrossRef]
- Krepstakies, M.; Lucifora, J.; Nagel, C.H.; Zeisel, M.B.; Holstermann, B.; Hohenberg, H.; Kowalski, I.; Gutsmann, T.; Baumert, T.F.; Brandenburg, K.; et al. A new class of synthetic peptide inhibitors blocks attachment and entry of human pathogenic viruses. J. Infect. Dis. 2012, 205, 1654–1664. [Google Scholar] [CrossRef]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Falth, M.; Stindt, J.; Koniger, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D Viruses Exploit Sodium Taurocholate Co-transporting Polypeptide for Species-Specific Entry into Hepatocytes. Gastroenterology 2014, 146, 1070–1083. [Google Scholar] [CrossRef]
- Schulze, A.; Gripon, P.; Urban, S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007, 46, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Liu, Y.; Sui, J.; Li, W. NTCP opens the door for hepatitis B virus infection. Antivir. Res. 2015, 121, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Michailidis, E.; Pabon, J.; Xiang, K.; Park, P.; Ramanan, V.; Hoffmann, H.H.; Schneider, W.M.; Bhatia, S.N.; de Jong, Y.P.; Shlomai, A.; et al. A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci. Rep. 2017, 7, 16616. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Si-Tayeb, K.; Noto, F.K.; Nagaoka, M.; Li, J.; Battle, M.A.; Duris, C.; North, P.E.; Dalton, S.; Duncan, S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 2010, 51, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Khetani, S.R. Micropatterned Co-Cultures of Human Hepatocytes and Stromal Cells for the Assessment of Drug Clearance and Drug-Drug Interactions. Curr. Protoc. Toxicol. 2017, 72, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Gural, N.; Mancio-Silva, L.; He, J.; Bhatia, S.N. Engineered Livers for Infectious Diseases. Cell. Mol. Gastroenter. 2018, 5, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Prior, N.; Inacio, P.; Huch, M. Liver organoids: From basic research to therapeutic applications. Gut 2019, 68, 2228–2237. [Google Scholar] [CrossRef]
- Gunther, S.; Li, B.C.; Miska, S.; Kruger, D.H.; Meisel, H.; Will, H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 1995, 69, 5437–5444. [Google Scholar] [CrossRef]
- Ren, S.; Nassal, M. Hepatitis B virus (HBV) virion and covalently closed circular DNA formation in primary tupaia hepatocytes and human hepatoma cell lines upon HBV genome transduction with replication-defective adenovirus vectors. J. Virol. 2001, 75, 1104–1116. [Google Scholar] [CrossRef]
- Sprinzl, M.F.; Oberwinkler, H.; Schaller, H.; Protzer, U. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: Crossing the species barrier. J. Virol. 2001, 75, 5108–5118. [Google Scholar] [CrossRef]
- Sugiyama, M.; Tanaka, Y.; Kato, T.; Orito, E.; Ito, K.; Acharya, S.K.; Gish, R.G.; Kramvis, A.; Shimada, T.; Izumi, N.; et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 2006, 44, 915–924. [Google Scholar] [CrossRef]
- Lucifora, J.; Durantel, D.; Belloni, L.; Barraud, L.; Villet, S.; Vincent, I.E.; Margeridon-Thermet, S.; Hantz, O.; Kay, A.; Levrero, M.; et al. Initiation of hepatitis B virus genome replication and production of infectious virus following delivery in HepG2 cells by novel recombinant baculovirus vector. J. Gen. Virol 2008, 89, 1819–1828. [Google Scholar] [CrossRef] [PubMed]
- Clippinger, A.J.; Gearhart, T.L.; Bouchard, M.J. Hepatitis B virus X protein modulates apoptosis in primary rat hepatocytes by regulating both NF-κB and the mitochondrial permeability transition pore. J. Virol. 2009, 83, 4718–4731. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Ueno, Y.; Nagasaki, F.; Wakui, Y.; Kondo, Y.; Fukushima, K.; Niitsuma, H.; Shimosegawa, T. Enhanced intracellular retention of a hepatitis B virus strain associated with fulminant hepatitis. Virology 2009, 395, 202–209. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Ye, S.; Wang, T.; Zhao, R.; Chen, F.; Abe, K.; Jin, X. The response to interferon is influenced by hepatitis B virus genotype in vitro and in vivo. Virus Res. 2013, 171, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Bhoola, N.H.; Reumann, K.; Kew, M.C.; Will, H.; Kramvis, A. Construction of replication competent plasmids of hepatitis B virus subgenotypes A1, A2 and D3 with authentic endogenous promoters. J. Virol. Methods 2014, 203, 54–64. [Google Scholar] [CrossRef]
- Zhang, Z.; Xia, J.; Sun, B.; Dai, Y.; Li, X.; Schlaak, J.F.; Lu, M. In vitro and in vivo replication of a chemically synthesized consensus genome of hepatitis B virus genotype B. J. Virol. Methods 2015, 213, 57–64. [Google Scholar] [CrossRef]
- Sozzi, V.; Walsh, R.; Littlejohn, M.; Colledge, D.; Jackson, K.; Warner, N.; Yuen, L.; Locarnini, S.A.; Revill, P.A. In Vitro Studies Show that Sequence Variability Contributes to Marked Variation in Hepatitis B Virus Replication, Protein Expression, and Function Observed across Genotypes. J. Virol. 2016, 90, 10054–10064. [Google Scholar] [CrossRef]
- Berke, J.M.; Tan, Y.; Verbinnen, T.; Dehertogh, P.; Vergauwen, K.; Vos, A.; Lenz, O.; Pauwels, F. Antiviral profiling of the capsid assembly modulator BAY41-4109 on full-length HBV genotype A-H clinical isolates and core site-directed mutants in vitro. Antivir. Res. 2017, 144, 205–215. [Google Scholar] [CrossRef]
- Jia, H.; Qin, Y.; Chen, C.; Zhang, F.; Li, C.; Zong, L.; Wang, Y.; Zhang, J.; Li, J.; Wen, Y.; et al. The Envelope Gene of Hepatitis B Virus Is Implicated in Both Differential Virion Secretion and Genome Replication Capacities between Genotype B and Genotype C Isolates. Viruses 2017, 9, 62. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, M.; Gong, M.; Xu, Y.; Xie, C.; Deng, H.; Li, X.; Wu, H.; Wang, Z. Inhibition of hepatitis B virus replication via HBV DNA cleavage by Cas9 from Staphylococcus aureus. Antivir. Res. 2018, 152, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Bannister, E.; Sozzi, V.; Mason, H.; Locarnini, S.A.; Hardiker, W.; Revill, P.A. Analysis of the in vitro replication phenotype of African hepatitis B virus (HBV) genotypes and subgenotypes present in Australia identifies marked differences in DNA and protein expression. Virology 2019, 54, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kostyushev, D.; Brezgin, S.; Kostyusheva, A.; Zarifyan, D.; Goptar, I.; Chulanov, V. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell Mol. Life Sci. 2019, 76, 1779–1794. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Kornepati, A.V.; Cullen, B.R. Targeting hepatitis B virus cccDNA using CRISPR/Cas9. Antivir. Res. 2015, 123, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Hasegawa, I.; Kato, T.; Orito, E.; Hirashima, N.; Acharya, S.K.; Gish, R.G.; Kramvis, A.; Kew, M.C.; Yoshihara, N.; et al. A case-control study for differences among hepatitis B virus infections of genotypes A (subtypes Aa and Ae) and D. Hepatology 2004, 40, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Kew, M.C. Molecular characterization of subgenotype A1 (subgroup Aa) of hepatitis B virus. Hepatol. Res. 2007, 37, S27–S32. [Google Scholar] [CrossRef]
- Kramvis, A. Molecular characteristics and clinical relevance of African genotypes and subgenotypes of hepatitis B virus. S. Afr. Med. J. 2018, 108, 17–21. [Google Scholar] [CrossRef]
- Kramvis, A.; Paraskevis, D. Subgenotype A1 of HBV—Tracing human migrations in and out of Africa. Antivir. Ther. 2013, 18, 513–521. [Google Scholar] [CrossRef]
- The NIH Hepatitis B Cure Strategic Plan Working Group. Strategic Plan for Trans-NIH Research to Cure Hepatitis B; Department of Health and Human Service—USA: Washington, DC, USA, 2019. [Google Scholar]
- ICE-HBV. HBV Research Protocols. Available online: https://ice-hbv.org/protocol/ (accessed on 2 March 2020).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).