A Turkey-origin H9N2 Avian Influenza Virus Shows Low Pathogenicity but Different Within-host Diversity in Experimentally Infected Turkeys, Quail and Ducks
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Propagation
2.2. Birds and Infection Experiments
2.3. Evaluation of Virus Shedding and Seroconversion
2.4. Deep Sequencing
2.5. Sequencing Data Analysis
2.6. Statistical Analysis
3. Results
3.1. Clinical Signs and Mortality
3.2. Virus Shedding
3.2.1. Turkeys
3.2.2. Quail
3.2.3. Ducks
3.2.4. Comparison of Shedding in Inoculated Turkeys, Quail and Ducks
3.3. Antibody Response
3.4. Diversity of Virus Population in Swabs
3.5. Synonymous and Nonsynonymous Mutations
3.6. Changes in the Variant Frequency and Consensus-Level Substitutions
3.7. Correlation Between Viral Load and Virus Diversity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Fouchier, R.A.; Munster, V.J. Epidemiology of low pathogenic avian influenza viruses in wild birds. Rev. Sci. Tech. 2009, 28, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Probst, C.; Gethmann, J.M.; Petermann, H.J.; Neudecker, J.; Jacobsen, K.; Conraths, F.J. Low pathogenic avian influenza H7N7 in domestic poultry in Germany in 2011. Vet. Rec. 2012, 171, 624. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.M.; Brookes, S.M.; Núñez, A.; Banks, J.; Parker, C.D.; Ceeraz, V.; Russell, C.; Seekings, A.; Thomas, S.S.; Puranik, A.; et al. Detection of non-notifiable H4N6 avian influenza virus in poultry in Great Britain. Vet. Microbiol. 2018, 224, 107–115. [Google Scholar] [CrossRef]
- Landman, W.J.M.; Germeraad, E.A.; Kense, M.J. An avian influenza virus H6N1 outbreak in commercial layers: Case report and reproduction of the disease. Avian Pathol. 2019, 48, 98–110. [Google Scholar] [CrossRef]
- Kuiken, T. Is low pathogenic avian influenza virus virulent for wild waterbirds? Proc. Biol. Sci. 2013, 280, 20130990. [Google Scholar] [CrossRef]
- Swayne, D.E.; Slemons, R.D. Using mean infectious dose of high- and low-pathogenicity avian influenza viruses originating from wild duck and poultry as one measure of infectivity and adaptation to poultry. Avian Dis. 2008, 52, 455–460. [Google Scholar] [CrossRef]
- Corrand, L.; Delverdier, M.; Lucas, M.N.; Croville, G.; Facon, C.; Balloy, D.; Ducatez, M.; Guérin, J.L. A low-pathogenic avian influenza H6N1 outbreak in a turkey flock in France: A comprehensive case report. Avian Pathol. 2012, 41, 569–577. [Google Scholar] [CrossRef]
- Spackman, E.; Gelb, J., Jr.; Preskenis, L.A.; Ladman, B.S.; Pope, C.R.; Pantin-Jackwood, M.J.; McKinley, E.T. The pathogenesis of low pathogenicity H7 avian influenza viruses in chickens, ducks and turkeys. Virol. J. 2010, 7, 331. [Google Scholar] [CrossRef]
- Śmietanka, K.; Minta, Z.; Świętoń, E.; Olszewska, M.; Jóźwiak, M.; Domańska-Blicharz, K.; Wyrostek, K.; Tomczyk, G.; Pikuła, A. Avian influenza H9N2 subtype in Poland – characterization of the isolates and evidence of concomitant infections. Avian Pathol. 2014, 43, 427–436. [Google Scholar] [CrossRef]
- Bonfante, F.; Patrono, L.V.; Aiello, R.; Beato, M.S.; Terregino, C.; Capua, I. Susceptibility and intra-species transmission of the H9N2 G1 prototype lineage virus in Japanese quail and turkeys. Vet. Microbiol. 2013, 165, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Makarova, N.V.; Ozaki, H.; Kida, H.; Webster, R.G.; Perez, D.R. Replication and transmission of influenza viruses in Japanese quail. Virology. 2003, 310, 8–15. [Google Scholar] [CrossRef]
- McDonald, S.M.; Nelson, M.I.; Turner, P.E.; Patton, J.T. Reassortment in segmented RNA viruses: Mechanisms and outcomes. Nat. Rev. Microbiol. 2016, 14, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Peck, K.M.; Lauring, A.S. Complexities of viral mutation rates. J. Virol. 2018, 92, e01031-17. [Google Scholar] [CrossRef]
- McCrone, J.T.; Woods, R.J.; Martin, E.T.; Malosh, R.E.; Monto, A.S.; Lauring, A.S. Stochastic processes constrain the within and between host evolution of influenza virus. Elife. 2018, 7, e35962. [Google Scholar] [CrossRef]
- Zhao, L.; Abbasi, A.B.; Illingworth, C.J.R. Mutational load causes stochastic evolutionary outcomes in acute RNA viral infection. Virus Evol. 2019, 5, vez008. [Google Scholar] [CrossRef]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef]
- Zaraket, H.; Baranovich, T.; Kaplan, B.S.; Carter, R.; Song, M.S.; Paulson, J.C.; Rehg, J.E.; Bahl, J.; Crumpton, J.C.; Seiler, J.; et al. Mammalian adaptation of influenza A(H7N9) virus is limited by a narrow genetic bottleneck. Nat. Commun. 2015, 6, 6553. [Google Scholar] [CrossRef]
- Xue, K.S.; Moncla, L.H.; Bedford, T.; Bloom, J.D. Within-host evolution of human influenza virus. Trends Microbiol. 2018, 26, 781–793. [Google Scholar] [CrossRef]
- Sobel Leonard, A.; McClain, M.T.; Smith, G.J.; Wentworth, D.E.; Halpin, R.A.; Lin, X.; Ransier, A.; Stockwell, T.B.; Das, S.R.; Gilbert, A.S.; et al. Deep sequencing of influenza A virus from a human challenge study reveals a selective bottleneck and only limited intrahost genetic diversification. J. Virol. 2016, 90, 11247–11258. [Google Scholar] [CrossRef]
- Barbezange, C.; Jones, L.; Blanc, H.; Isakov, O.; Celniker, G.; Enouf, V.; Shomron, N.; Vignuzzi, M.; van der Werf, S. Seasonal genetic drift of human influenza A virus quasispecies revealed by deep sequencing. Front. Microbiol. 2018, 9, 2596. [Google Scholar] [CrossRef] [PubMed]
- Welkers, M.R.A.; Pawestri, H.A.; Fonville, J.M.; Sampurno, O.D.; Pater, M.; Holwerda, M.; Han, A.X.; Russell, C.A.; Jeeninga, R.E.; Setiawaty, V.; et al. Genetic diversity and host adaptation of avian H5N1 influenza viruses during human infection. Emerg. Microbes. Infect. 2019, 8, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Simon, B.; Pichon, M.; Valette, M.; Burfin, G.; Richard, M.; Lina, B.; Josset, L. Whole genome sequencing of A(H3N2) influenza viruses reveals variants associated with severity during the 2016-2017 season. Viruses 2019, 11, E108. [Google Scholar] [CrossRef] [PubMed]
- Moncla, L.H.; Zhong, G.; Nelson, C.W.; Dinis, J.M.; Mutschler, J.; Hughes, A.L.; Watanabe, T.; Kawaoka, Y.; Friedrich, T.C. Selective bottlenecks shape evolutionary pathways taken during mammalian adaptation of a 1918-like avian influenza virus. Cell Host Microbe. 2016, 19, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Murcia, P.R.; Baillie, G.J.; Stack, J.C.; Jervis, C.; Elton, D.; Mumford, J.A.; Daly, J.; Kellam, P.; Grenfell, B.T.; Holmes, E.C.; et al. Evolution of equine influenza virus in vaccinated horses. J. Virol. 2013, 87, 4768–4771. [Google Scholar] [CrossRef]
- Nagy, A.; Vostinakova, V.; Pirchanova, Z.; Cernikova, L.; Dirbakova, Z.; Mojzis, M.; Jirincova, H.; Havlickova, M.; Dan, A.; Ursu, K.; et al. Development and evaluation of a one-step real-time RT-PCR assay for universal detection of influenza A viruses from avian and mammal species. Arch. Virol. 2010, 155, 665–673. [Google Scholar] [CrossRef]
- OIE. Manual of diagnostic tests and vaccines for terrestrial animals, chapter 3.3.4. “Avian influenza (infection with avian influenza viruses)”. World Organization for Animal Health: Paris, France, 2018. [Google Scholar]
- Zhou, B.; Donnelly, M.E.; Scholes, D.T.; St George, K.; Hatta, M.; Kawaoka, Y.; Wentworth, D.E. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza A viruses. J. Virol. 2009, 83, 10309–10313. [Google Scholar] [CrossRef]
- Watson, S.J.; Welkers, M.R.; Depledge, D.P.; Coulter, E.; Breuer, J.M.; de Jong, M.D.; Kellam, P. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci. 2013, 368, 20120205. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Wilm, A.; Aw, P.P.; Bertrand, D.; Yeo, G.H.; Ong, S.H.; Wong, C.H.; Khor, C.C.; Petric, R.; Hibberd, M.L.; Nagarajan, N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; McLellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding, L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009, 25, 2283–2285. [Google Scholar] [CrossRef] [PubMed]
- Milani, A.; Fusaro, A.; Bonfante, F.; Zamperin, G.; Salviato, A.; Mancin, M.; Mastrorilli, E.; Hughes, J.; Hussein, H.A.; Hassan, M.; et al. Vaccine immune pressure influences viral population complexity of avian influenza virus during infection. Vet. Microbiol. 2017, 203, 88–94. [Google Scholar] [CrossRef]
- Sanjuán, R. From molecular genetics to phylodynamics: Evolutionary relevance of mutation rates across viruses. PLoS Pathog. 2012, 8, e1002685. [Google Scholar] [CrossRef]
- Peck, K.M.; Chan, C.H.; Tanaka, M.M. Connecting within-host dynamics to the rate of viral molecular evolution. Virus Evol. 2015, 1, vev013. [Google Scholar] [CrossRef][Green Version]
- Hanada, K.; Suzuki, Y.; Gojobori, T. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol. Biol. Evol. 2004, 21, 1074–1080. [Google Scholar] [CrossRef]
- Scholle, S.O.; Ypma, R.J.; Lloyd, A.L.; Koelle, K. Viral substitution rate variation can arise from the interplay between within-host and epidemiological dynamics. Am. Nat. 2013, 182, 494–513. [Google Scholar] [CrossRef]
- Fourment, M.; Holmes, E.C. Avian influenza virus exhibits distinct evolutionary dynamics in wild birds and poultry. BMC Evol. Biol. 2015, 15, 120. [Google Scholar] [CrossRef]
- Germeraad, E.A.; Sanders, P.; Hagenaars, T.J.; Jong, M.C.M.; Beerens, N.; Gonzales, J.L. Virus shedding of avian influenza in poultry: A systematic review and meta-analysis. Viruses 2019, 11, E812. [Google Scholar] [CrossRef]
- Slomka, M.J.; Seekings, A.H.; Mahmood, S.; Thomas, S.; Puranik, A.; Watson, S.; Byrne, A.M.P.; Hicks, D.; Nunez, A.; Brown, I.H.; et al. Unexpected infection outcomes of China-origin H7N9 low pathogenicity avian influenza virus in turkeys. Sci. Rep. 2018, 8, 7322. [Google Scholar] [CrossRef] [PubMed]
- Thontiravong, A.; Wannaratana, S.; Tantilertcharoen, R.; Prakairungnamthip, D.; Tuanudom, R.; Sasipreeyajan, J.; Pakpinyo, S.; Amonsin, A.; Kitikoon, P.; Oraveerakul, K. Comparative study of pandemic (H1N1) 2009, swine H1N1, and avian H3N2 influenza viral infections in quails. J. Vet. Sci. 2012, 13, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Vidaña, B.; Dolz, R.; Busquets, N.; Ramis, A.; Sánchez, R.; Rivas, R.; Valle, R.; Cordón, I.; Solanes, D.; Martínez, J.; et al. Transmission and immunopathology of the avian influenza virus A/Anhui/1/2013 (H7N9) human isolate in three commonly commercialized avian species. Zoonoses Public Health. 2018, 65, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.R.; Lim, W.; Seiler, J.P.; Yi, G.; Peiris, M.; Shortridge, K.F.; Webster, R.G. Role of quail in the interspecies transmission of H9 influenza A viruses: Molecular changes on HA that correspond to adaptation from ducks to chickens. J. Virol. 2003, 77, 3148–3156. [Google Scholar] [CrossRef] [PubMed]
- Sorrell, E.M.; Perez, D.R. Adaptation of influenza A/Mallard/Potsdam/178-4/83 H2N2 virus in Japanese quail leads to infection and transmission in chickens. Avian Dis. 2007, 51, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.J.; Hickman, D.; Perez, D.R. Evidence of expanded host range and mammalian-associated genetic changes in a duck H9N2 influenza virus following adaptation in quail and chickens. PLoS One. 2008, 3, e3170. [Google Scholar] [CrossRef]
- Giannecchini, S.; Clausi, V.; Di Trani, L.; Falcone, E.; Terregino, C.; Toffan, A.; Cilloni, F.; Matrosovich, M.; Gambaryan, A.S.; Bovin, N.V.; et al. Molecular adaptation of an H7N3 wild duck influenza virus following experimental multiple passages in quail and turkey. Virology 2010, 408, 167–173. [Google Scholar] [CrossRef]
- Yamada, S.; Shinya, K.; Takada, A.; Ito, T.; Suzuki, T.; Suzuki, Y.; Le, Q.M.; Ebina, M.; Kasai, N.; Kida, H.; et al. Adaptation of a duck influenza A virus in quail. J. Virol. 2012, 86, 1411–1420. [Google Scholar] [CrossRef]
- Wan, H.; Perez, D.R. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 2006, 346, 278–286. [Google Scholar] [CrossRef]
- Bertran, K.; Lee, D.H.; Pantin-Jackwood, M.J.; Spackman, E.; Balzli, C.; Suarez, D.L.; Swayne, D.E. Pathobiology of clade 2.3.4.4 H5Nx high-pathogenicity avian influenza virus infections in minor gallinaceous poultry supports early backyard flock introductions in the Western United States in 2014-2015. J. Virol. 2017, 91, e00960-17. [Google Scholar] [CrossRef]
- Leyson, C.; Youk, S.S.; Smith, D.; Dimitrov, K.; Lee, D.H.; Larsen, L.E.; Swayne, D.E.; Pantin-Jackwood, M.J. Pathogenicity and genomic changes of a 2016 European H5N8 highly pathogenic avian influenza virus (clade 2.3.4.4) in experimentally infected mallards and chickens. Virology 2019, 537, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, M.E.; Brooke, C.B.; Ke, R.; Koelle, K. Causes and consequences of spatial within-host viral spread. Viruses 2018, 10, E627. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Xiao, H.; Baillie, G.; Warry, A.; Essen, S.C.; Londt, B.; Brookes, S.M.; Brown, I.H.; McCauley, J.W. Within-host variation of avian influenza viruses. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 2009, 364, 2739–2747. [Google Scholar] [CrossRef] [PubMed]
- Hulse-Post, D.J.; Sturm-Ramirez, K.M.; Humberd, J.; Seiler, P.; Govorkova, E.A.; Krauss, S.; Scholtissek, C.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; et al. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc. Natl. Acad. Sci. USA 2005, 102, 10682–10687. [Google Scholar] [CrossRef] [PubMed]
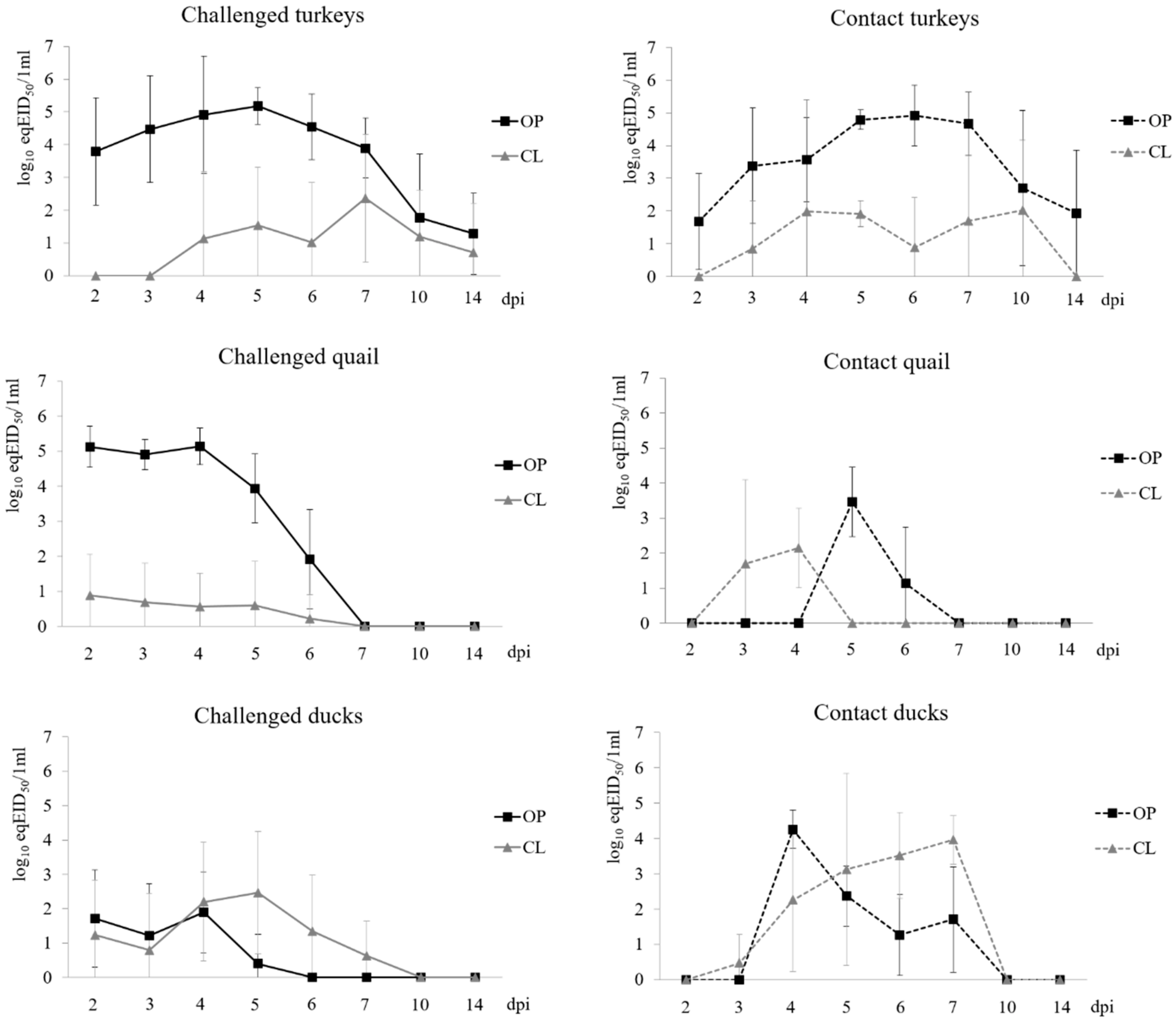
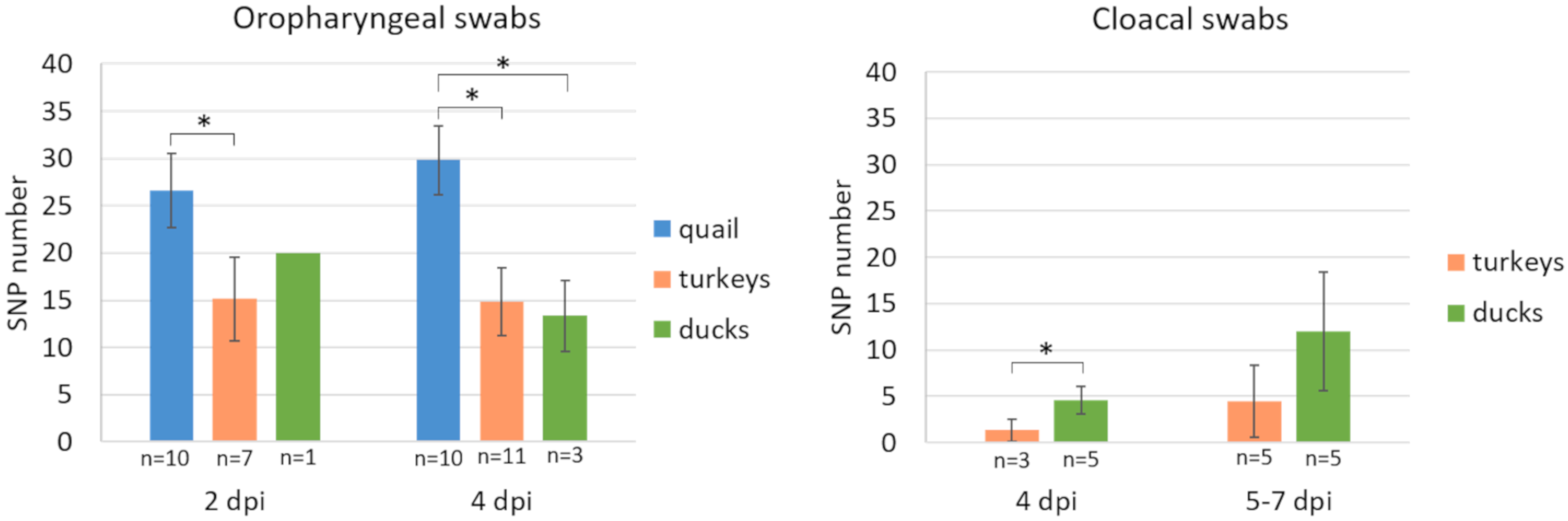
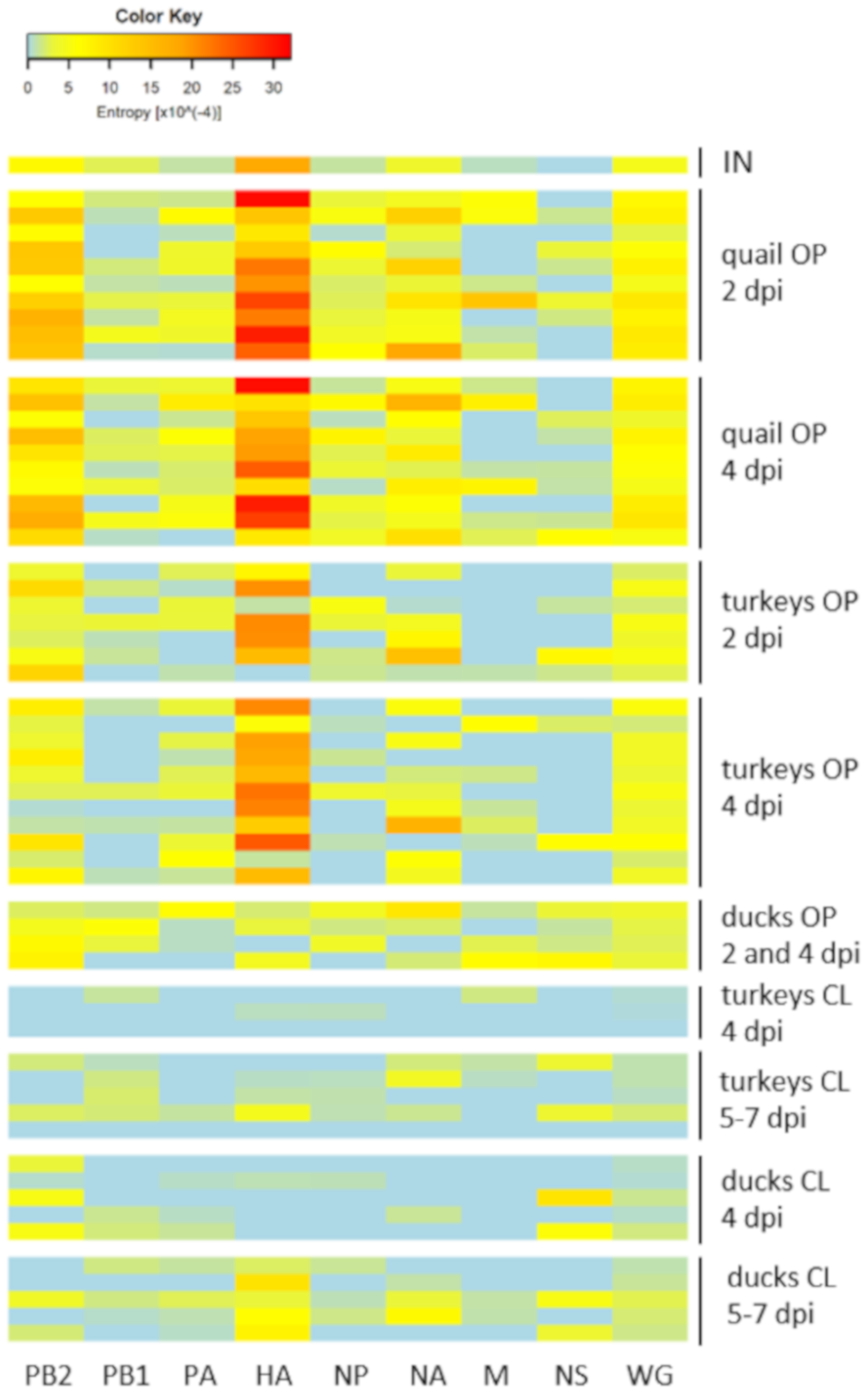
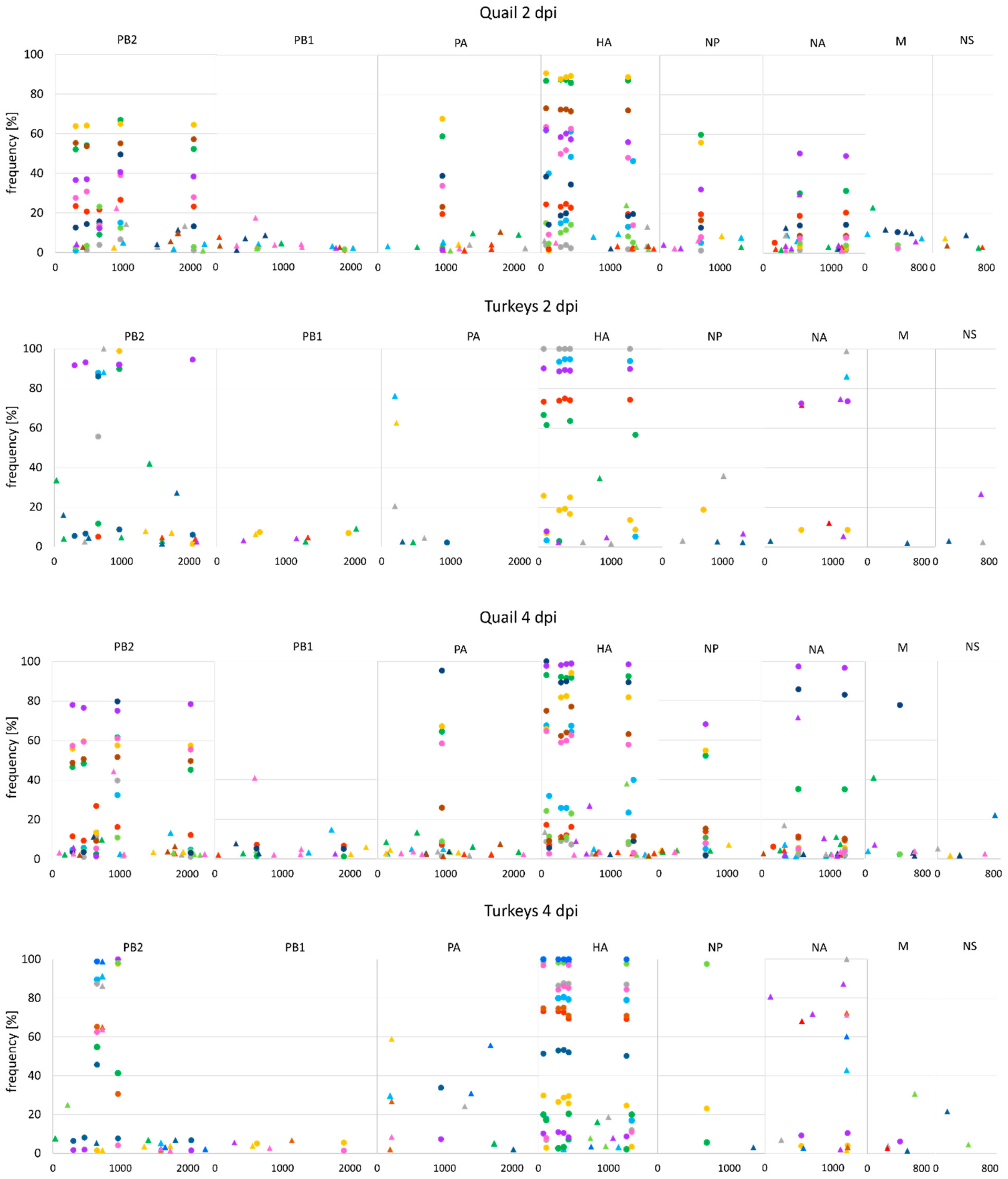
| dpi/dpc | Turkeys | Quail | Ducks | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inoculated | Contact | Inoculated | Contact | Inoculated | Contact | |||||||
| OP | CL | OP | CL | OP | CL | OP | CL | OP | CL | OP | CL | |
| 2/1 | 9/10 | 0/10 | 2/3 | 0/3 | 10/10 | 4/10 | 0/2 | 0/2 | 7/10 | 5/10 | 0/3 | 0/3 |
| 3/2 | 9/10 | 0/10 | 3/3 | 1/3 | 10/10 | 3/10 | 0/2 | 1/2 | 5/10 | 3/10 | 0/3 | 1/3 |
| 4/3 | 9/10 | 3/10 | 3/3 | 1/3 | 10/10 | 3/10 | 0/2 | 2/2 | 8/10 | 7/10 | 3/3 | 2/3 |
| 5/4 | 10/10 | 7/10 | 3/3 | 3/3 | 10/10 | 2/10 | 2/2 | 0/2 | 2/10 | 7/10 | 3/3 | 2/3 |
| 6/5 | 10/10 | 3/10 | 3/3 | 1/3 | 7/10 | 1/10 | 1/2 | 0/2 | 0/10 | 5/10 | 2/3 | 3/3 |
| 7/6 | 10/10 | 7/10 | 3/3 | 1/3 | 0/10 | 0/10 | 0/2 | 0/2 | 0/10 | 3/10 | 2/3 | 3/3 |
| 10/9 | 5/9 | 4/9 | 2/3 | 2/3 | 0/10 | 0/10 | 0/2 | 0/2 | 0/10 | 0/10 | 0/3 | 0/3 |
| 14/13 | 5/9 | 2/9 | 2/3 | 0/3 | 0/10 | 0/10 | 0/2 | 0/2 | 0/10 | 0/10 | 0/3 | 0/3 |
| Total | 10/10 | 10/10 | 3/3 | 3/3 | 10/10 | 6/10 | 2/2 | 2/2 | 10/10 | 8/10 | 3/3 | 3/3 |
| Latency period [days (SD)] | 2.3 (0.9) | 5.3 (1.3) | 2.0 (0.0) | 2.5 (0.8) | 2.5 (0.8) | 2.6 (0.9) | ||||||
| Duration of shedding [days (SD)] | 10.3 (4.2) | 4.2 (3.3) | 4.7 (0.5) | 1.3 (1.6) | 2.2 (0.9) | 3.0 (2.0) | ||||||
| Peak shedding [log10 eqEID50/1mL (SD)] | 5.5 (0.5) | 3.0 (1.7) | 5.4 (0.4) | 1.5 (1.4) | 2.4 (0.9) | 3.4 (1.9) | ||||||
| Mean daily shedding [log10 eqEID50/1mL (SD)] | 4.4 (0.6) | 2.5 (1.3) | 4.4 (0.5) | 1.3 (1.1) | 2.2 (0.8) | 2.4 (1.3) | ||||||
| Quail – Oropharyngeal Swabs | |||
|---|---|---|---|
| Gene | Position | Amino acid Change | No. of Birds with Frequency >50% at Any Day |
| PB2 | 303 † | - | 5 |
| 467 † | A156V | 5 | |
| 966 † | - | 6 | |
| 2052 † | - | 5 | |
| PA | 948 † | - | 4 |
| HA | 73 † | Y17H | 7 |
| 285 † | - | 6 | |
| 363 † | - | 6 | |
| 433 † | L133R L133S | 7 | |
| 434 † | 7 | ||
| 1260 † | - | 6 | |
| NP | 687 † | - | 3 |
| NA | 534 | - | 1 |
| 540 † | - | 2 | |
| 1218 † | - | 2 | |
| M | 486 † | - | 1 |
| † SNPs of inoculum origin | |||
| Turkeys | ||||
|---|---|---|---|---|
| Gene | Position | Amino Acid Change | No. of Birds with Frequency >50% at Any Day | |
| Oropharyngeal Swabs | Cloacal Swabs | |||
| PB2 | 303† | - | 1 | - |
| 467† | A156V | 1 | - | |
| 654† | - | 6 | 3 | |
| 735 | - | 5 | 2 | |
| 966† | - | 4 | 2 | |
| 1355 | E452V | - | 1 | |
| 1740 | - | - | 1 | |
| 2052† | - | 1 | - | |
| PB1 | 560 | R187K | - | 1 |
| 623† | K208R | - | 1 | |
| 1911† | - | - | 1 | |
| PA | 192 | - | 1 | - |
| 215 | L72S | 1 | - | |
| 1680 | - | 1 | - | |
| HA | 73† | Y17H | 10 | 4 |
| 113† | I30T | 1 | 1 | |
| 285† | - | 9 | 3 | |
| 363† | - | 9 | 3 | |
| 433† | L133S | 9 | 3 | |
| 434† | L133R | 10 | 4 | |
| 1260† | - | 9 | 3 | |
| 1334† | T436I | 1 | 1 | |
| NP | 687† | - | 1 | 1 |
| 1396 | L466I | - | 1 | |
| NA | 84 | - | 1 | - |
| 540† | - | 1 | 1 | |
| 544 | G182S | 1 | - | |
| 702 | - | 1 | - | |
| 1114 | V372I | 1 | - | |
| 1156 | V386I | 1 | - | |
| 1203 | - | 5 | 2 | |
| 1218† | - | 1 | 1 | |
| M | 699 | - | - | 1 |
| † SNPs of inoculum origin | ||||
| Ducks | ||||
|---|---|---|---|---|
| Gene | Position | Amino Acid Change | No. of Birds with Frequency >50% at Any Day | |
| Oropharyngeal Swabs | Cloacal Swabs | |||
| PB2 | 60 | - | - | 1 |
| 1139 | R380K | - | 1 | |
| HA | 73† | Y17H | 1 | 2 |
| 285† | - | 1 | 2 | |
| 363† | - | 1 | 2 | |
| 433† | L133S | 1 | 2 | |
| 434† | 1 | 2 | ||
| 1260† | - | 1 | 2 | |
| † SNPs of inoculum origin | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Świętoń, E.; Tarasiuk, K.; Olszewska-Tomczyk, M.; Iwan, E.; Śmietanka, K. A Turkey-origin H9N2 Avian Influenza Virus Shows Low Pathogenicity but Different Within-host Diversity in Experimentally Infected Turkeys, Quail and Ducks. Viruses 2020, 12, 319. https://doi.org/10.3390/v12030319
Świętoń E, Tarasiuk K, Olszewska-Tomczyk M, Iwan E, Śmietanka K. A Turkey-origin H9N2 Avian Influenza Virus Shows Low Pathogenicity but Different Within-host Diversity in Experimentally Infected Turkeys, Quail and Ducks. Viruses. 2020; 12(3):319. https://doi.org/10.3390/v12030319
Chicago/Turabian StyleŚwiętoń, Edyta, Karolina Tarasiuk, Monika Olszewska-Tomczyk, Ewelina Iwan, and Krzysztof Śmietanka. 2020. "A Turkey-origin H9N2 Avian Influenza Virus Shows Low Pathogenicity but Different Within-host Diversity in Experimentally Infected Turkeys, Quail and Ducks" Viruses 12, no. 3: 319. https://doi.org/10.3390/v12030319
APA StyleŚwiętoń, E., Tarasiuk, K., Olszewska-Tomczyk, M., Iwan, E., & Śmietanka, K. (2020). A Turkey-origin H9N2 Avian Influenza Virus Shows Low Pathogenicity but Different Within-host Diversity in Experimentally Infected Turkeys, Quail and Ducks. Viruses, 12(3), 319. https://doi.org/10.3390/v12030319





