Genetic Characterization of Avian Influenza A (H11N9) Virus Isolated from Mandarin Ducks in South Korea in 2018
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Virus Isolation from the Samples
2.3. RNA Extraction and Subtyping
2.4. Sequencing Using Illumina HiSeq X Method for Next-Generation Sequencing (NGS)
2.5. Phylogenetic Analysis
2.6. Measurement of 50% Tissue Culture Infectious Dose (TCID50) and 50% Egg Infectious Dose (EID50)
2.7. Animal Infection
2.8. Statistics
3. Results
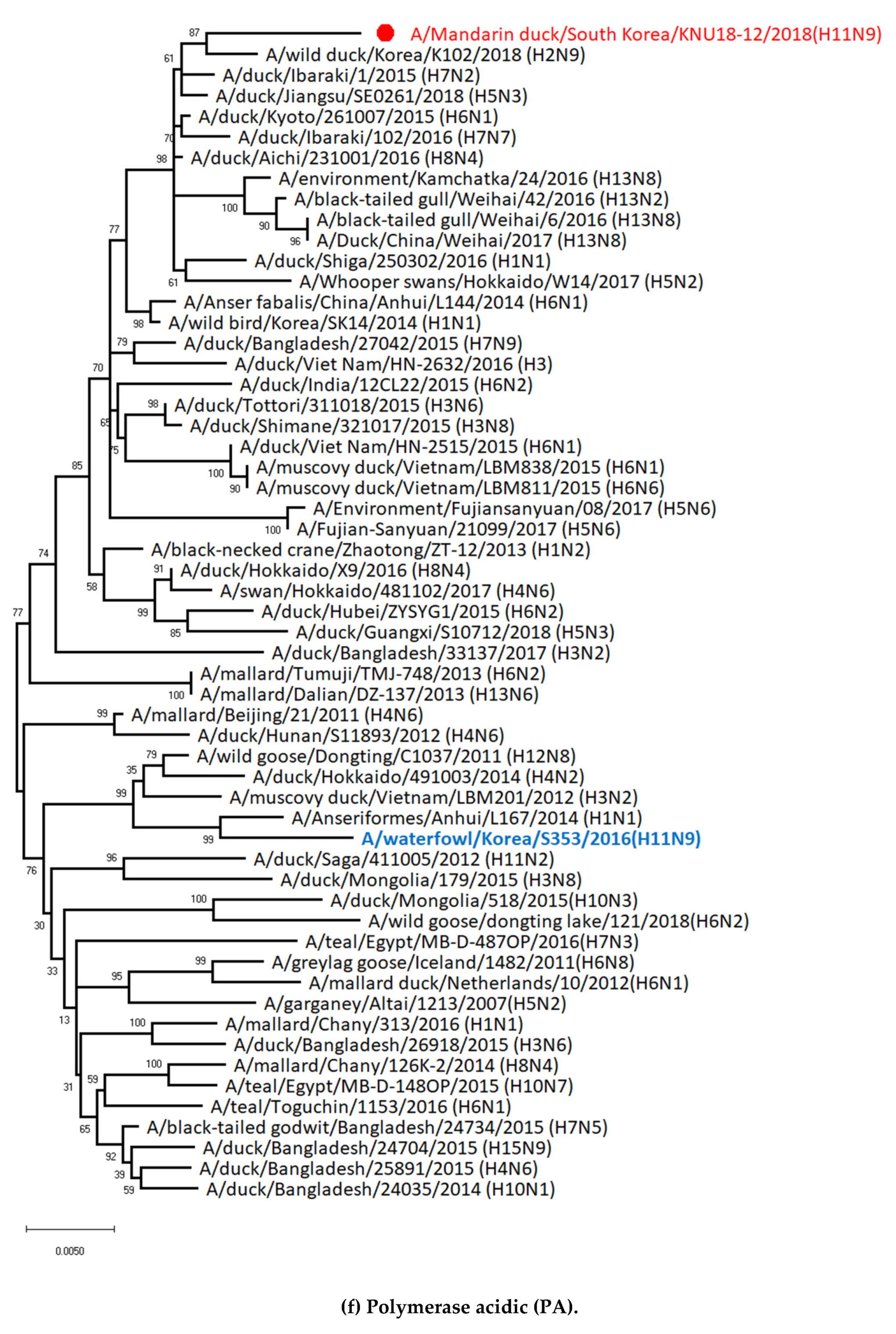
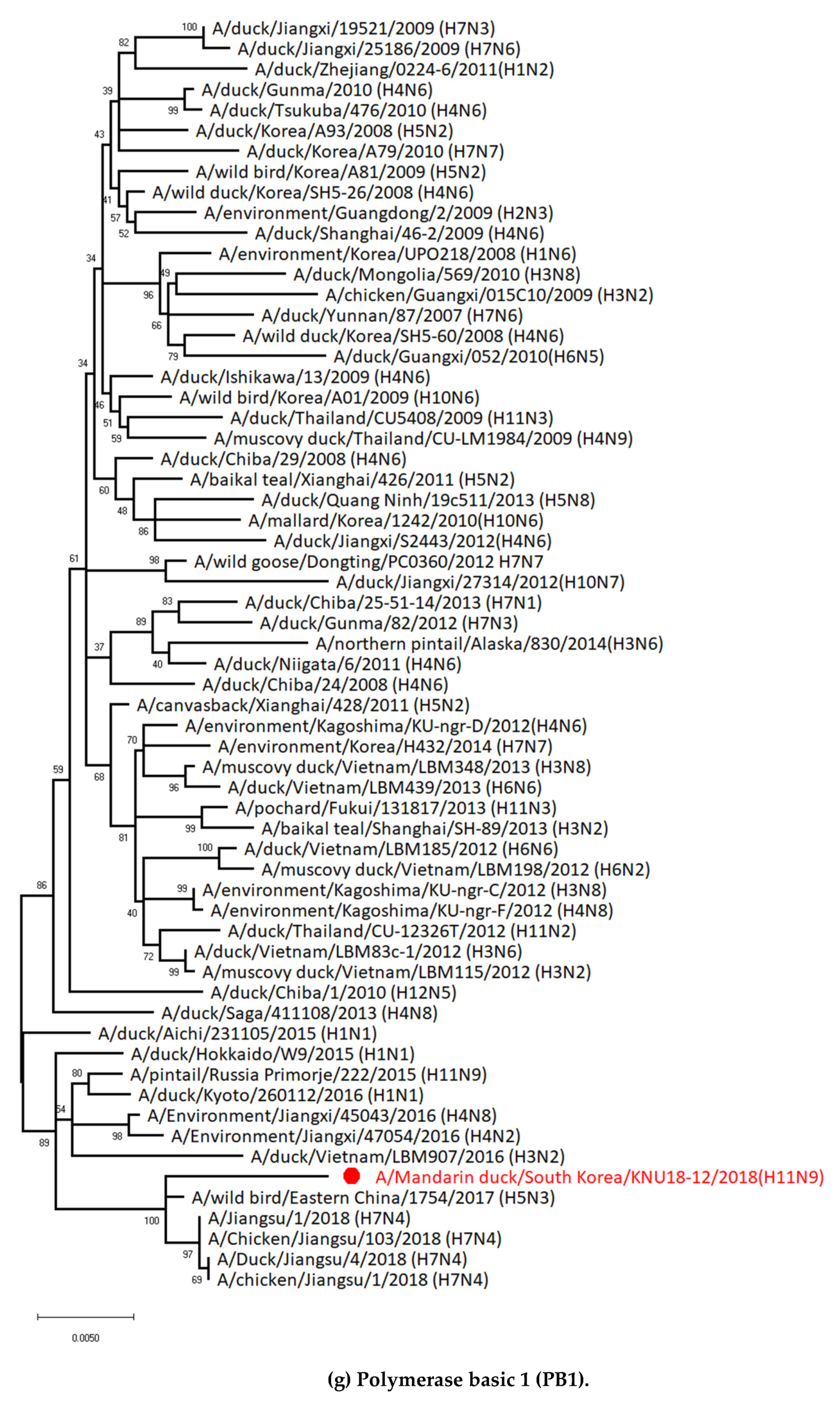
3.1. Genetic Characterization of Novel Avian Influenza A (H11N9) in 2018
3.2. Molecular Characterization of H11N9 Isolate
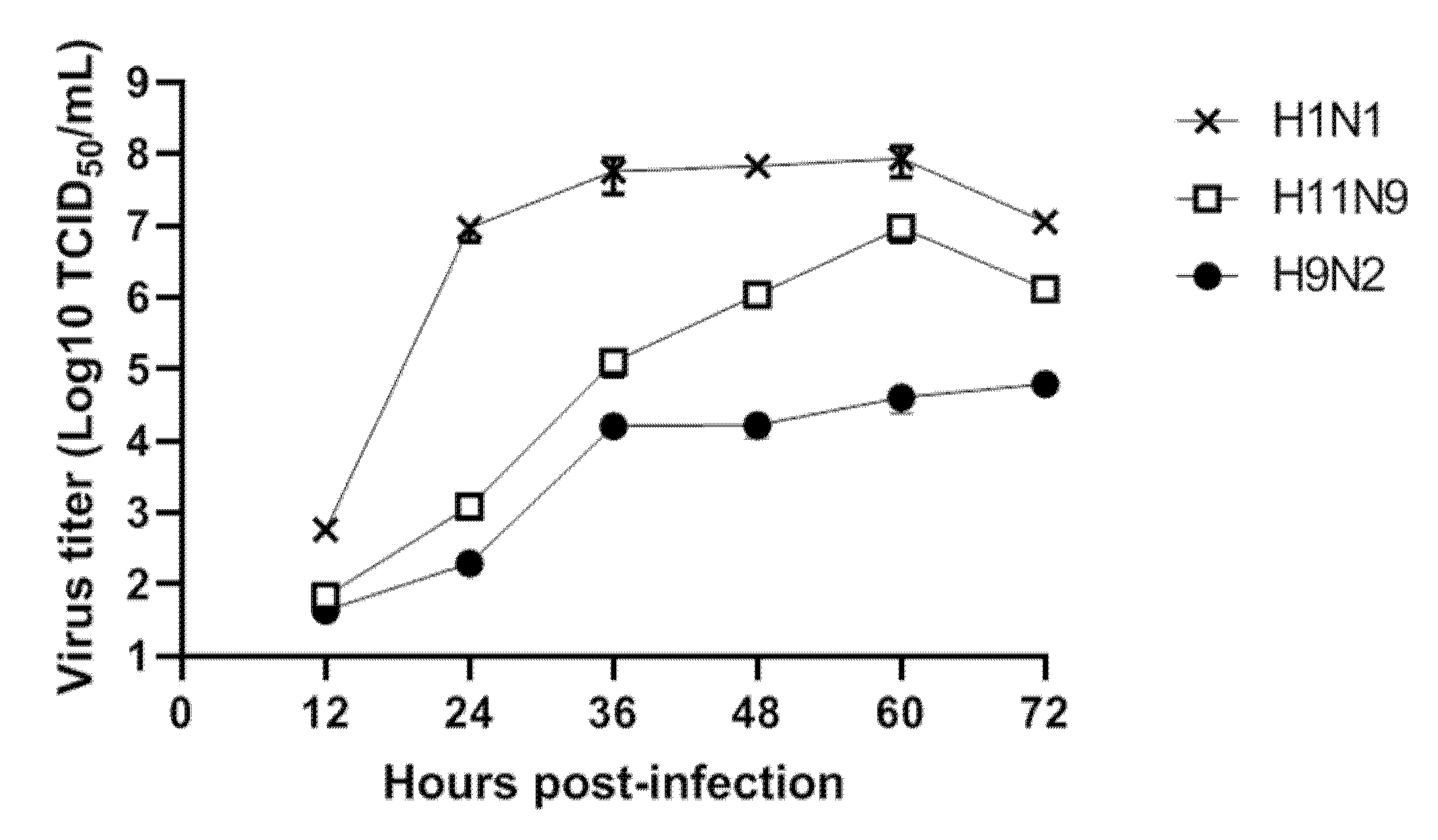
3.3. Replication of H11N9 in Mammalian Cells
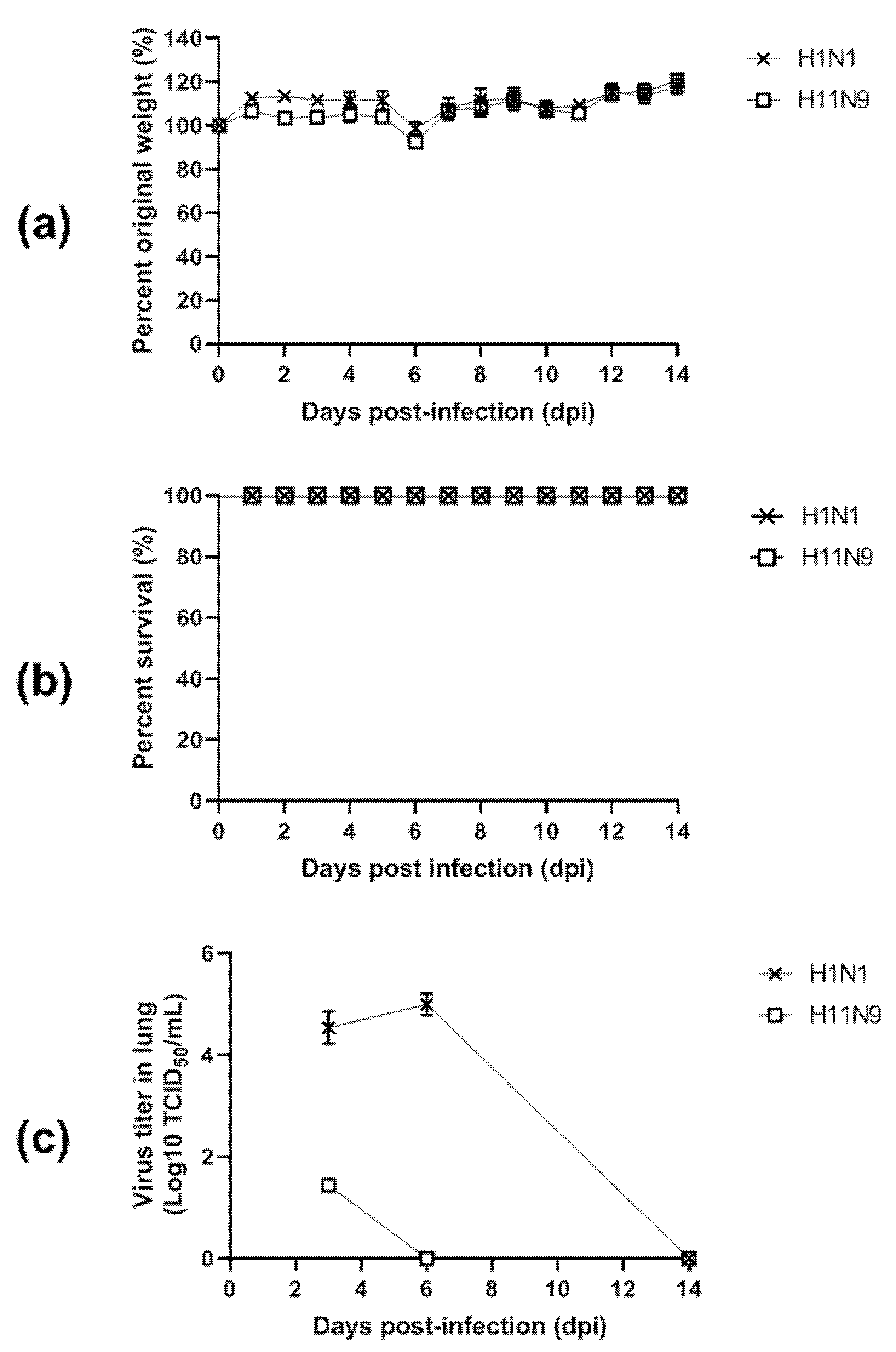
3.4. Pathogenicity in Mice
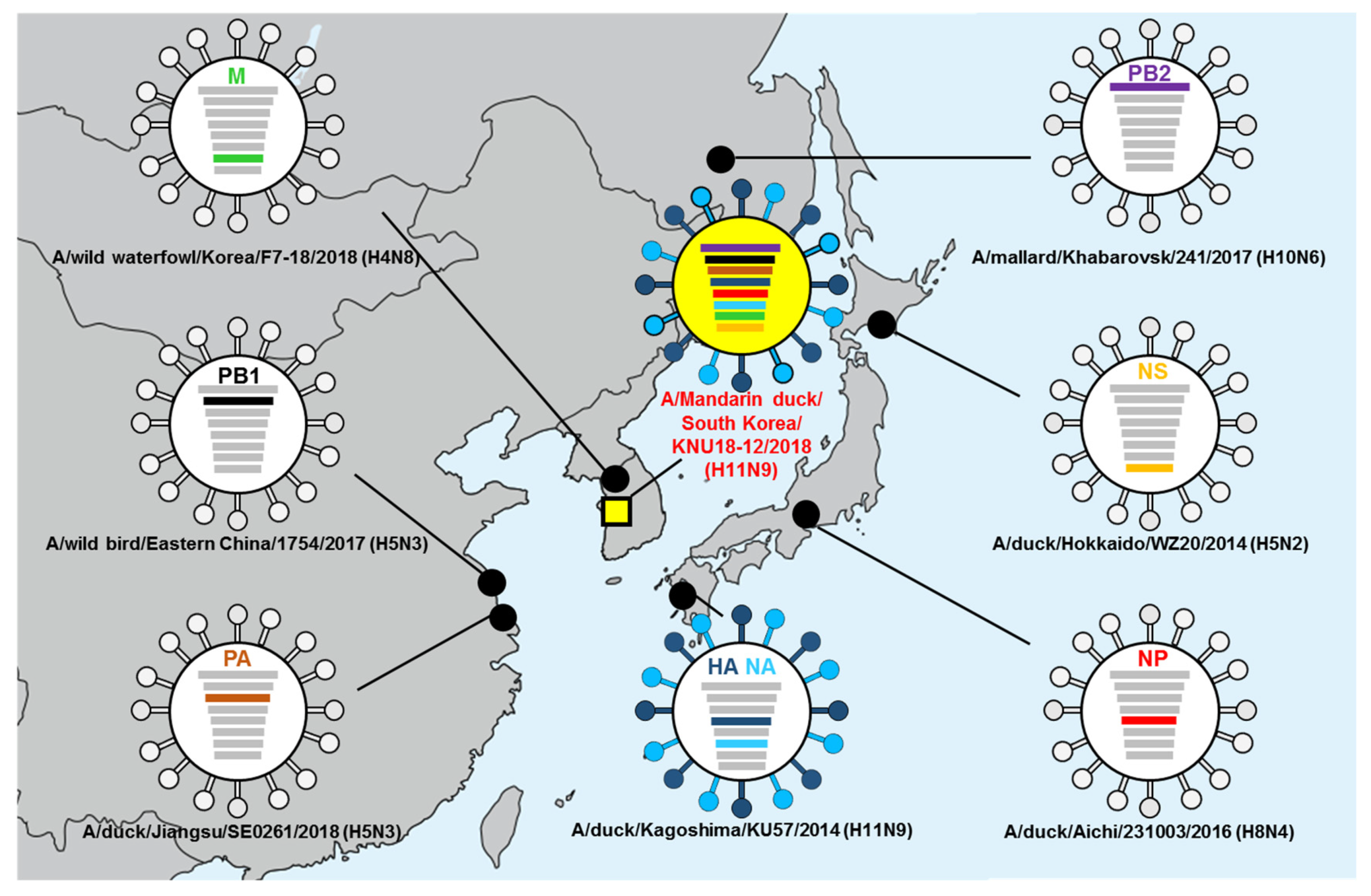
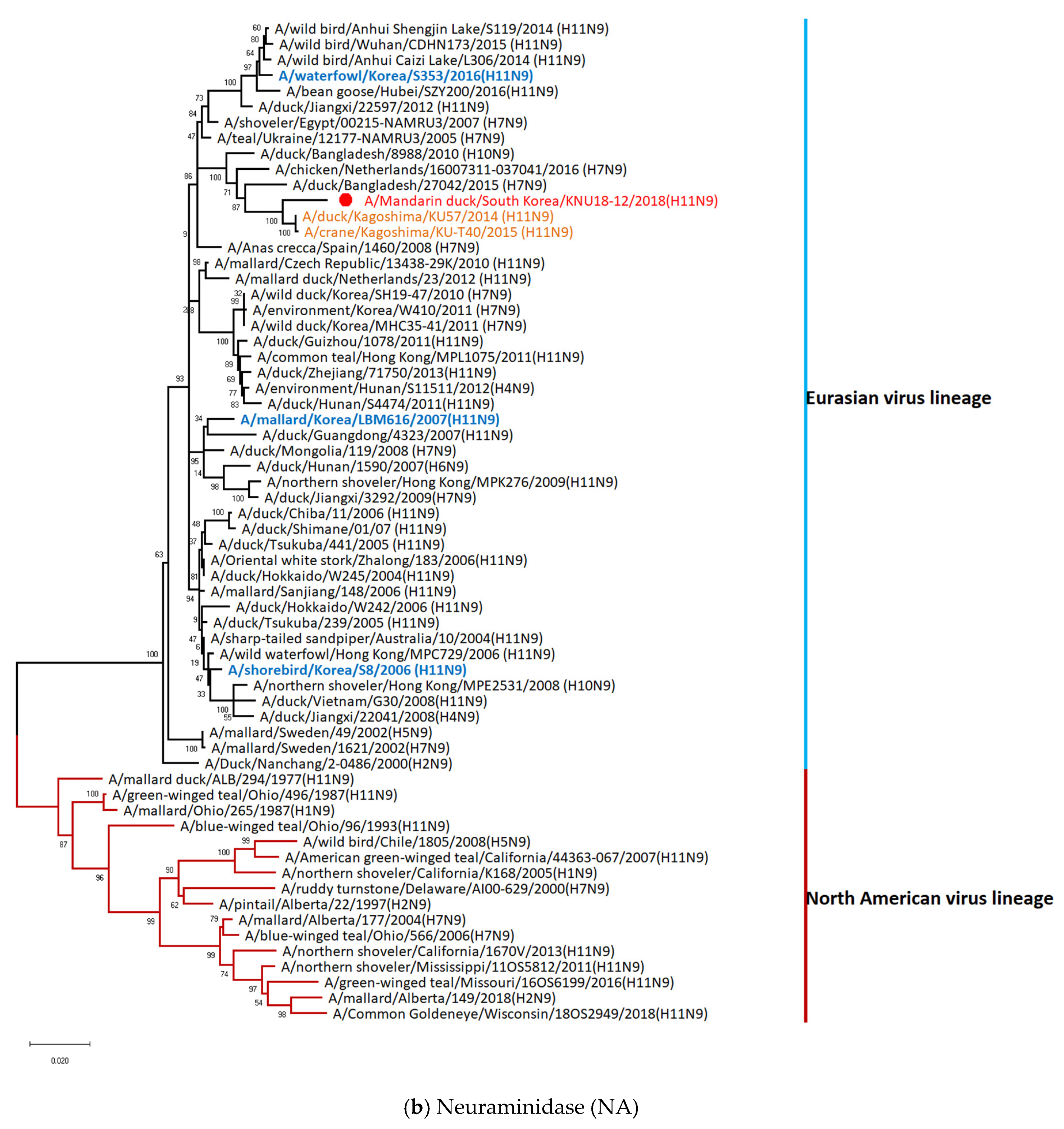
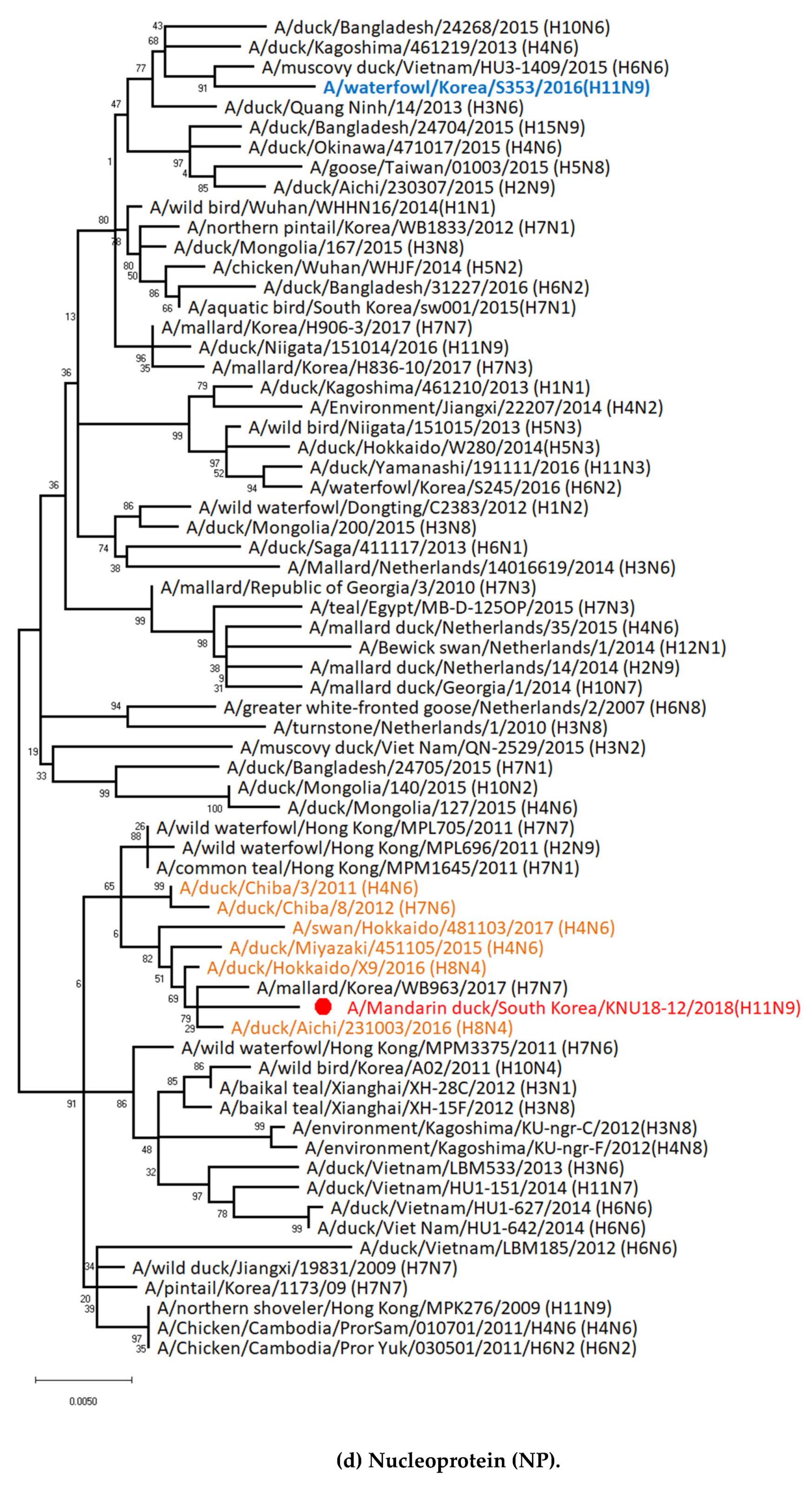
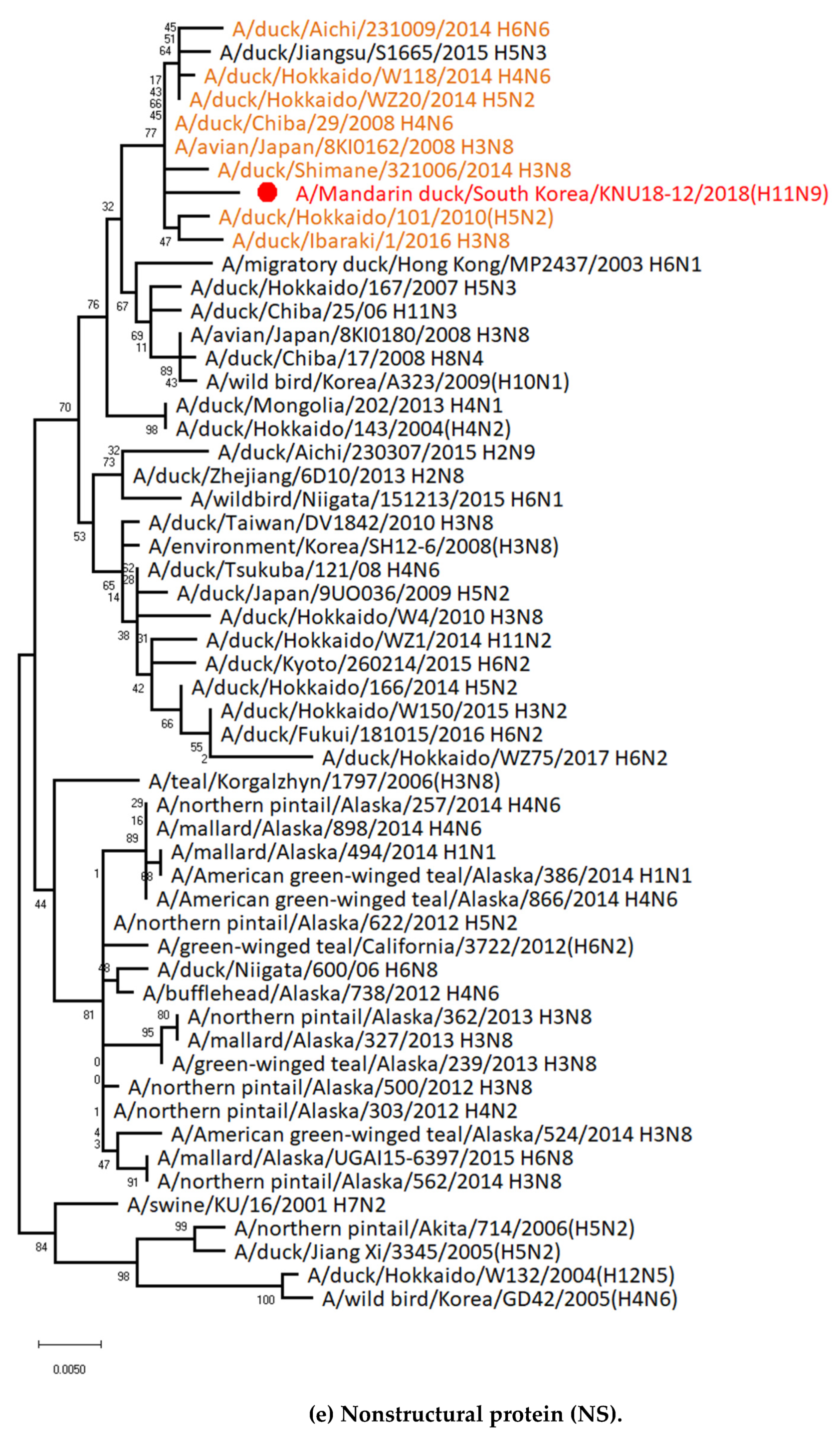
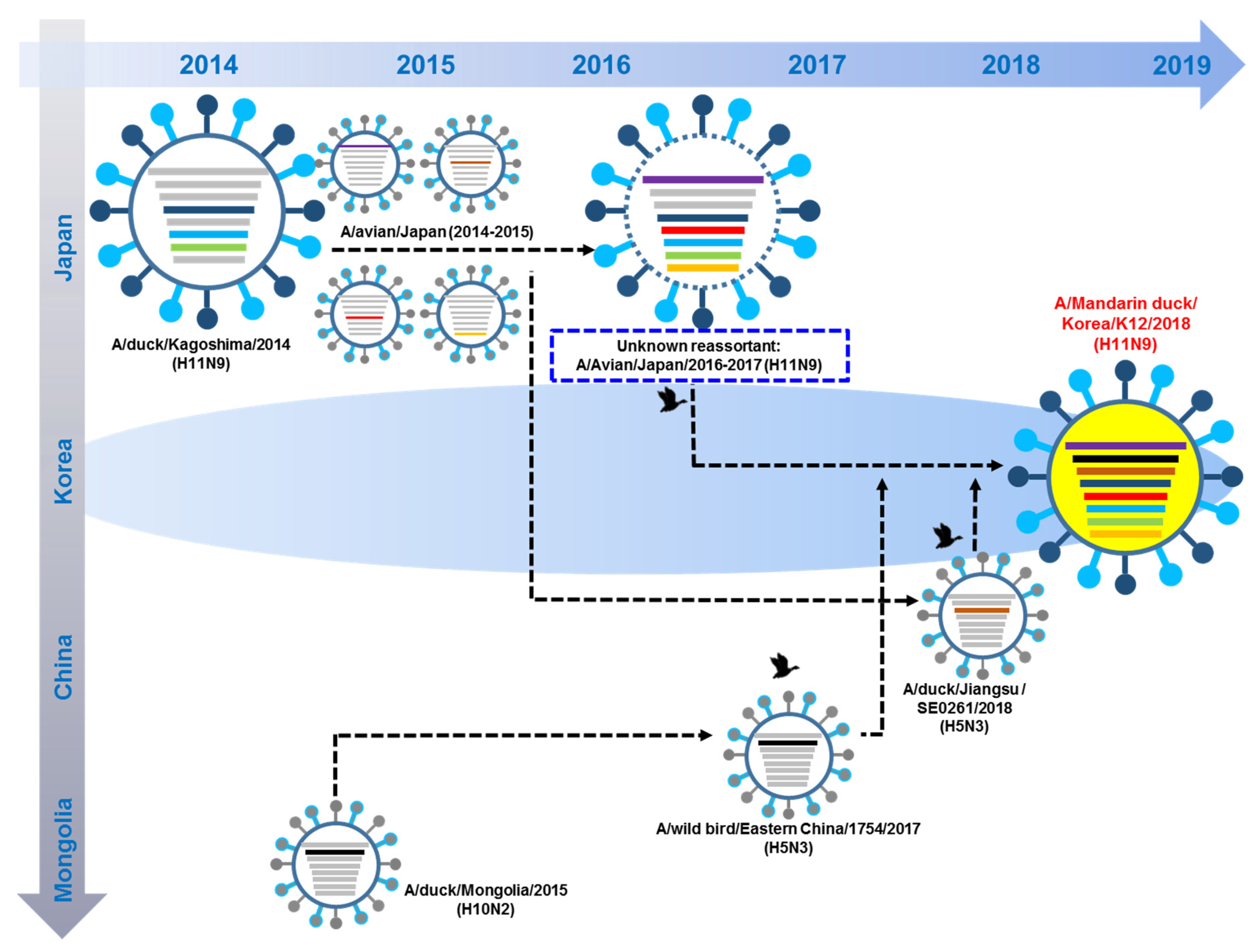
3.5. Hypothesis of Reassortment Event of A/Mandarin Duck/South Korea/KNU18-12/2018(H11N9)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hofer, U. Viral evolution: Past, present and future of influenza viruses. Nat. Rev. Microbiol. 2014, 12, 237. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, T.; Mamahit, A.; Cox, N.J. 65 years of influenza surveillance by a World Health Organization-coordinated global network. Influenza Respir. Viruses 2018, 12, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-K.; Lee, Y.-N.; Kye, S.-J.; Lewis, N.S.; Brown, I.H.; Sagong, M.; Heo, G.-B.; Kang, Y.-M.; Cho, H.-K.; Kang, H.-M. Characterization of a novel reassortant H5N6 highly pathogenic avian influenza virus clade 2.3. 4.4 in Korea, 2017. Emerg. Microbes Infect. 2018, 7, 1–3. [Google Scholar]
- Centers for Disease Control and Prevention. Influenza a Subtypes and the Species Affected; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2018. [Google Scholar]
- Alexander, D.J. Isolation of influenza A viruses from birds in Great Britain during 1980 and 1981. Vet. Rec. 1982, 111, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Jonassen, C.M.; Handeland, K. Avian influenza virus screening in wild waterfowl in Norway, 2005. Avian Dis. 2007, 51, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Perez-Ramirez, E.; Gerrikagoitia, X.; Barral, M.; Hofle, U. Detection of low pathogenic avian influenza viruses in wild birds in Castilla-La Mancha (south central Spain). Vet. Microbiol. 2010, 146, 200–208. [Google Scholar] [CrossRef]
- Van Borm, S.; Rosseel, T.; Vangeluwe, D.; van den bussche, F.; van den Berg, T.; Lambrecht, B. Phylogeographic analysis of avian influenza viruses isolated from Charadriiformes in Belgium confirms intercontinental reassortment in gulls. Arch. Virol. 2012, 157, 1509–1522. [Google Scholar] [CrossRef]
- Latorre-Margalef, N.; Tolf, C.; Grosbois, V.; Avril, A.; Bengtsson, D.; Wille, M.; Osterhaus, A.D.; Fouchier, R.A.; Olsen, B.; Waldenstrom, J. Long-term variation in influenza a virus prevalence and subtype diversity in migratory mallards in northern Europe. Proc. Biol. Sci. 2014, 281, 20140098. [Google Scholar] [CrossRef]
- Nagy, A.; Cernikova, L.; Jirincova, H.; Havlickova, M.; Hornickova, J. Local-scale diversity and between-year "frozen evolution" of avian influenza a viruses in nature. PLoS ONE 2014, 9, e103053. [Google Scholar] [CrossRef]
- Simulundu, E.; Ishii, A.; Igarashi, M.; Mweene, A.S.; Suzuki, Y.; Hang’ombe, B.M.; Namangala, B.; Moonga, L.; Manzoor, R.; Ito, K.; et al. Characterization of influenza a viruses isolated from wild waterfowl in Zambia. J. Gen. Virol. 2011, 92 Pt 6, 1416–1427. [Google Scholar] [CrossRef]
- Karamendin, K.; Kydyrmanov, A.; Zhumatov, K.; Asanova, S.; Ishmukhametova, N.; Sayatov, M. Phylogenetic analysis of avian influenza viruses of H11 subtype isolated in Kazakhstan. Virus Genes 2011, 43, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Nomura, N.; Sakoda, Y.; Endo, M.; Yoshida, H.; Yamamoto, N.; Okamatsu, M.; Sakurai, K.; Hoang, N.V.; Nguyen, L.V.; Chu, H.D.; et al. Characterization of avian influenza viruses isolated from domestic ducks in Vietnam in 2009 and 2010. Arch. Virol. 2012, 157, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Zou, S.M.; Li, X.D.; Dong, L.B.; Bo, H.; Gao, R.B.; Wang, D.Y.; Shu, Y.L. Detection of reassortant avian influenza A (H11N9) virus in environmental samples from live poultry markets in China. Infect. Dis. Poverty 2016, 5, 59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wilcox, B.R.; Knutsen, G.A.; Berdeen, J.; Goekjian, V.; Poulson, R.; Goyal, S.; Sreevatsan, S.; Cardona, C.; Berghaus, R.D.; Swayne, D.E.; et al. Influenza-A viruses in ducks in northwestern Minnesota: Fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS ONE 2011, 6, e24010. [Google Scholar] [CrossRef]
- De Araujo, J.; de Azevedo, S.M., Jr.; Gaidet, N.; Hurtado, R.F.; Walker, D.; Thomazelli, L.M.; Ometto, T.; Seixas, M.M.; Rodrigues, R.; Galindo, D.B.; et al. Avian influenza virus (H11N9) in migratory shorebirds wintering in the Amazon Region, Brazil. PLoS ONE 2014, 9, e110141. [Google Scholar] [CrossRef]
- Hurtado, R.; Fabrizio, T.; Vanstreels, R.E.; Krauss, S.; Webby, R.J.; Webster, R.G.; Durigon, E.L. Molecular Characterization of Subtype H11N9 Avian Influenza Virus Isolated from Shorebirds in Brazil. PLoS ONE 2015, 10, e0145627. [Google Scholar] [CrossRef]
- Kim, H.M.; Oh, J.H.; Seo, S.H. Genetic characterization of avian influenza viruses isolated from waterfowl in southern part of South Korea in 2006. Virus Genes 2008, 37, 49–51. [Google Scholar] [CrossRef]
- Le, T.B.; Lee, I.H.; Kim, H.S.; Oh, S.K.; Seo, S.H. Genetic analysis of a novel reassortant H11N9 Isolated from waterfowl in South Korea in 2016. Virus Genes 2017, 53, 656–660. [Google Scholar] [CrossRef]
- Li, J.; Cardona, C.J.; Xing, Z.; Woolcock, P.R. Genetic and phenotypic characterization of a low-pathogenicity avian influenza H11N9 virus. Arch. Virol. 2008, 153, 1899–1908. [Google Scholar] [CrossRef]
- Wu, H.; Peng, X.; Peng, X.; Wu, N. Molecular characterization of a reassortant H11N9 subtype avian influenza virus isolated from a domestic duck in Eastern China. Arch. Virol. 2015, 160, 2595–2601. [Google Scholar] [CrossRef]
- Yao, Y.; Shao, Z.; He, B.; Yang, W.; Chen, J.; Zhang, T.; Chen, X.; Chen, J. Characterization of a reassortant H11N9 subtype avian influenza virus isolated from bean goose along the East Asian—Australian flyway. Virus Genes 2017, 53, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Lebarbenchon, C.; Brown, J.D.; Stallknecht, D.E. Evolution of influenza a virus H7 and N9 subtypes, Eastern Asia. Emerg. Infect. Dis. 2013, 19, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Luo, J.; Wang, J.; Su, W.; Gao, S.; Zhang, M.; Xie, L.; Ding, H.; Liu, S.; Liu, X. Novel human H7N9 influenza virus in China. Integr. Zool. 2014, 9, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.S.; Webby, R.; Gilchrist, M.J.R.; Gray, G.C. Avian influenza among waterfowl hunters and wildlife professionals. Emerg. Infect. Dis. 2006, 12, 1284–1286. [Google Scholar] [CrossRef]
- World Health Organization. CDC Protocol of Realtime RTPCR for Influenza A (H1N1); WHO: Geneva, Switzerland, 2009. [Google Scholar]
- Hoffmann, B.; Hoffmann, D.; Henritzi, D.; Beer, M.; Harder, T.C. Riems influenza a typing array (RITA): An RT-qPCR-based low density array for subtyping avian and mammalian influenza a viruses. Sci. Rep. 2016, 3, 27211. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of Birds through DNA Barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef]
- Ambardar, S.; Gupta, R.; Trakroo, D.; Lal, R.; Vakhlu, J. High Throughput Sequencing: An Overview of Sequencing Chemistry. Indian J. Microbiol. 2016, 56, 394–404. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Global Influenza Surveillance Network. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; World Health Organization Global Influenza Surveillance Network: Geneva, Switzerland, 2011. [Google Scholar]
- Reed, L.J.; Muench, A.H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Jimenez-Bluhm, P.; Karlsson, E.A.; Ciuoderis, K.A.; Cortez, V.; Marvin, S.A.; Hamilton-West, C.; Schultz-Cherry, S.; Osorio, J.E. Avian H11 influenza virus isolated from domestic poultry in a Colombian live animal market. Emerg. Microbes Infect. 2016, 5, e121. [Google Scholar] [CrossRef]
- Killian, M.L. Hemagglutination assay for influenza virus. Methods Mol. Biol. 2014, 1161, 3–9. [Google Scholar] [PubMed]
- The University of IOWA. 2018. Available online: https://animal.research.uiowa.edu/iacuc-guidelines-anesthesia (accessed on 10 October 2018).
- Jeong, S.; Lee, D.-H.; Kim, Y.-J.; Lee, S.-H.; Cho, A.Y.; Noh, J.-Y.; Tseren-Ochir, E.-O.; Jeong, J.-H.; Song, C.-S.; Jeong, S. Introduction of Avian Influenza A (H6N5) Virus into Asia from North America by Wild Birds. Emerg. Infect. Dis. 2019, 25. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Gambaryan, A.S.; Teneberg, S.; Piskarev, V.E.; Yamnikova, S.S.; Lvov, D.K.; Robertson, J.S.; Karlsson, K.-A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 1997, 233, 224–234. [Google Scholar] [CrossRef]
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human Infection with a Novel Avian-Origin Influenza A (H7N9) Virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef] [PubMed]
- Husain, M. Avian influenza A (H7N9) virus infection in humans: Epidemiology, evolution, and pathogenesis. Infect. Genet. Evol. 2014, 28, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Hatta, M.; Gao, P.; Halfmann, P.; Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza a viruses. Science 2001, 293, 1840–1842. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, D.B.; Mukatira, S.; Mehta, P.K.; Obenauer, J.C.; Su, X.; Webster, R.G.; Naeve, C.W. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 2007, 81, 10292–10299. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Reid, A.H.; Lourens, R.M.; Wang, R.; Jin, G.; Fanning, T.G. Characterization of the 1918 influenza virus polymerase genes. Nature 2005, 437, 889–893. [Google Scholar] [CrossRef]
- El-Shesheny, R.; Kandeil, A.; Bagato, O.; Maatouq, A.M.; Moatasim, Y.; Rubrum, A.; Song, M.S.; Webby, R.J.; Ali, M.A.; Kayali, G. Molecular characterization of avian influenza H5N1 virus in Egypt and the emergence of a novel endemic subclade. J. Gen. Virol. 2014, 95, 1444–1463. [Google Scholar] [CrossRef]
- To, K.K.; Song, W.; Lau, S.Y.; Que, T.L.; Lung, D.C.; Hung, I.F.; Chen, H.; Yuen, K.Y. Unique reassortant of influenza A (H7N9) virus associated with severe disease emerging in Hong Kong. J. Infect. 2014, 69, 60–68. [Google Scholar] [CrossRef]
- Li, J.; Ishaq, M.; Prudence, M.; Xi, X.; Hu, T.; Liu, Q.; Guo, D. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009, 144, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.; Cooper, L.; Xu, X.; Thompson, W.; Krauss, S.; Guan, Y.; Zhou, N.; Klimov, A.; Cox, N.; Webster, R.; et al. Molecular changes associated with the transmission of avian influenza a H5N1 and H9N2 viruses to humans. J. Med. Virol. 2002, 66, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhang, X.; Gao, W.; Wang, C.; Wang, J.; Sun, H.; Sun, Y.; Guo, L.; Zhang, R.; Chang, K.C.; et al. Prevailing PA mutation K356R in avian influenza H9N2 virus increases mammalian replication and pathogenicity. J. Virol. 2016, 90, 8105–8114. [Google Scholar] [CrossRef] [PubMed]
- Leung, B.W.; Chen, H.; Brownlee, G.G. Correlation between polymerase activity and pathogenicity in two duck H5N1 influenza viruses suggests that the polymerase contributes to pathogenicity. Virology 2010, 401, 96–106. [Google Scholar] [CrossRef]
- Hulse-Post, D.J.; Franks, J.; Boyd, K.; Salomon, R.; Hoffmann, E.; Yen, H.L.; Webby, R.J.; Walker, D.; Nguyen, T.D.; Webster, R.G. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 2007, 81, 8515–8524. [Google Scholar] [CrossRef]
- Lycett, S.J.; Ward, M.J.; Lewis, F.I.; Poon, A.F.; Kosakovsky Pond, S.L.; Brown, A.J. Detection of mammalian virulence determinants in highly pathogenic avian influenza H5N1 viruses: Multivariate analysis of published data. J. Virol. 2009, 83, 9901–9910. [Google Scholar] [CrossRef]
- Jiao, P.; Tian, G.; Li, Y.; Deng, G.; Jiang, Y.; Liu, C.; Liu, W.; Bu, Z.; Kawaoka, Y.; Chen, H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008, 82, 1146–1154. [Google Scholar] [CrossRef]
- Lee, M.S.; Deng, M.C.; Lin, Y.J.; Chang, C.Y.; Shieh, H.K.; Shiau, J.Z.; Huang, C.C. Characterization of an H5N1 avian influenza virus from Taiwan. Vet. Microbiol. 2007, 124, 193–201. [Google Scholar] [CrossRef]
- Seo, S.H.; Hoffmann, E.; Webster, R.G. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 2002, 8, 950–954. [Google Scholar] [CrossRef]
- Li, M.; Wang, B. Homology modeling and examination of the effect of the D92E mutation on the H5N1 nonstructural protein NS1 effector domain. J. Mol. Modeling 2007, 13, 1237–1244. [Google Scholar] [CrossRef]
- Fan, S.; Deng, G.; Song, J.; Tian, G.; Suo, Y.; Jiang, Y.; Guan, Y.; Bu, Z.; Kawaoka, Y.; Chen, H. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 2009, 384, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, T.; Fujisaki, S.; Takashita, E.; Xu, H.; Yamada, S.; Uchida, Y.; Neumann, G.; Saito, T.; Kawaoka, Y.; Tashiro, M. Genetic analysis of novel avian A (H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro surveill. 2013, 18, 20453. [Google Scholar] [PubMed]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza virus M2 protein has ion channel activity. Cell 1992, 69, 517–528. [Google Scholar] [CrossRef]
- Cappelle, J.; Zhao, D.; Gilbert, M.; Nelson, M.I.; Newman, S.H.; Takekawa, J.Y.; Gaidet, N.; Prosser, D.J.; Liu, Y.; Li, P.; et al. Risks of avian influenza transmission in areas of intensive free-ranging duck production with wild waterfowl. EcoHealth 2014, 11, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Andreas, L.B.; Eddy, M.T.; Chou, J.J.; Griffin, R.G. Magic-angle-spinning NMR of the drug resistant S31N M2 proton transporter from influenza A. J. Am. Chem. Soc. 2012, 134, 7215–7218. [Google Scholar] [CrossRef]
- Conenello, G.M.; Zamarin, D.; Perrone, L.A.; Tumpey, T.; Palese, P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007, 3, 1121–1141. [Google Scholar] [CrossRef]
- Lock, M.; Korn, M.; Wilson, J.; Sena-Esteves, M.; Gao, G. Measuring the Infectious Titer of Recombinant Adenovirus Using Tissue Culture Infection Dose 50% (TCID50) End-Point Dilution and Quantitative Polymerase Chain Reaction (qPCR). Cold Spring Harb. Protoc. 2019, 2019, pdb–prot095562. [Google Scholar] [CrossRef]
- Yeo, S.J.; Cuc, B.T.; Sung, H.W.; Park, H. Evaluation of a smartphone-based rapid fluorescent diagnostic system for H9N2 virus in specific-pathogen-free chickens. Arch. Virol. 2016, 161, 2249–2256. [Google Scholar] [CrossRef]
- Schrauwen, E.J.; Fouchier, R.A. Host adaptation and transmission of influenza a viruses in mammals. Emerg. Microbes Infect. 2014, 3, 1–10. [Google Scholar] [CrossRef]
- Kim, H.R.; Lee, Y.J.; Park, C.K.; Oem, J.K.; Lee, O.S.; Kang, H.M.; Choi, J.G.; Bae, Y.C. Highly pathogenic avian influenza (H5N1) outbreaks in wild birds and poultry, South Korea. Emerg. Infect. Dis. 2012, 18, 480–483. [Google Scholar] [CrossRef]
- Mase, M.; Kim, J.H.; Lee, Y.J.; Tsukamoto, K.; Imada, T.; Imai, K.; Yamaguchi, S. Genetic comparison of H5N1 influenza a viruses isolated from chickens in Japan and Korea. Microbiol. Immunol. 2005, 49, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Shivakoti, S.; Ito, H.; Otsuki, K.; Ito, T. Characterization of H5N1 highly pathogenic avian influenza virus isolated from a mountain hawk eagle in Japan. J. Vet. Med. Sci. 2010, 72, 459–463. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Banks, J.; Speidel, E.S.; Moore, E.; Plowright, L.; Piccirillo, A.; Capua, I.; Cordioli, P.; Fioretti, A.; Alexander, D.J. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 2001, 146, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Horimoto, T.; Rivera, E.; Pearson, J.; Senne, D.; Krauss, S.; Kawaoka, Y.; Webster, R.G. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 1995, 213, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Yao, Q.; Wang, X.; Chai, H.; Deng, G.; Chen, H.; Hua, Y. Detection of reassortant avian influenza A (H11N9) virus in wild birds in China. Transbound. Emerg. Dis. 2019, 66, 1142–1157. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Okuya, K.; Kawabata, T.; Matsuu, A.; Takase, K.; Kuwahara, M.; Toda, S.; Ozawa, M. Genetic characterization of low-pathogenic avian influenza viruses isolated on the Izumi plain in Japan: Possible association of dynamic movements of wild birds with AIV evolution. Arch. Virol. 2018, 163, 911–923. [Google Scholar] [CrossRef]
- Onuma, M.; Kakogawa, M.; Yanagisawa, M.; Haga, A.; Okano, T.; Neagari, Y.; Okano, T.; Goka, K.; Asakawa, M. Characterizing the temporal patterns of avian influenza virus introduction into Japan by migratory birds. J. Vet. Med. Sci. 2017, 5, 943–951. [Google Scholar] [CrossRef]
- Uraki, R.; Kiso, M.; Shinya, K.; Goto, H.; Takano, R.; Iwatsuki-Horimoto, K.; Takahashi, K.; Daniels, R.S.; Hungnes, O.; Watanabe, T.; et al. Virulence determinants of pandemic A (H1N1)2009 influenza virus in a mouse model. J. Virol. 2013, 87, 2226–2233. [Google Scholar] [CrossRef]
- Ilyushina, N.A.; Khalenkov, A.M.; Seiler, J.P.; Forrest, H.L.; Bovin, N.V.; Marjuki, H.; Barman, S.; Webster, R.G.; Webby, R.J. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 2010, 84, 8607–8616. [Google Scholar] [CrossRef]
- Mi, C.; Møller, A.P.; Guo, Y. Annual spatio-temporal migration patterns of Hooded Cranes wintering in Izumi based on satellite tracking and their implications for conservation. Avian Res. 2018, 9, 23. [Google Scholar] [CrossRef]
- Brown, J.D.; Stallknecht, D.E.; Beck, J.R.; Suarez, D.L.; Swayne, D.E. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg. Infect. Dis. 2006, 12, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.C.; Xing, Z.; Li, J.; Cardona, C.J. Immune-related gene expression in response to H11N9 low pathogenic avian influenza virus infection in chicken and Pekin duck peripheral blood mononuclear cells. Mol. Immunol. 2009, 46, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Hurt, A.C.; Vijaykrishna, D.; Butler, J.; Baas, C.; Maurer-Stroh, S.; Silva-de-la-Fuente, M.C.; Medina-Vogel, G.; Olsen, B.; Kelso, A.; Barr, I.G. Detection of evolutionarily distinct avian influenza a viruses in Antarctica. MBio 2014, 5, e01098-14. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, M.; Matsuu, A.; Tokorozaki, K.; Horie, M.; Masatani, T.; Nakagawa, H.; Okuya, K.; Kawabata, T.; Toda, S. Genetic diversity of highly pathogenic H5N8 avian influenza viruses at a single overwintering site of migratory birds in Japan, 2014/15. Eurosurveillance 2015, 20, 21132. [Google Scholar] [CrossRef] [PubMed]
- Son, K.; Kim, Y.-K.; Oem, J.-K.; Jheong, W.-H.; Sleeman, J.M.; Jeong, J. Experimental infection of highly pathogenic avian influenza viruses, clade 2.3. 4.4 H5N6 and H5N8, in mandarin ducks from South Korea. Transbound. Emerg. Dis. 2018, 65, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.-H.; Lee, D.-H.; Swayne, D.E.; Noh, J.-Y.; Yuk, S.-S.; Erdene-Ochir, T.-O.; Hong, W.-T.; Jeong, J.-H.; Jeong, S.; Gwon, G.-B. Reassortant clade 2.3. 4.4 avian influenza A (H5N6) virus in a wild mandarin duck, South Korea, 2016. Emerg. Infect. Dis. 2017, 23, 822. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cui, P.; Zeng, X.; Jiang, Y.; Li, Y.; Yang, J.; Pan, Y.; Gao, X.; Zhao, C.; Wang, J. Characterization of avian influenza H5N3 reassortants isolated from migratory waterfowl and domestic ducks in China from 2015 to 2018. Transbound. Emerg. Dis. 2019, 66, 2605–2610. [Google Scholar] [CrossRef]
- Yeo, S.-J.; Than, D.-D.; Park, H.-S.; Sung, H.W.; Park, H. Molecular Characterization of a Novel Avian Influenza A (H2N9) Strain Isolated from Wild Duck in Korea in 2018. Viruses 2019, 11, 1046. [Google Scholar] [CrossRef]



| Virus Strain. | HA Receptor Binding Residues (H3 Numbering). | NA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage Sites | A138S | E190D | G225D | Q226L | G228S | 69–73 (QISNT) | 152 | 274 | 292 | |
| A/Mandarin duck/South Korea/KNU18-12/2018(H11N9) | PAIASR↓GLF | A | E | G | Q | G | No deletion | R | H | R |
| A/waterfowl/Korea/S353/2016 (H11N9) | PAIASR↓GLF | A | E | G | Q | G | No deletion | R | H | R |
| A/duck/Vietnam/OIE-2386/2009 (H11N9) | PAIASR↓GLF | A | E | G | Q | G | No deletion | R | H | R |
| A/goose/Zambia/09/2009 (H11N9) | PAIASR↓GLF | A | E | G | Q | G | No deletion | R | H | R |
| A/duck/Vietnam/LBM81/2012 (H11N9) | PAIASR↓GLF | A | E | G | Q | G | No deletion | R | H | R |
| A/duck/Kagoshima/KU57/2014 (H11N9) | PAIASR↓GLF | A | E | G | Q | G | No deletion | R | H | R |
| A/swine/KU/2/2001 (H11N6) | PAIASR↓GLF | A | E | G | Q | G | Deletion | R | H | R |
| Viral Protein. | Amino Acid | K/2018 a | K /2016 b | Za/2009 c | Vi/2012 d | Sh/2013 e | An/2013 f | Ka /2014 g | Comments | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | E627K | E | E | E | E | K | K | E | Mammalian host adaptation | [39,40,41,42,43] |
| D701N | D | D | D | D | D | D | D | Increase polymerase activity and viral replication in mammalian cells | [39,40,41,42,43] | |
| L89V | V | V | V | V | V | V | V | Enhanced polymerase activity, Increased virulence in mice | [44,45] | |
| G309D | D | D | D | D | D | D | D | Enhanced polymerase activity, Increased virulence in mice | [45] | |
| T339K | K | K | K | K | K | K | K | Enhanced polymerase activity, Increased virulence in mice | [45] | |
| PA | V100A | V | V | V | V | A | A | V | Contributed to the virulence and mammalian adaptation | [41,42,46] |
| K356R | K | K | K | K | R | R | K | Contributed to the virulence and mammalian adaptation | [47] | |
| S409N | S | S | S | S | N | N | S | Contributed to the virulence and mammalian adaptation | [41,46] | |
| A515T | T | T | T | T | T | T | T | Increased polymerase activity, Increased virulence in mammals and birds | [48,49] | |
| PB1 | H436Y | Y | Y | Y | Y | Y | Y | Y | Increased polymerase activity and virulence in mallards, ferrets and mice | [49] |
| NS1 | P/A42S | A | S | S | S | S | S | S | Increased virulence in mice (most avian influenza A viruses encode 42S) | [50,51] |
| T/D92E | D | D | D | D | D | D | D | Increased virulence in mammals, Escape of antiviral host response | [50,52,53,54] | |
| M1 | N30D | D | D | D | D | D | D | D | Increased virulence in mice (most influenza A viruses encode 30D) | [55,56] |
| T215A | A | A | A | A | A | A | A | Increased virulence in mice (most influenza A viruses encode 215A) | [55,56] | |
| M2 | L26P | L | L | L | L | L | L | L | Reduced susceptibility to amantadine and rimantadine | [57] |
| V27A/I | V | V | V | I | V | V | V | Reduced susceptibility to amantadine and rimantadine | [57] | |
| A30T | A | A | A | A | A | A | A | Reduced susceptibility to amantadine and rimantadine | [57] | |
| S31N | S | S | S | S | N | N | S | Reduced susceptibility to amantadine and rimantadine | [57,58,59] | |
| PB1-F2 | N66S | N | N | N | N | N | N | S | Increased virulence in mammals | [60] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuong, H.T.; Nguyen, N.M.; Sung, H.W.; Park, H.; Yeo, S.-J. Genetic Characterization of Avian Influenza A (H11N9) Virus Isolated from Mandarin Ducks in South Korea in 2018. Viruses 2020, 12, 203. https://doi.org/10.3390/v12020203
Tuong HT, Nguyen NM, Sung HW, Park H, Yeo S-J. Genetic Characterization of Avian Influenza A (H11N9) Virus Isolated from Mandarin Ducks in South Korea in 2018. Viruses. 2020; 12(2):203. https://doi.org/10.3390/v12020203
Chicago/Turabian StyleTuong, Hien Thi, Ngoc Minh Nguyen, Haan Woo Sung, Hyun Park, and Seon-Ju Yeo. 2020. "Genetic Characterization of Avian Influenza A (H11N9) Virus Isolated from Mandarin Ducks in South Korea in 2018" Viruses 12, no. 2: 203. https://doi.org/10.3390/v12020203
APA StyleTuong, H. T., Nguyen, N. M., Sung, H. W., Park, H., & Yeo, S.-J. (2020). Genetic Characterization of Avian Influenza A (H11N9) Virus Isolated from Mandarin Ducks in South Korea in 2018. Viruses, 12(2), 203. https://doi.org/10.3390/v12020203




