The Antimalarial Compound Atovaquone Inhibits Zika and Dengue Virus Infection by Blocking E Protein-Mediated Membrane Fusion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Reagents
2.2. Construction of Expression Vectors
2.3. DSP Assay in the 384-Well Format
2.4. Western Blot
2.5. Flow Cytometric Analysis
2.6. Viral Infection Assay
2.7. Trypan Blue Dye-Exclusion Assay
2.8. Statistical Analysis
3. Results
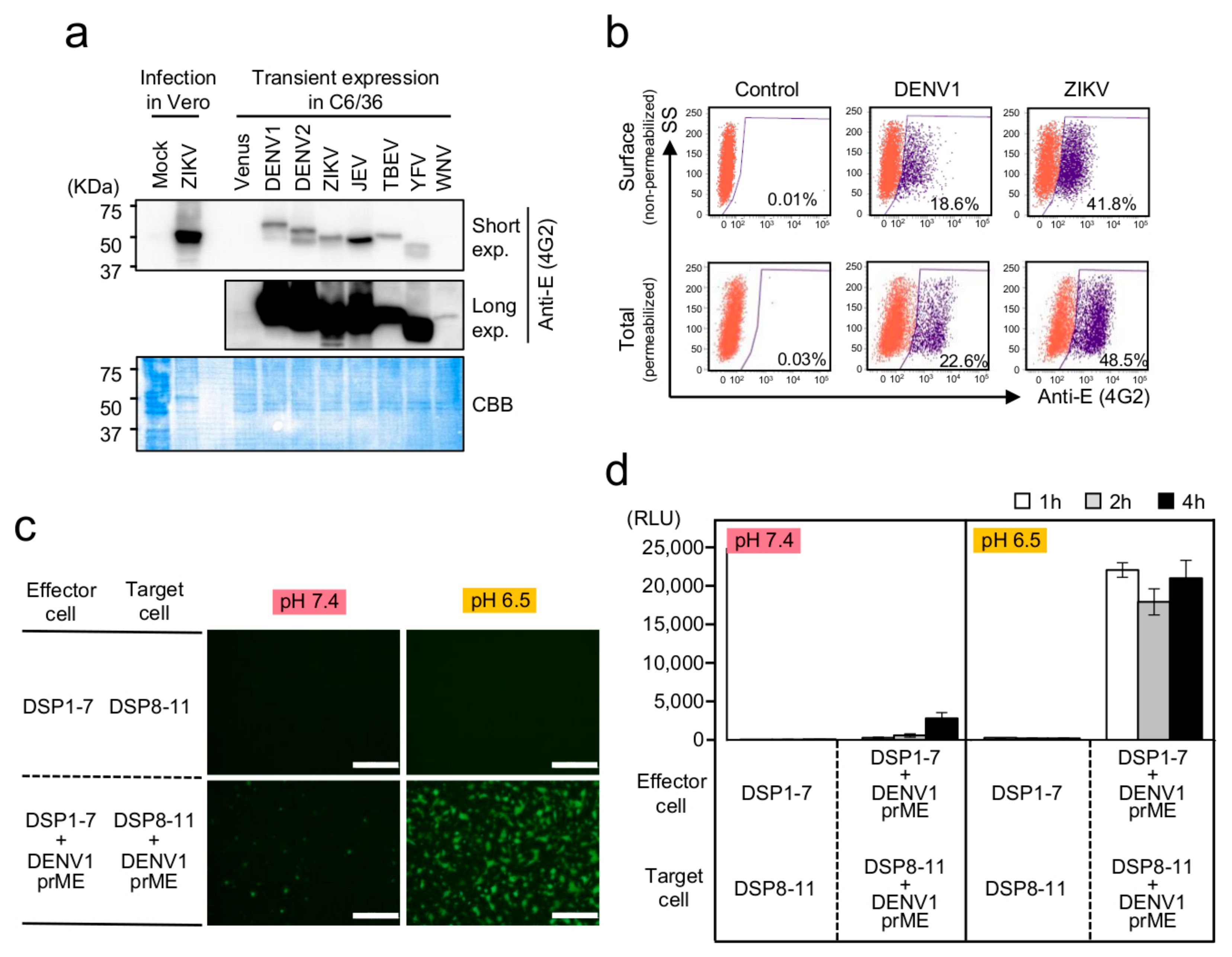
3.1. Establishment of a Membrane-Fusion Assay for Flavivirus E Proteins Based on the DSP Reporter in Mosquito-Derived C6/36 Cells
3.2. Optimization of the C6/36-Based DSP Assay for HTS of Membrane-Fusion Inhibitors
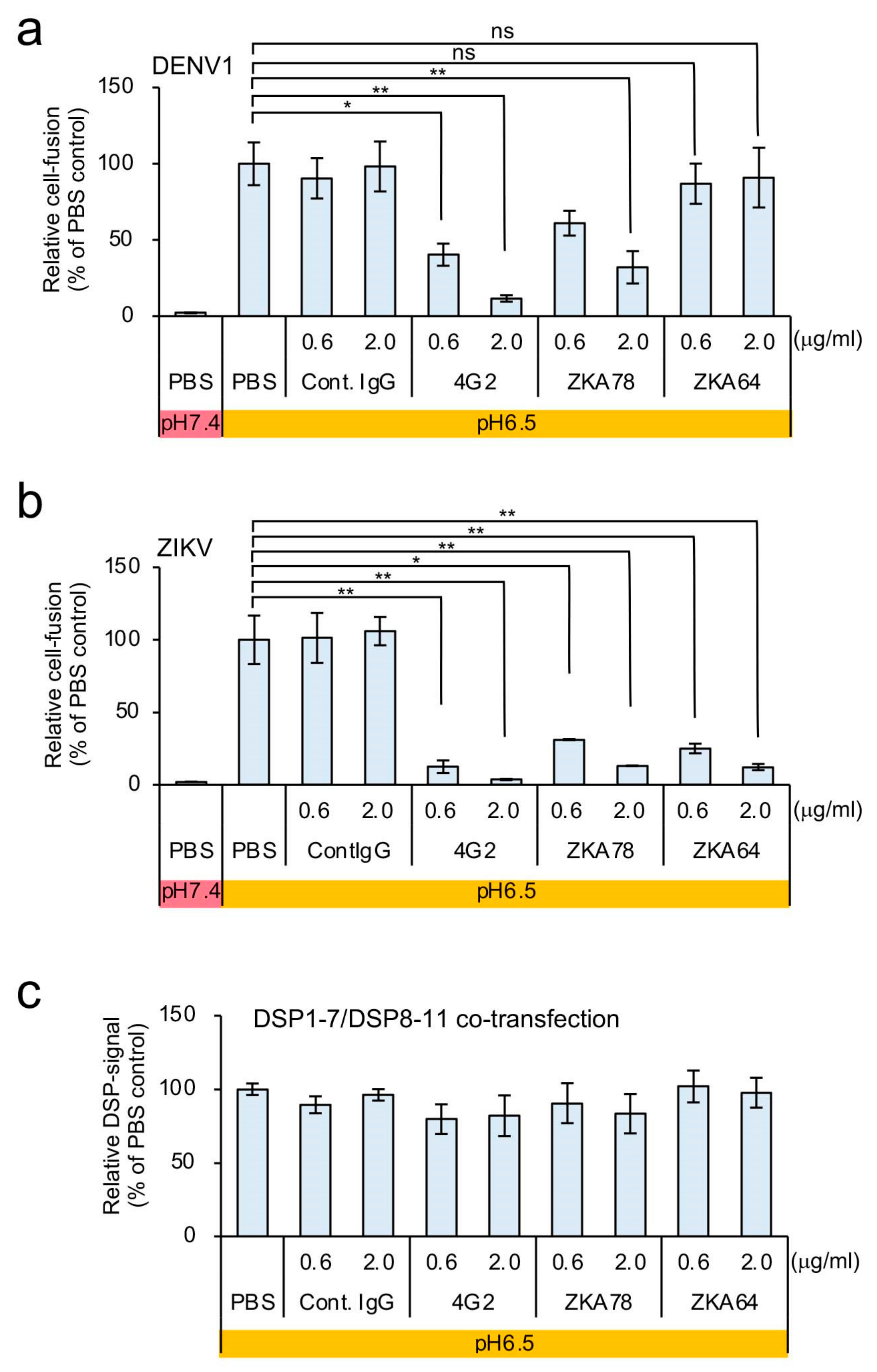
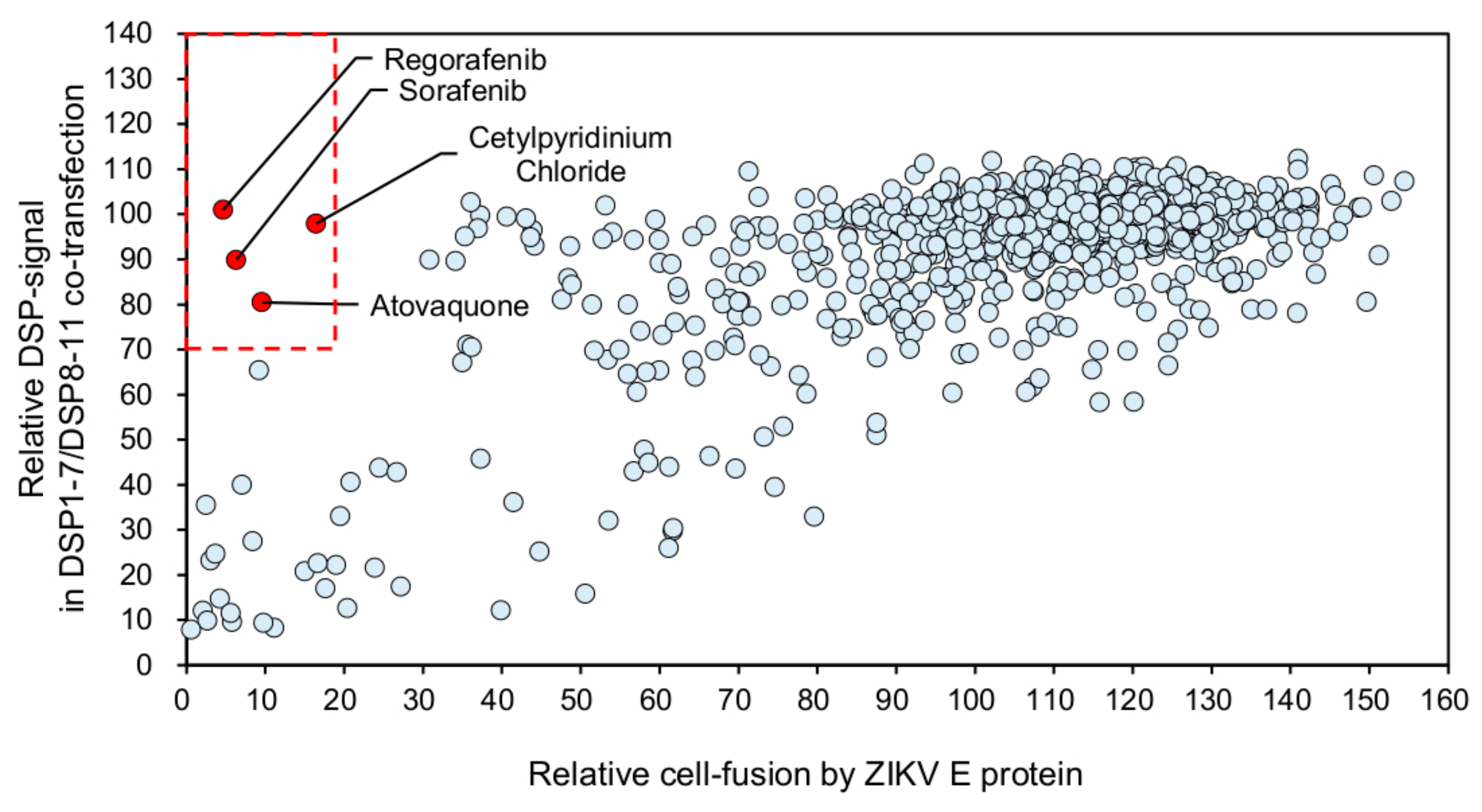
3.3. Identification of Atovaquone as a Potent Inhibitor of ZIKV E-Protein-Induced Membrane Fusion
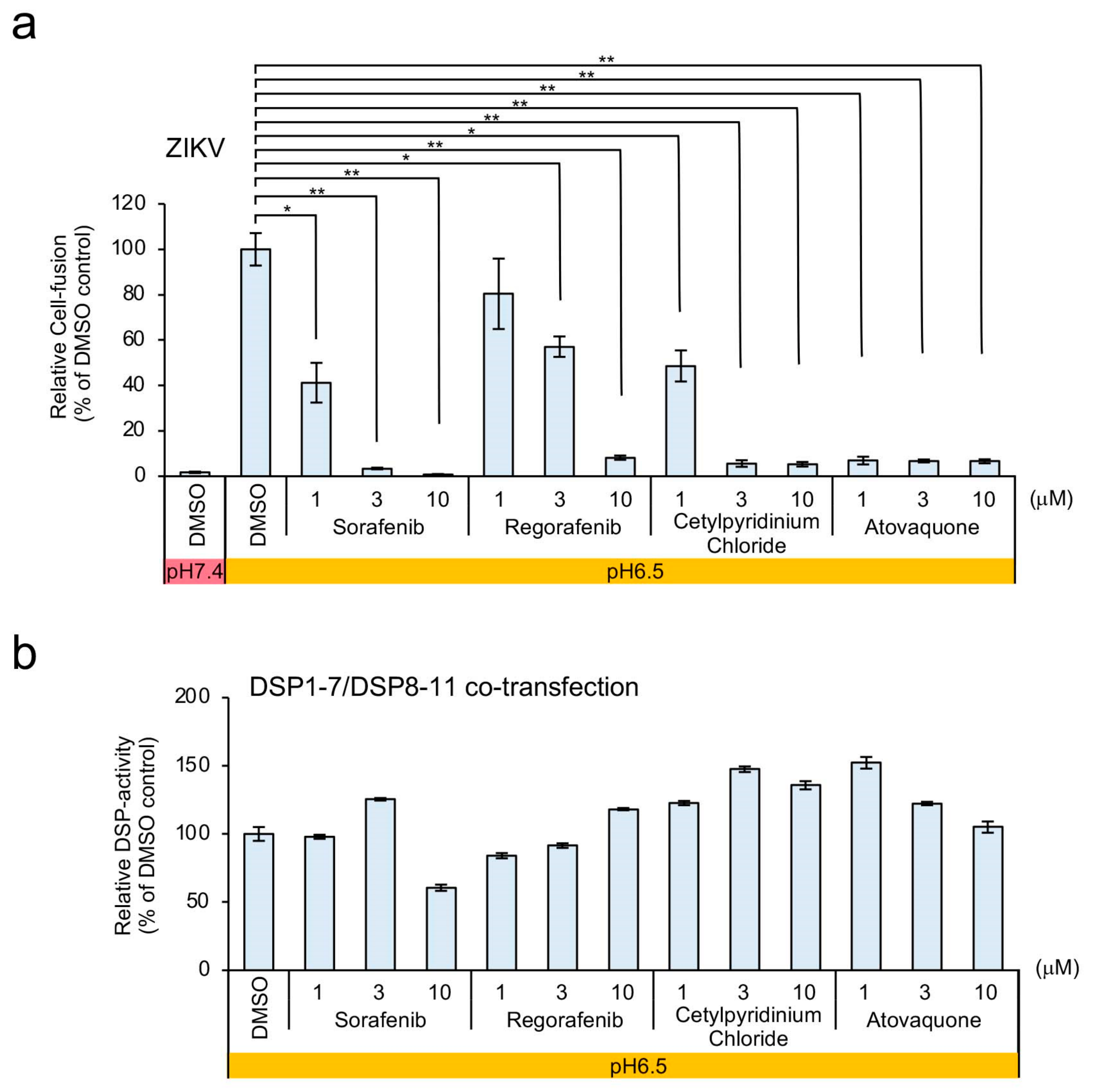
3.4. Efficient Inhibition of ZIKV and DENV1-4 Infection in Mosquito and Mammalian Cells by Atovaquone
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vigant, F.; Santos, N.C.; Lee, B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol. 2015, 13, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.C. Viral membrane fusion. Virology 2015, 479–480, 498–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melikyan, G.B. Common principles and intermediates of viral protein-mediated fusion: The HIV-1 paradigm. Retrovirology 2008, 5, 111. [Google Scholar] [CrossRef] [Green Version]
- Eckert, D.M.; Kim, P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001, 70, 777–810. [Google Scholar] [CrossRef] [Green Version]
- White, J.M.; Delos, S.E.; Brecher, M.; Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 189–219. [Google Scholar] [CrossRef] [Green Version]
- Hasan, S.S.; Sevvana, M.; Kuhn, R.J.; Rossmann, M.G. Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 2018, 25, 13–20. [Google Scholar] [CrossRef]
- Luo, H.; Wang, T. Recent advances in understanding West Nile virus host immunity and viral pathogenesis. F1000Res 2018, 7, 338. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.S.; Zhou, Y.; Takagi, T.; Kameoka, M.; Kawashita, N. Dengue Virus and Its Inhibitors: A Brief Review. Chem. Pharm. Bull. 2018, 66, 191–206. [Google Scholar] [CrossRef] [Green Version]
- Turtle, L.; Solomon, T. Japanese encephalitis—The prospects for new treatments. Nat. Rev. Neurol. 2018, 14, 298–313. [Google Scholar] [CrossRef]
- Yoshii, K. Tick-borne encephalitis. Uirusu 2017, 67, 143–150. [Google Scholar] [CrossRef]
- Epelboin, Y.; Talaga, S.; Epelboin, L.; Dusfour, I. Zika virus: An updated review of competent or naturally infected mosquitoes. PLoS Negl. Trop. Dis. 2017, 11, e0005933. [Google Scholar] [CrossRef] [Green Version]
- Heald-Sargent, T.; Muller, W. Zika Virus: A Review for Pediatricians. Pediatr. Ann. 2017, 46, e428–e432. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Nguyen, D.; Debs, L.H.; Patel, A.P.; Liu, G.; Jhaveri, V.M.; SI, S.K.; Mittal, J.; Bandstra, E.S.; Younis, R.T.; et al. Zika Virus: An Emerging Global Health Threat. Front. Cell Infect. Microbiol. 2017, 7, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, M.; Matsuyama, S.; Li, X.; Takeda, M.; Kawaguchi, Y.; Inoue, J.I.; Matsuda, Z. Identification of Nafamostat as a Potent Inhibitor of Middle East Respiratory Syndrome Coronavirus S Protein-Mediated Membrane Fusion Using the Split-Protein-Based Cell-Cell Fusion Assay. Antimicrob. Agents Chemother. 2016, 60, 6532–6539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, M.; Kiso, M.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; Imai, M.; Takeda, M.; Kinoshita, N.; Ohmagari, N.; Gohda, J.; Semba, K.; et al. The Anticoagulant Nafamostat Potently Inhibits SARS-CoV-2 S Protein-Mediated Fusion in a Cell Fusion Assay System and Viral Infection In Vitro in a Cell-Type-Dependent Manner. Viruses 2020, 12. [Google Scholar] [CrossRef]
- Ishikawa, H.; Meng, F.; Kondo, N.; Iwamoto, A.; Matsuda, Z. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng. Des. Sel. 2012, 25, 813–820. [Google Scholar] [CrossRef] [Green Version]
- Kondo, N.; Miyauchi, K.; Meng, F.; Iwamoto, A.; Matsuda, Z. Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J. Biol. Chem. 2010, 285, 14681–14688. [Google Scholar] [CrossRef] [Green Version]
- Kondo, N.; Miyauchi, K.; Matsuda, Z. Monitoring viral-mediated membrane fusion using fluorescent reporter methods. Curr. Protoc. Cell Biol. 2011. [Google Scholar] [CrossRef]
- Nakane, S.; Matsuda, Z. Dual Split Protein (DSP) Assay to Monitor Cell-Cell Membrane Fusion. Methods Mol. Biol. 2015, 1313, 229–236. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, R.; Shen, H.; Wang, M.; Yin, Z.; Cheng, A. Structures and Functions of the Envelope Glycoprotein in Flavivirus Infections. Viruses 2017, 9, 338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaitseva, E.; Yang, S.T.; Melikov, K.; Pourmal, S.; Chernomordik, L.V. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 2010, 6, e1001131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coil, D.A.; Miller, A.D. Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J. Virol. 2004, 78, 10920–10926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haile, L.G.; Flaherty, J.F. Atovaquone: A review. Ann. Pharmacother. 1993, 27, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Kain, K.C. Chemotherapy of drug-resistant malaria. Can. J. Infect. Dis. 1996, 7, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, M.; Abe, C.; Wakinaga, S.; Sakane, K.; Yumiketa, Y.; Taguchi, Y.; Matsumura, T.; Ishikawa, K.; Fujimoto, J.; Semba, K.; et al. TRAF6 maintains mammary stem cells and promotes pregnancy-induced mammary epithelial cell expansion. Commun. Biol. 2019, 2, 292. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.P.; Hsieh, S.C.; King, C.C.; Wang, W.K. Characterization of retrovirus-based reporter viruses pseudotyped with the precursor membrane and envelope glycoproteins of four serotypes of dengue viruses. Virology 2007, 368, 376–387. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, X.; Nakane, S.; Liu, S.; Ishikawa, H.; Iwamoto, A.; Matsuda, Z. Co-expression of foreign proteins tethered to HIV-1 envelope glycoprotein on the cell surface by introducing an intervening second membrane-spanning domain. PLoS ONE 2014, 9, e96790. [Google Scholar] [CrossRef]
- Hsieh, S.C.; Liu, I.J.; King, C.C.; Chang, G.J.; Wang, W.K. A strong endoplasmic reticulum retention signal in the stem-anchor region of envelope glycoprotein of dengue virus type 2 affects the production of virus-like particles. Virology 2008, 374, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igarashi, A. Isolation of a Singh's Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 1978, 40, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.A.; Gross, T.L.; Myles, K.M.; Adelman, Z.N. Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect. Mol. Biol. 2010, 19, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Henchal, E.A.; Gentry, M.K.; McCown, J.M.; Brandt, W.E. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 1982, 31, 830–836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes Kottkamp, A.; De Jesus, E.; Grande, R.; Brown, J.A.; Jacobs, A.R.; Lim, J.K.; Stapleford, K.A. Atovaquone Inhibits Arbovirus Replication through the Depletion of Intracellular Nucleotides. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randolph, V.B.; Stollar, V. Low pH-induced cell fusion in flavivirus-infected Aedes albopictus cell cultures. J. Gen. Virol. 1990, 71, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Komano, J.; Yokomaku, Y.; Sugiura, W.; Yamamoto, N.; Matsuda, Z. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J. Virol. 2005, 79, 4720–4729. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.S.; Patel, C.J. A standard database for drug repositioning. Sci. Data 2017, 4, 170029. [Google Scholar] [CrossRef] [Green Version]
- Kessl, J.J.; Hill, P.; Lange, B.B.; Meshnick, S.R.; Meunier, B.; Trumpower, B.L. Molecular basis for atovaquone resistance in Pneumocystis jirovecii modeled in the cytochrome bc(1) complex of Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 2817–2824. [Google Scholar] [CrossRef] [Green Version]
- Basco, L.K.; Ramiliarisoa, O.; Le Bras, J. In vitro activity of atovaquone against the African isolates and clones of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 1995, 53, 388–391. [Google Scholar] [CrossRef]
- Ino, H.; Takahashi, N.; Doi, Y.; Hashimoto, H.; Hirama, T. Pharmacokinetics of atovaquone suspension and atovaquone/proguanil combination tablet in healthy Japanese subjects. Jpn. J. Chemother. 2013, 61, 335–342. [Google Scholar]
- Salam, N.; Mustafa, S.; Hafiz, A.; Chaudhary, A.A.; Deeba, F.; Parveen, S. Global prevalence and distribution of coinfection of malaria, dengue and chikungunya: A systematic review. BMC Public Health 2018, 18, 710. [Google Scholar] [CrossRef] [PubMed]
- Cushion, M.T.; Collins, M.; Hazra, B.; Kaneshiro, E.S. Effects of atovaquone and diospyrin-based drugs on the cellular ATP of Pneumocystis carinii f. sp. carinii. Antimicrob. Agents Chemother. 2000, 44, 713–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, M.; Pudney, M. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4'-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80). Biochem. Pharmacol. 1992, 43, 1545–1553. [Google Scholar] [CrossRef]
- Knecht, W.; Löffler, M. Redoxal as a new lead structure for dihydroorotate dehydrogenase inhibitors: A kinetic study of the inhibition mechanism. FEBS Lett. 2000, 467, 27–30. [Google Scholar] [CrossRef] [Green Version]
- Ashton, T.M.; Fokas, E.; Kunz-Schughart, L.A.; Folkes, L.K.; Anbalagan, S.; Huether, M.; Kelly, C.J.; Pirovano, G.; Buffa, F.M.; Hammond, E.M.; et al. The anti-malarial atovaquone increases radiosensitivity by alleviating tumour hypoxia. Nat. Commun. 2016, 7, 12308. [Google Scholar] [CrossRef] [Green Version]
- Yeo, A.E.; Seymour, K.K.; Rieckmann, K.H.; Christopherson, R.I. Effects of dual combinations of antifolates with atovaquone or dapsone on nucleotide levels in Plasmodium falciparum. Biochem. Pharmacol. 1997, 53, 943–950. [Google Scholar] [CrossRef]
- Baggish, A.L.; Hill, D.R. Antiparasitic agent atovaquone. Antimicrob. Agents Chemother. 2002, 46, 1163–1173. [Google Scholar] [CrossRef] [Green Version]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Amara, A. Flavivirus entry receptors: An update. Viruses 2013, 6, 69–88. [Google Scholar] [CrossRef] [Green Version]
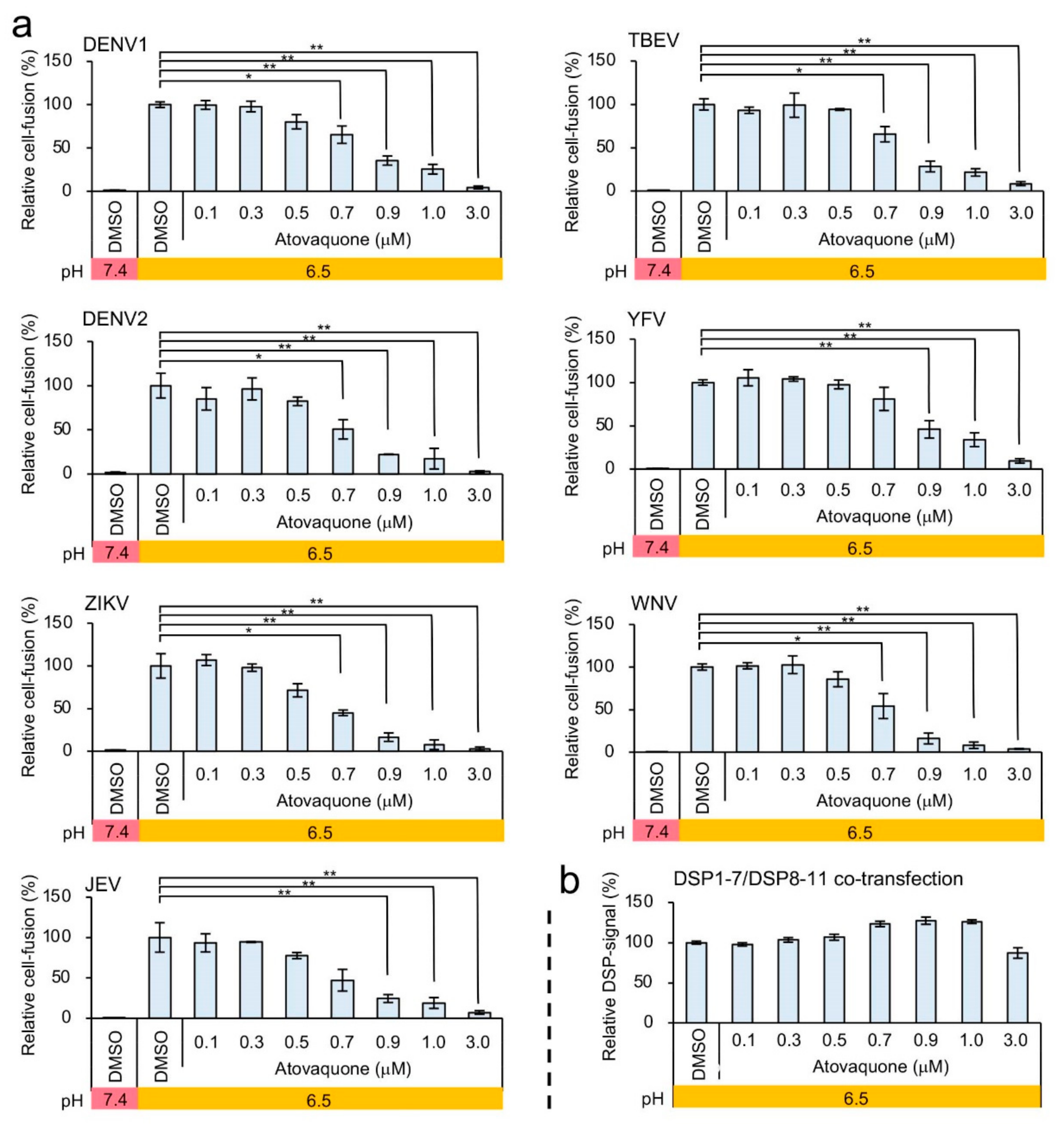
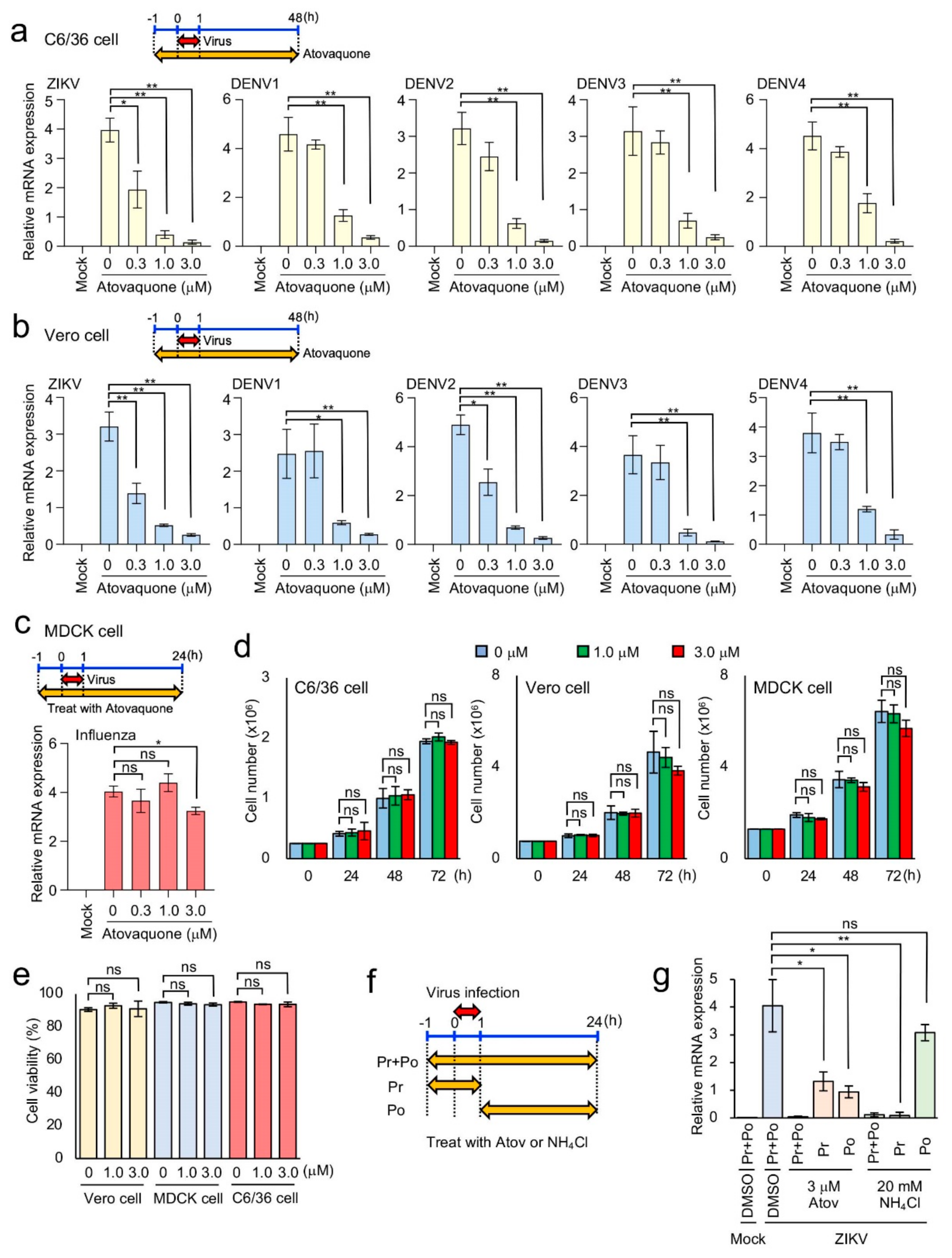
| Virus | Signal/Background Ratio | Z Factor |
|---|---|---|
| DENV1 | 136.2 | 0.76 |
| DENV2 | 15.4 | 0.58 |
| ZIKV | 35.2 | 0.82 |
| JEV | 390.5 | 0.75 |
| TBEV | 31.1 | 0.67 |
| YFV | 220.4 | 0.69 |
| WNV | 13.0 | 0.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, M.; Ichinohe, T.; Watanabe, A.; Kobayashi, A.; Zhang, R.; Song, J.; Kawaguchi, Y.; Matsuda, Z.; Inoue, J.-i. The Antimalarial Compound Atovaquone Inhibits Zika and Dengue Virus Infection by Blocking E Protein-Mediated Membrane Fusion. Viruses 2020, 12, 1475. https://doi.org/10.3390/v12121475
Yamamoto M, Ichinohe T, Watanabe A, Kobayashi A, Zhang R, Song J, Kawaguchi Y, Matsuda Z, Inoue J-i. The Antimalarial Compound Atovaquone Inhibits Zika and Dengue Virus Infection by Blocking E Protein-Mediated Membrane Fusion. Viruses. 2020; 12(12):1475. https://doi.org/10.3390/v12121475
Chicago/Turabian StyleYamamoto, Mizuki, Takeshi Ichinohe, Aya Watanabe, Ayako Kobayashi, Rui Zhang, Jiping Song, Yasushi Kawaguchi, Zene Matsuda, and Jun-ichiro Inoue. 2020. "The Antimalarial Compound Atovaquone Inhibits Zika and Dengue Virus Infection by Blocking E Protein-Mediated Membrane Fusion" Viruses 12, no. 12: 1475. https://doi.org/10.3390/v12121475
APA StyleYamamoto, M., Ichinohe, T., Watanabe, A., Kobayashi, A., Zhang, R., Song, J., Kawaguchi, Y., Matsuda, Z., & Inoue, J.-i. (2020). The Antimalarial Compound Atovaquone Inhibits Zika and Dengue Virus Infection by Blocking E Protein-Mediated Membrane Fusion. Viruses, 12(12), 1475. https://doi.org/10.3390/v12121475






