1. Introduction
Cattle populations are exposed to a wide variety of acute and chronic diseases caused by pathogens derived from many different members of the animal kingdom including viruses, bacteria, protozoa, Platyhelminthes, and nematodes. Some of these are zoonotic (e.g., Cryptosporidiosis) [
1]. Others (e.g., bovine viral diarrhea) reduce immunity, thus making cattle more susceptible to other diseases and increasing antibiotic usage [
2]. Many bovine diseases are endemic in the population where they cause financial losses through mortality, decreased outputs of milk and meat, reductions in fertility, premature culling, and costs of veterinary treatment and prevention strategies [
3]. The complexity, variety, and interaction of cattle pathogens have therefore made it difficult to fully understand their impact on the animals’ health and which effective measures to undertake to achieve eradication, or at least acceptable control, at both the herd and national levels. At present, bovine pathogen diagnostic procedures are based on a variety of methodologies including post mortems, antibiotic susceptibility testing, serological antibody profiles, targeted PCR, histopathology, and microbiological culture. These techniques may be time-consuming and expensive, and they may vary in their sensitivity and specificity. Furthermore, in many cases, each individual disease requires a separate test, and latent infection among susceptible animals may render serology alone inconclusive evidence of current disease.
For many diseases, however, distinctive transcripts of the pathogen RNA may be present in blood samples of the host [
4,
5]. Recent developments in RNA sequencing technology mean that it is now possible to perform highly sensitive whole transcriptome sequencing with the accurate detection of only a few transcript copies per cell [
6]. The RNA reads produced from next-generation sequencing are usually mapped against a reference genome using alignment algorithms [
7]. Following this process, a significant number of reads remain unmapped due, in part, to sequencing errors and sequence variants. These unmapped reads were previously considered as “junk.” Recent work has, however, shown that they also contain transcripts derived from, and specific to, a variety of pathogens [
8,
9,
10]. Using appropriate bioinformatic pipelines, it is therefore possible not only to look for evidence of particular microbes but also to discover novel viruses in circulation.
We used this approach to mine for viral transcripts in blood samples collected from six European dairy herds. This analysis revealed the widespread presence of bovine herpes virus 6 (BoHV-6). This belongs to the subfamily Gammaherpesvirinae within the genus
Macavirus, which includes viruses that infect lymphoid cells of both domestic and wild ruminants [
11,
12]. BoHV-6 was first isolated from the leukocytes of cattle with lymphosarcoma [
13] and was originally known as bovine lymphotropic herpesvirus [
11]. It has since been reported in dairy cow populations in a variety of countries including the USA [
11,
12], Canada [
14], New Zealand [
15], United Kingdom [
16], Belgium [
17], and Poland [
18]. Care in diagnosis is necessary, however, as BoHV-6 shares 65% sequence homology with BoHV-4 [
18], and infected cows were found to test positive in a commercial ELISA directed against BoHV-4 [
15].
There is much that remains unknown about this virus, including the mode of transmission and its relevance, if any, to clinical disease. The cows used in our study were part of GplusE, an FP7-Project funded by the European Union (
http://www.gpluse.eu) to investigate links between the phenotypes and genotypes of commercial dairy cows in order to improve their breeding and management. Extensive phenotype and blood transcriptome data from cows in early lactation were therefore already available. This enabled us to investigate the associations between the presence of BoHV-6, clinical disease, metabolic status, and gene expression in circulating leucocytes in relation to the viral load detected, as described in the present paper.
4. Discussion
Our results suggest that exploring whole blood transcriptomic unmapped reads from cows has huge and currently untapped potential to extract useful information regarding the presence of potential pathogens in domestic livestock. The RNA-Seq technology is highly sensitive and provides the precise measurement of levels of transcripts. It is now possible to detect many varied microbial sequences from a single sample, providing evidence that could be used for surveillance programs ranging from the individual herd to the national level.
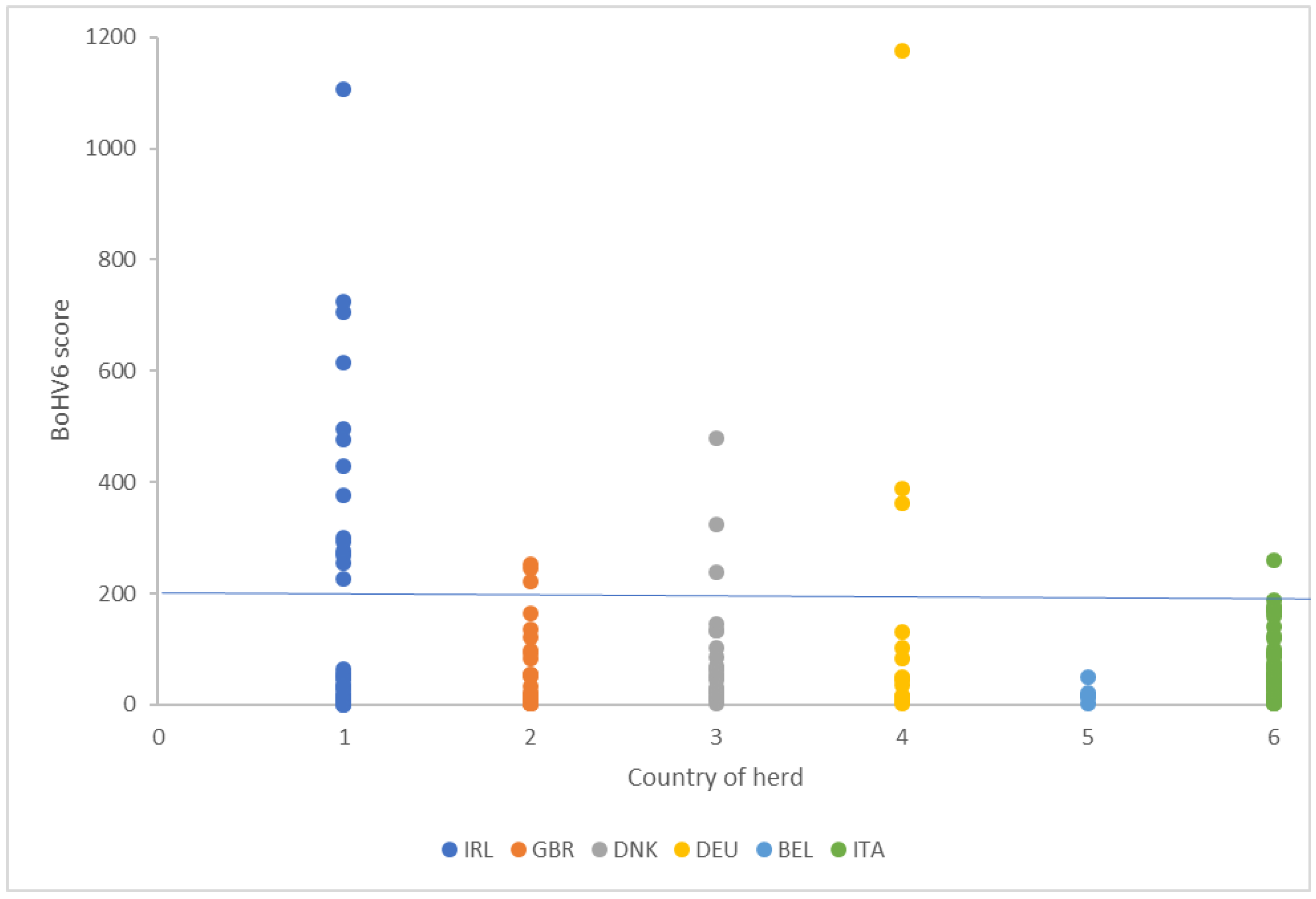
Within the six herds in our study, BoHV-6 was the most prevalent virus detected. The pathogen load is correlated with the PathSeq score, with a higher score providing more robust evidence that a taxon is indeed present based on the number of reads that aligned to the taxon reference. Walker et al. [
25] developed the PathSeq tool within their genome analysis toolkit (GATK) and previously reported read counts of 1984 and 9882 for the human papillomavirus HPV-16 in mRNA and WGS libraries, respectively, derived from a patient with cervical cancer. The type of tissue used and its storage, the technique used to isolate the RNA, and the depth and quality of sequencing could all influence viral detection. In our study, all of these parameters were kept consistent. Our analysis was based on RNA-Seq data originally intended for traditional gene expression profiling, so pathogen detection was not the primary project aim and the methodology was not optimized with this in mind. Nevertheless, the highest PathSeq score for BoHV-6 was 1175, a similar figure to that reported in the HPV-16 study [
25], and the reads gave repeatable values in pairs of samples taken from the same cow after a three week interval. For 16.4% of the cows in our study, the unaligned reads did not map to the BoHV-6 genome, although other pathogens were detectable in these samples at a low frequency. It is always difficult with this type of data to be completely confident at setting a value to discriminate animals that were definitely positive or negative. For an analysis of the phenotypic data, we therefore decided to omit the cows with no reads and to use a threshold of 200 to discriminate between basal levels and animals that had clearly elevated BoHV-6 scores (
Figure 1). The gene transcription data provided sound evidence that leukocytes from animals in the high and low score groups showed real differences in viral detection and response pathways, thus validating this analytical approach.
Using a cut-off of 1, 85.5% cows tested positive for BoHV-6 across the six analyzed herds, with an incidence of high score >200 cows of between 0% and 44% between herds. This was in accordance with previous assessments. Within the USA, BoHV-6-specific DNA sequences have been found in peripheral blood mononuclear cells from 91% of adult samples tested [
11] and 52–87% of cows in four dairies [
37], while in Europe, 64% of 92 Polish dairy cows tested positive [
18]. Rovnak et al. [
11] also reported that 38% (
n = 13) of two-week-old calves tested were positive for the presence of BoHV-6, suggesting that many animals become infected before or shortly after birth. Our study therefore supports previous data that this virus is indeed endemic in dairy cow populations worldwide.
The subfamily Gammaherpesvirinae preferentially infect lymphoid cells, where they establish latent infections and, for select viruses, cause malignant cell transformation. Reactivation can occur in immunosuppressed individuals. Within this subfamily, the genus
Macavirus currently includes around nine species that infect domestic and wild ruminants and swine. BoHV-6 is most similar to alcelaphine herpesvirus 1 (AlHV-1) and ovine herpesvirus 2 (OvHV-2), sharing an average of 50% amino acid identity [
12]. These two viruses cause asymptomatic infections in their reservoir hosts, which are wildebeest and sheep, respectively [
38]. They can, however, also infect cattle and other ruminants, thus causing malignant catarrhal fever. This is a severe lymphoproliferative disease with a high mortality rate in which the activation and proliferation of latently infected T lymphocytes causes the inflammation and necrosis of a variety of internal organs [
39,
40]. With respect to BoHV-6, the virus has been detected in lymphoma cells, leucocytes, and peripheral blood mononuclear cells (PBMCs) [
11,
18]. As far as the authors are aware, no one has yet determined if it has a preference for B- or T-lymphocytes or monocytes, although the in vitro experimental infection of a bovine B-lymphocyte cell line has been demonstrated [
11].
Though BoHV-6 was first isolated from the leukocytes of cattle with lymphosarcoma [
13], most of the studies referenced above considered that the virus was not associated with disease. On the other hand, BoHV-6 has been isolated from the vaginal discharge of cows with postpartum metritis [
15,
16,
17,
41], an aborted fetus [
14], and from buffaloes with lymphoproliferative disease [
42]. Its association with uterine disease does, however, need to be interpreted with caution because BoHV-6 tested positive in a commercial ELISA directed against BoHV-4 [
15]. BoHV-4 is cytopathic to endometrial epithelial and stromal cells [
43], and there is accumulating evidence that it can act as a co-factor with established uterine bacterial pathogens such as
Escherichia coli and
Arcanobacterium pyogenes to promote the development of endometritis [
44]. As cows infected with BoHV-4 appear more likely to have uterine disease after calving [
45,
46], there is a need to distinguish carefully which viruses are indeed present.
Our study provided the opportunity to examine associations between cows having a high BoHV-6 score with both milk yield and disease, as assessed over the first 50 days after calving. Over this time scale, there was no evidence that the virus had an adverse effect on milk production or quality. Indeed, LDH and NAGase, enzymes that act as inflammatory indicators of mastitis [
33,
47], were both lower in the milk of high BoVH-6 cows. This was in accordance with the slightly lower proportion of high BoHV-6 cows with a medium or high SCC reading compared with the low BoHV-6 cows (4/24 (16.7%) vs. 19/82 (23.2%), respectively). Metritis and endometritis are both extremely common in dairy cows, estimated to occur in around 40% and 20% of cows, respectively [
34]. This was confirmed by the results of this study in which 61% of the cows were diagnosed with either metritis or endometritis, but there was no evidence supporting any association between uterine disease and the presence of either BoHV-6 or BoHV-4. On the contrary, high BoHV-6 cows had proportionally fewer cases of both uterine disease and mastitis during the first 50 days of lactation. We show here, as discussed below, that there are changes in immunity associated with BoHV-6. While this is likely to have altered the immune response of cows to other infections, an increased infection rate seems more likely, so this may have been a chance finding.
Measurements of various blood and milk metabolites did, however, show significant differences between high and low BoHV-6 score cows. Our samples were collected at around 14 DIM, when cows experience major metabolic challenges as they transition into lactation [
48,
49]. This commonly causes a negative energy balance (NEB) [
50] that, in turn, is associated with raised circulating concentrations of NEFA and BHB and reductions in glucose and the metabolic hormone IGF-1 [
49,
51]. With respect to the measured metabolites, fructosamine concentrations were lower in the blood and glucose-6-phosphate was lower in milk in cows with high BoHV-6 scores. Circulating concentrations of fructosamine are thought to reflect the average plasma glucose concentration over the previous two-to-three weeks [
52] and have been used to identify disturbances in glucose metabolism in a variety of species including sheep and cattle [
53]. Within milk, glucose-6-phosphate reflects the nutrient availability and metabolic turnover in the mammary gland [
54]. BHB was higher in the milk in our study, and elevated milk BHB concentrations can been used as an indicator of ketosis [
32]. Urea concentrations were lower in both the blood and milk of high BoHV-6 cows, indicating a dietary protein deficiency. Uric acid was also lower in the milk, and this has been suggested as an indicator of microbial protein synthesis originating, in part, from ruminal digestion [
29]. Circulating IGF-1 was also significantly reduced, and this is strongly associated with a poor energy balance status [
55].
The metabolic profiles of the high BoHV-6 cows therefore suggested that even though they had a slightly higher BCS, they were more deficient in both energy and protein. There is mounting evidence that metabolic and disease status in early lactation are intimately linked. On the one hand, metabolic imbalance and a shortage of glucose reduce the ability of cows to mount a robust immune defense [
56,
57]. On the other hand, common infections such as
E. coli mastitis can impair hepatic function and thus influence metabolic activity [
58]. Herpes viruses acquired in young animals can remain latent after the initial infection and become reactivated at times of stress, mediated by increased glucocorticoid secretion [
59], as occurs in cows during calving and the periparturient period [
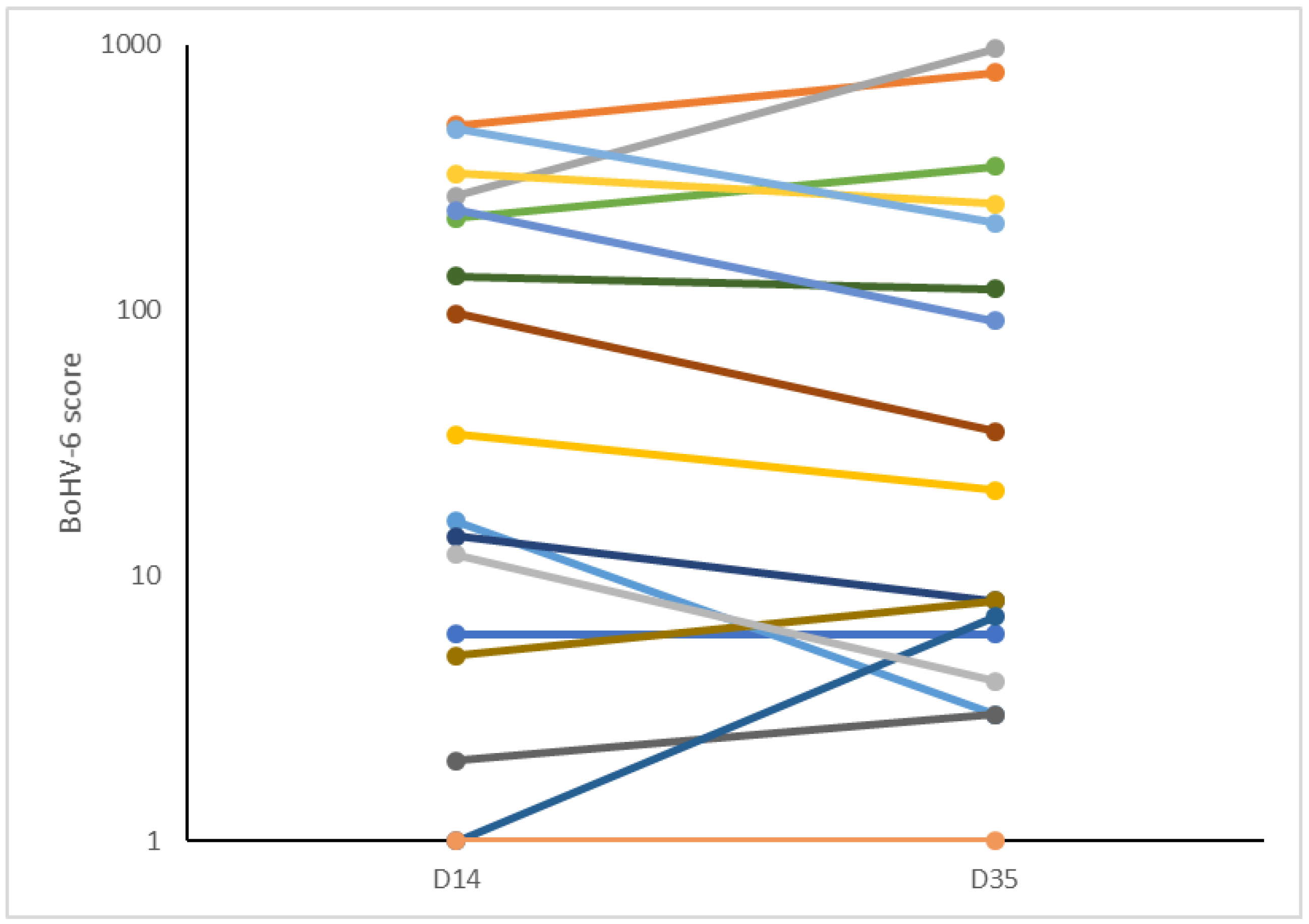
60]. In the present study, we found that the BoHV-6 scores remained stable between 14 and 35 DIM, but no pre-calving samples were taken for comparison. It is not therefore possible to determine from our data whether latent BoHV-6 infections were reactivated in cows with the worst metabolic status or whether being infected with BoHV-6 then caused the energy deficit to develop.
The leucocyte gene expression data provided clear evidence of differences in immune pathways between the high and low BoHV-6 cows. The most significant DEG was
AQP3. This encodes aquaporin-3, primarily known for its ability to transport water and non-ionic small solutes such as urea and glycerol across cell membranes. It can also facilitate the uptake of H
2O
2 into mammalian cells, thus potentially playing a role in host defense mechanisms [
61,
62]. Three other of the most significantly up-regulated genes in the high BoHV-6 cows are involved in protein folding within the endoplasmic reticulum (ER) (
TXNDC5,
PDIA4, and
XBP1). In times of high protein synthesis, the protein load may exceed the folding capacity of the ER, and misfolded proteins thus accumulate. The cellular stress causes an unfolded protein response (UPR) involving translational and transcriptional programming to help the cell return to homeostasis [
63]. Herpes viruses induce a burst of synthesis of envelope glycoproteins during lytic replication and they have evolved mechanisms to use the UPR to promote their own replication and to avoid immune surveillance [
64].
XBP1 encodes a transcription factor that regulates MHC class II genes. The IRE1-XBP1 pathway is central to the UPR and is also involved in inflammatory responses to infection. XBP1 may increase the expression of viral proteins by acting as the DNA binding partner of a viral transactivator.
Fibrinogen is an important acute phase protein that plays an essential role in coagulation, and its circulating concentration increases in various inflammatory conditions of cattle [
65]. The final secreted form is composed of two trimers, with each trimer made of polypeptide chains of fibrinogen alpha, beta, and gamma, encoded by
FGA,
FGB, and
FGG, respectively [
66]. All three of these genes were among the most up-regulated in leucocytes of the high BoHV-6 cows in terms of fold change increases. This was unexpected because the main source of secreted fibrinogen is liver hepatocytes, with little previous evidence supporting a leucocyte source. Genes encoding albumin, transferrin, and apolipoprotein A-II, major plasma proteins that are also primarily produced by the liver, were also up-regulated. Circulating albumin concentrations usually decline in the immediate postpartum period, particularly in cows with evidence of more severe inflammation [
67]. Transferrin is the main blood transport protein for iron, and its receptor TFR1 is commonly used as a viral entry point into the cell [
68]. Apolipoprotein A-II is the second most abundant protein contributing to high density lipoprotein particles, which are important for cholesterol transport. The leucocyte production of these various transport proteins is unlikely to be a major determinant of their circulating concentrations, so the relevance of these changes in gene expression to cow physiology remains to be determined.
Of particular interest were the DEGs encoding proteins known to be involved in viral detection and cellular responses to viral infection, which were mainly down-regulated in the high BoHV-6 cows. These included
DDX58, which encodes a key cytoplasmic receptor that detects both positive and negative strand RNA viruses and activates a downstream signaling cascade that leads to the production of type I interferons and pro-inflammatory cytokines. This ties in with the lower expression of genes encoding both interferon response genes (
IFIT2,
IFIT3, IFITM3, IFITM5, LOC112441484, LOC618409, and
OAS1Z) and cytokines (
IL1A, IL1B, IL15, and
IL17D).
IFIT3 (also called retinoic acid-induced gene G protein—RIGG) encodes an IFN-inducible protein that can form a cytoplasmic complex to recognize and destroy viral RNA, acting as an inhibitor of viral replication [
69]. Some viruses, e.g., hepatitis C virus, can escape the antiviral functions of the interferon-induced protein with tetratricopeptide repeats (IFIT) family by suppressing the expression of the IFIT family of genes [
70].
Interferons mediate a number of antiviral responses that include an ISG15 ubiquitin-like pathway and an OAS1-RNaseL pathway [
70,
71]. Within the present study, a number of genes encoding proteins with a potential involvement on the ubiquitin-like pathway were down-regulated in the high BoHV-6 cows. Of these,
HECW2, HERC1, HERC5, LNX1, LOC529930, MKRN3, RNF149, and
TRIM17 all encode E3 ubiquitin ligases, key components of ISGylation. This involves the ubiquitin-like modification of proteins that may then be targeted to lysosomes for destruction or used elsewhere. During this process, the C-terminus of ISG15 is conjugated to lysine residues in the target protein following consecutive catalysis with three enzymes E1, E2, and E3 [
72]. HERC5 is recognized as a major E3 ligase that is relatively non-specific, so it is able to block the IFN-mediated rise in the total level of ISGylated cellular proteins.
SKP1 encodes S-phase kinase-associated protein 1. This forms an essential component of the SCF (SKP1-CUL1-F-box protein) ubiquitin ligase complex.
UBA7 encodes ubiquitin-like modifier-activating enzyme 7. Both HERC5 and UBA7 have recognized roles in catalyzing the ISGylation of influenza A virus NS1 protein, which attenuates its virulence [
73]. The OAS1 gene family (including
OAS1Z) senses exogenous nucleic acid.
OAS1Z encodes a protein that synthesizes 2′,5′-oligoadenylates, which regulate the early stage of viral infection by activating latent RNase L, an endoribonuclease, resulting in viral RNA degradation and the inhibition of viral replication. Polymorphisms in this gene have been associated with susceptibility to viral infections [
74]. NLRP12 acts as an anti-inflammatory agent by inhibiting both canonical and non-canonical NF-kappa-B and ERK activation pathways [
75]. It also functions as a checkpoint for anti-viral DDX58 activation [
76].
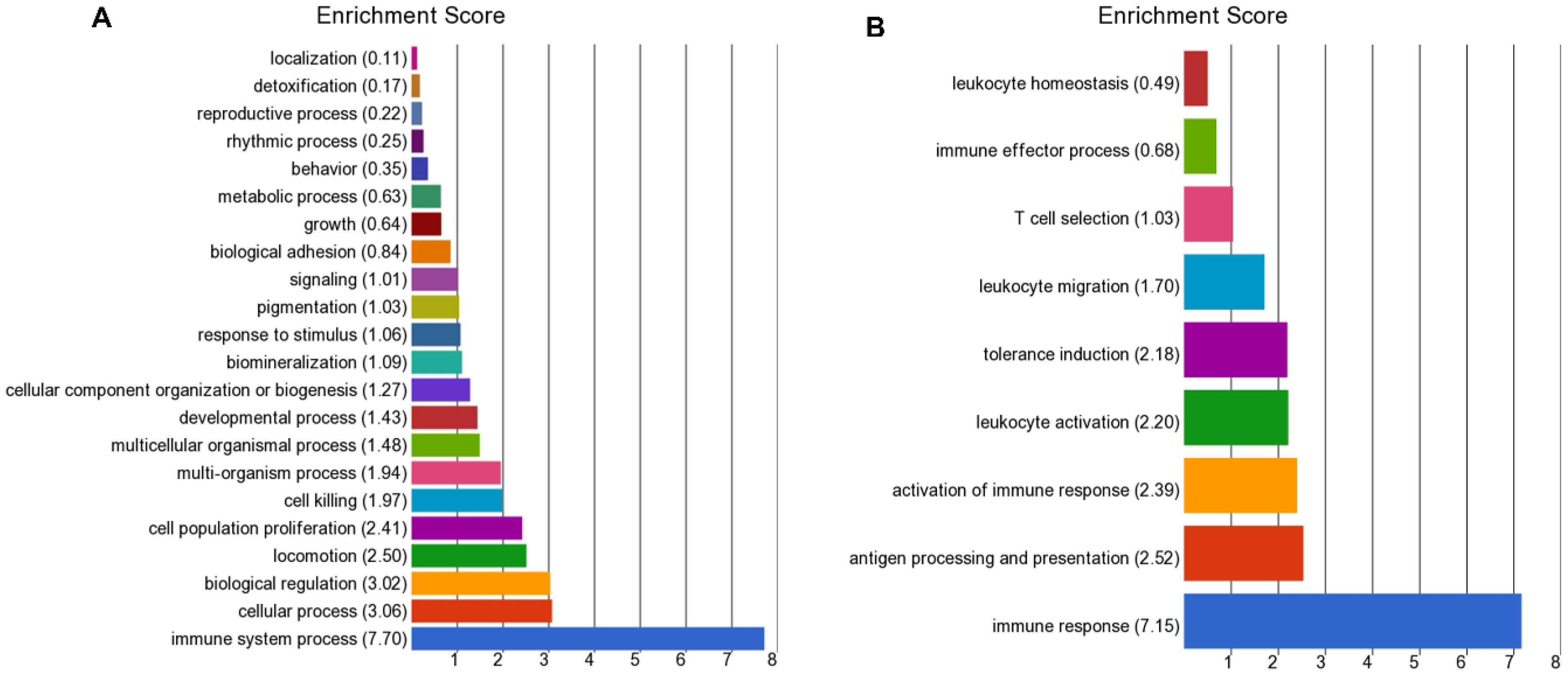
Other immune pathways were mainly down-regulated in the high BoHV-6 cows and included antigen processing, presentation (MHC molecules), and the activation of immune response and leukocyte activation. The latter involved genes encoding proteins involved in the detection of pathogen-associated molecular patterns (PAMPs) and TLR signaling (
ACOD1, ALPK1, LYST, TARM1, and
TIRAP), as well as several chemokines and chemokine receptors
(CCRL2, CXCL5, CXCR1, CXCR2, and
LOC504773). SEMA7A, another gene of interest, encodes semaphorin7A, a secreted glycoprotein that acts as an important immune regulator. Pox viruses and Gammaherpesviruses of the
Macavirus genus including AlHV-1 encode semaphorin 7A mimics. Viral semaphorins have been suggested to mediate immune evasion mechanisms and alter host T-cell and dendritic cell function [
77].
Viral response genes that were up-regulated in high BoHV-6 cows included
XBP1 (described above),
ADA, and
IRF4.
ADA encodes a DNA exonuclease. This gene is better characterized in humans, which have three adenosine deaminases, termed ADAR1-3, although ADAR3 lacks enzymatic activity [
78]. ADAR1 appears to mainly function as a pro-viral factor by suppressing dsRNA-dependent antiviral molecules including MDA5, PKR, and OASes/RNase L [
78]. It can, however, also display antiviral activities against a subset of viruses including paramyxoviruses and orthomyxoviruses by acting as a restriction factor to prevent their replication [
79].
IRF4 encodes interferon regulatory factor 4. This is a lymphocyte-specific transcription factor that is implicated in a number of hematological malignancies including B-cell and Hodgkin lymphoma and is highly expressed in transformed cells infected with Epstein–Barr virus, in which it promotes their proliferation [
80].