Bovine Adenovirus-3 Tropism for Bovine Leukocyte Sub-Populations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Virus
2.2. Culture Media and Reagents
2.3. Monoclonal Antibodies
2.4. BAdV-3 Protein-Specific Antibodies
2.5. Plasmid Construction
2.5.1. Plasmid pBAVNdA-10GS-CATRGD
2.5.2. Plasmid pUC304a-CATRGD
2.6. Isolation of Recombinant BAdV-3
2.7. Isolation of Bovine Blood Leukocytes
2.7.1. Isolation of Bovine Peripheral Blood Mononuclear Cells (PBMCs)
2.7.2. Bovine Polymorphonuclear (PMN) Cell Isolation
2.7.3. Bovine Monocyte Isolation
2.7.4. Bovine Dendritic Cell Isolation
2.8. Transduction of Bovine Leukocytes by BAV304a
2.9. Transduction of Bovine PBMC Subpopulations by BAV304a
2.10. Western Blot Analysis
2.11. Virus Titer Assay
2.12. qRT-PCR
2.13. qPCR Assay for Viral Quantification
2.14. Data Analysis
3. Results
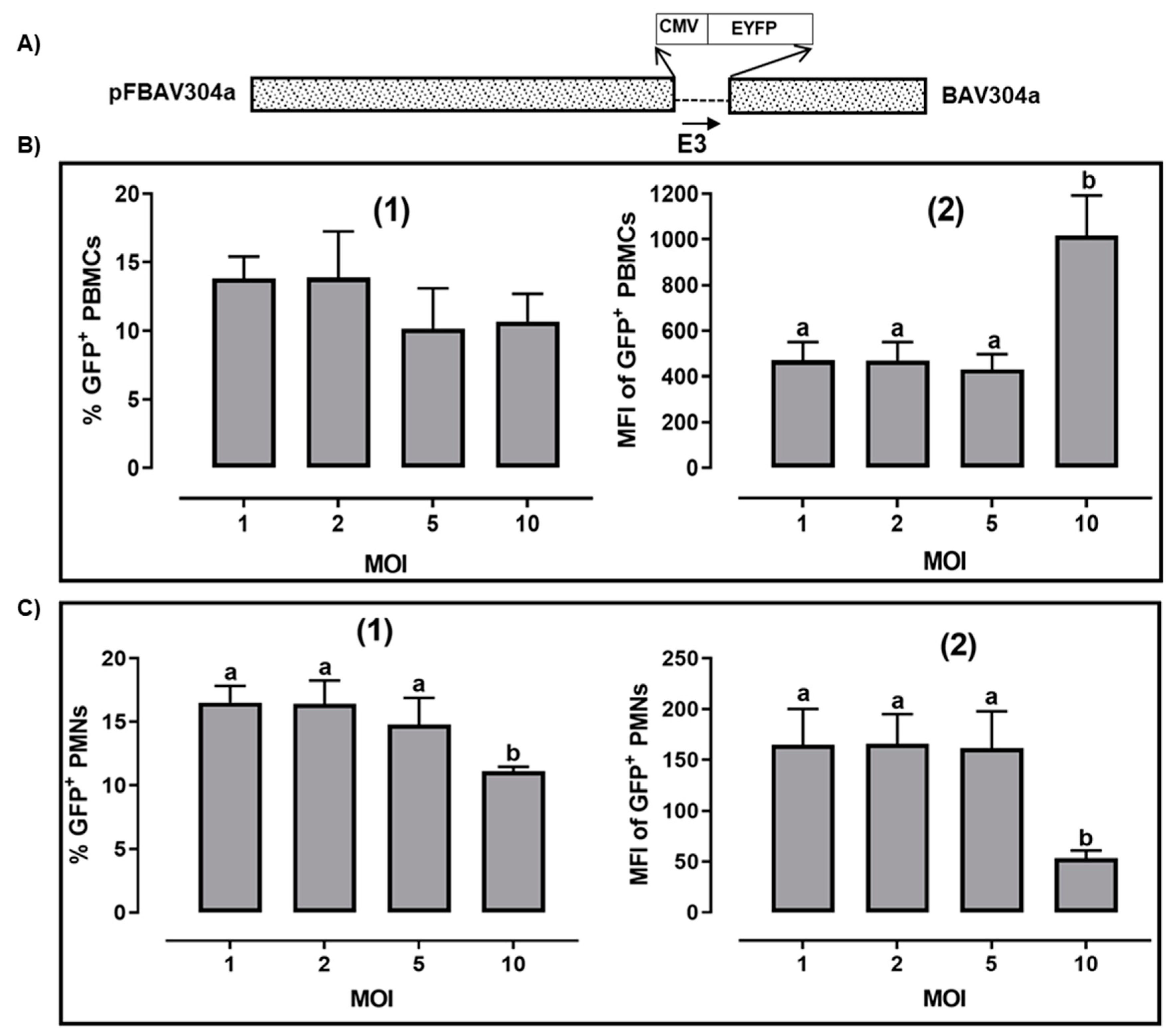
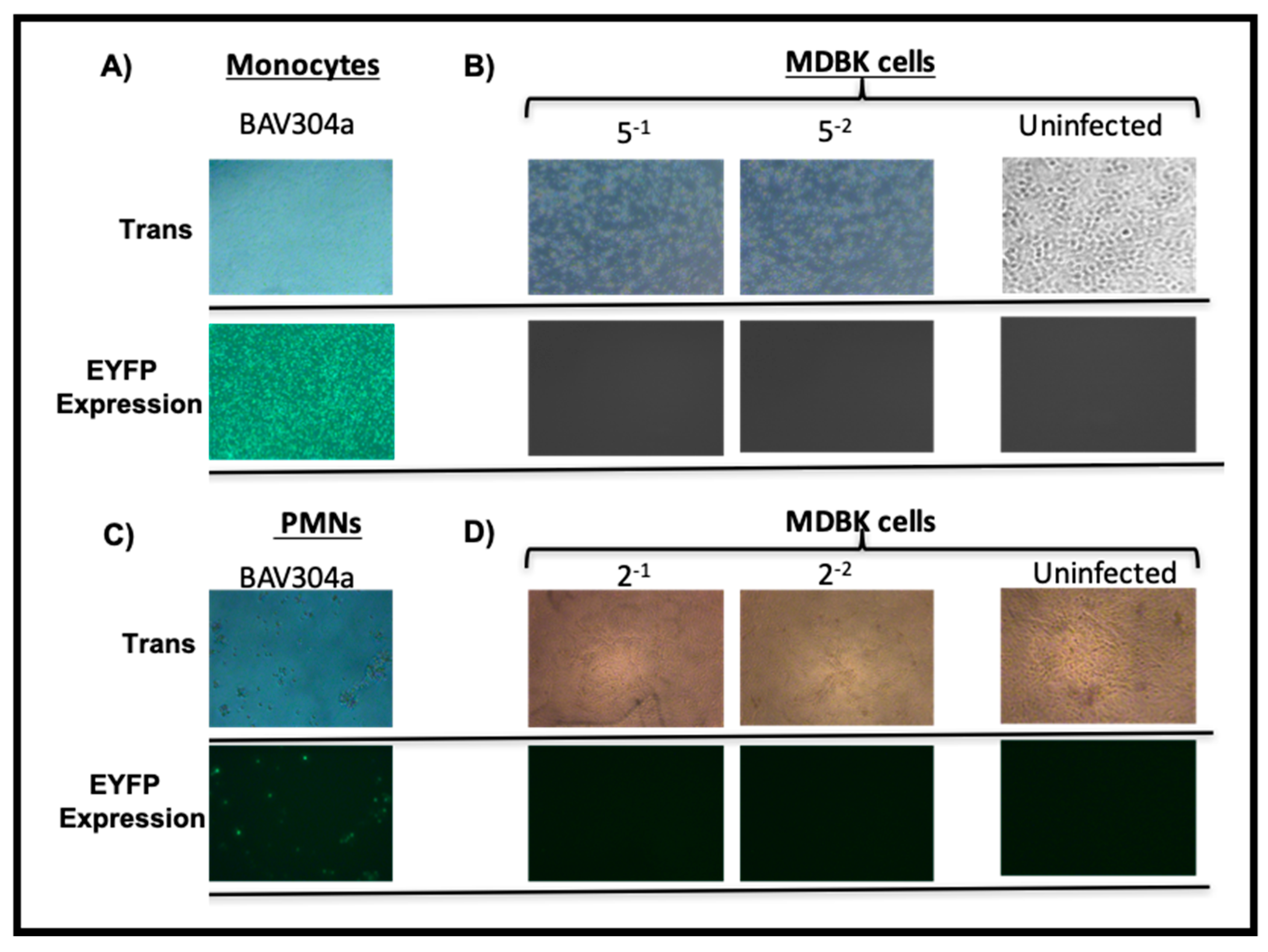
3.1. Transduction of Bovine PBMCs by BAV304a
3.2. Transduction of Bovine PMN Cells by BAV304a
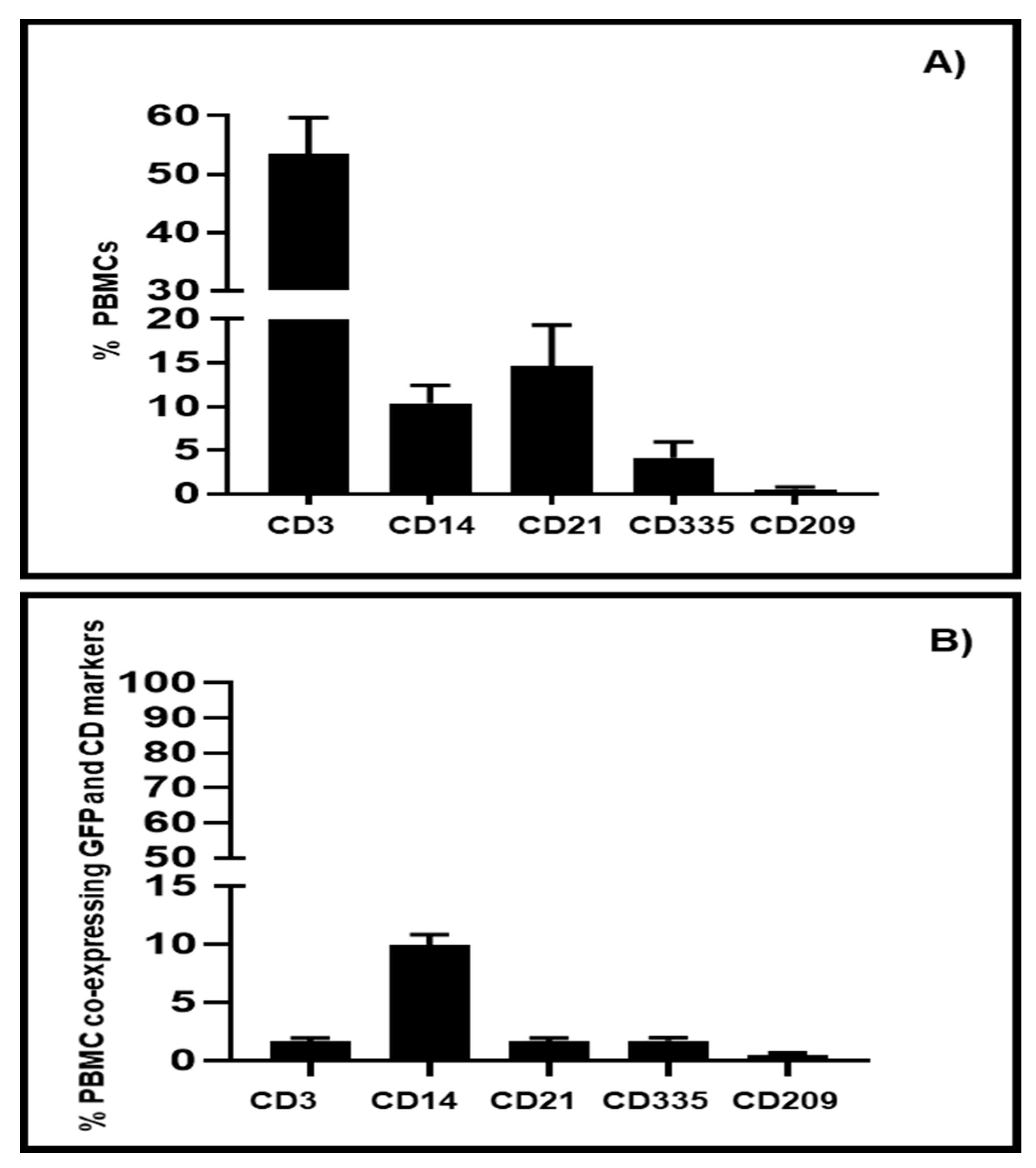
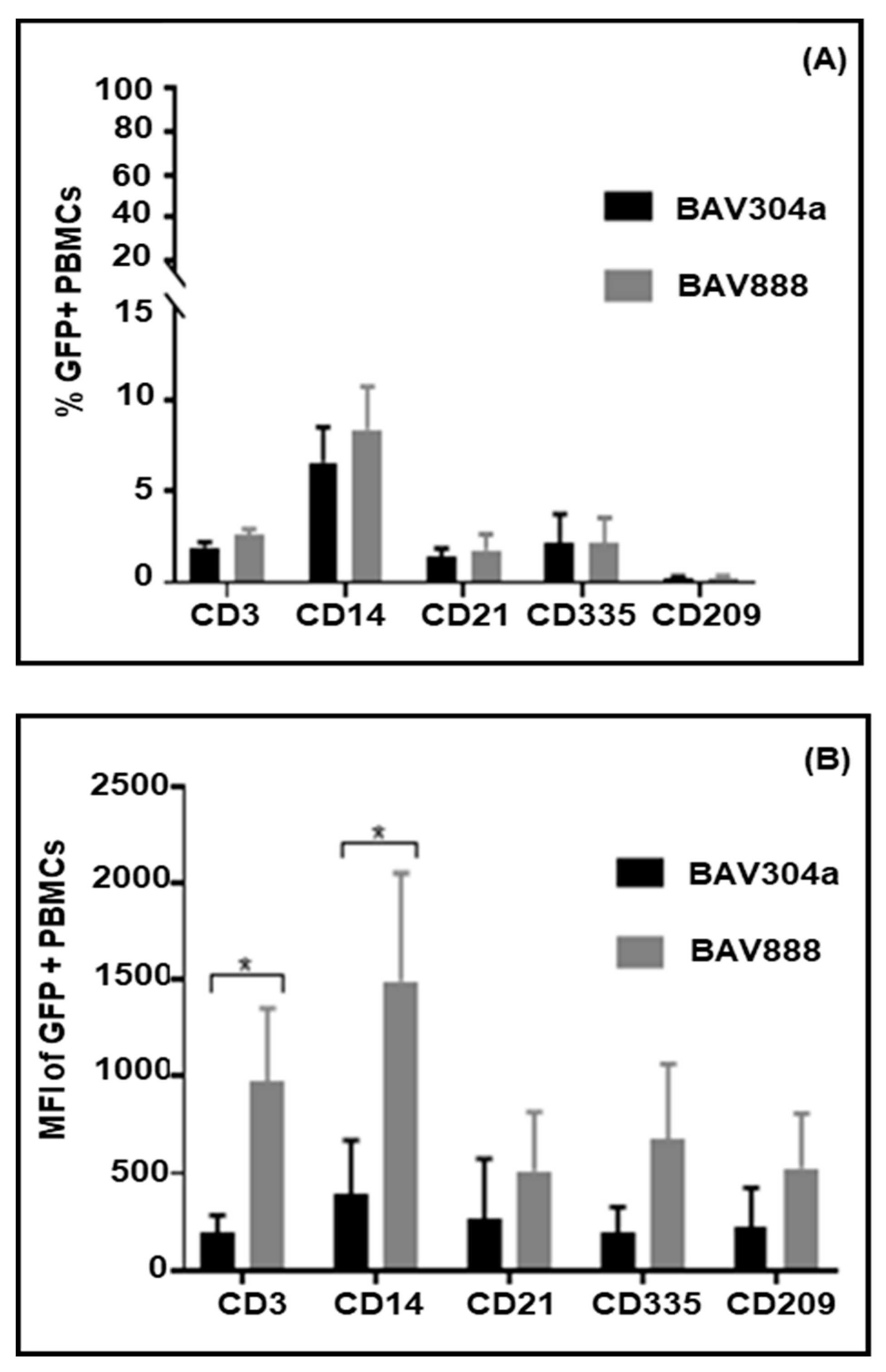
3.3. Transduction of PBMC Subpopulations by BAV304a
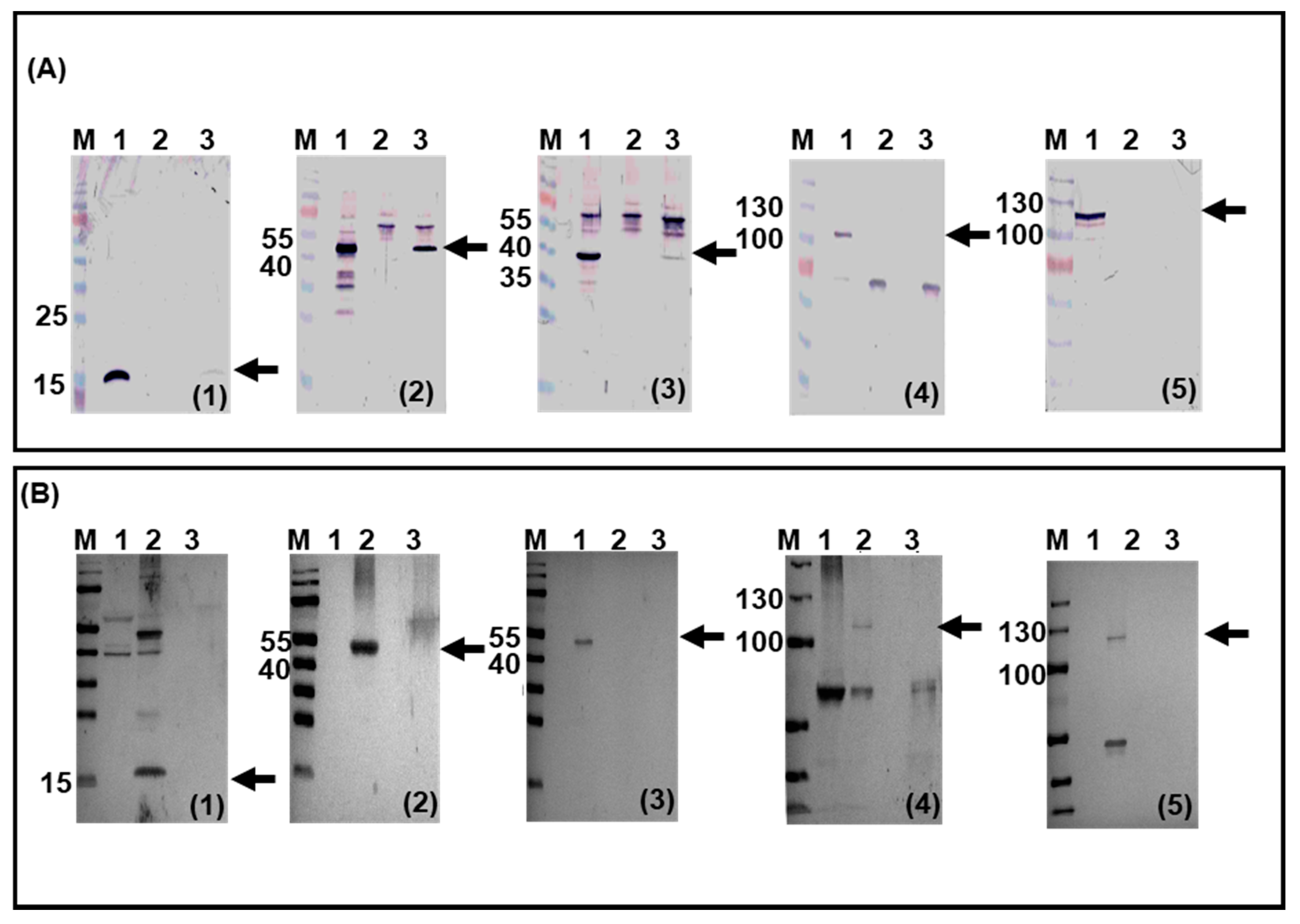
3.4. Viral Replication in Transduced Bovine Monocytes and PMNs Cells
3.5. BAdV-3 Viral Gene Expression in Transduced Bovine Monocytes and PMN Cells
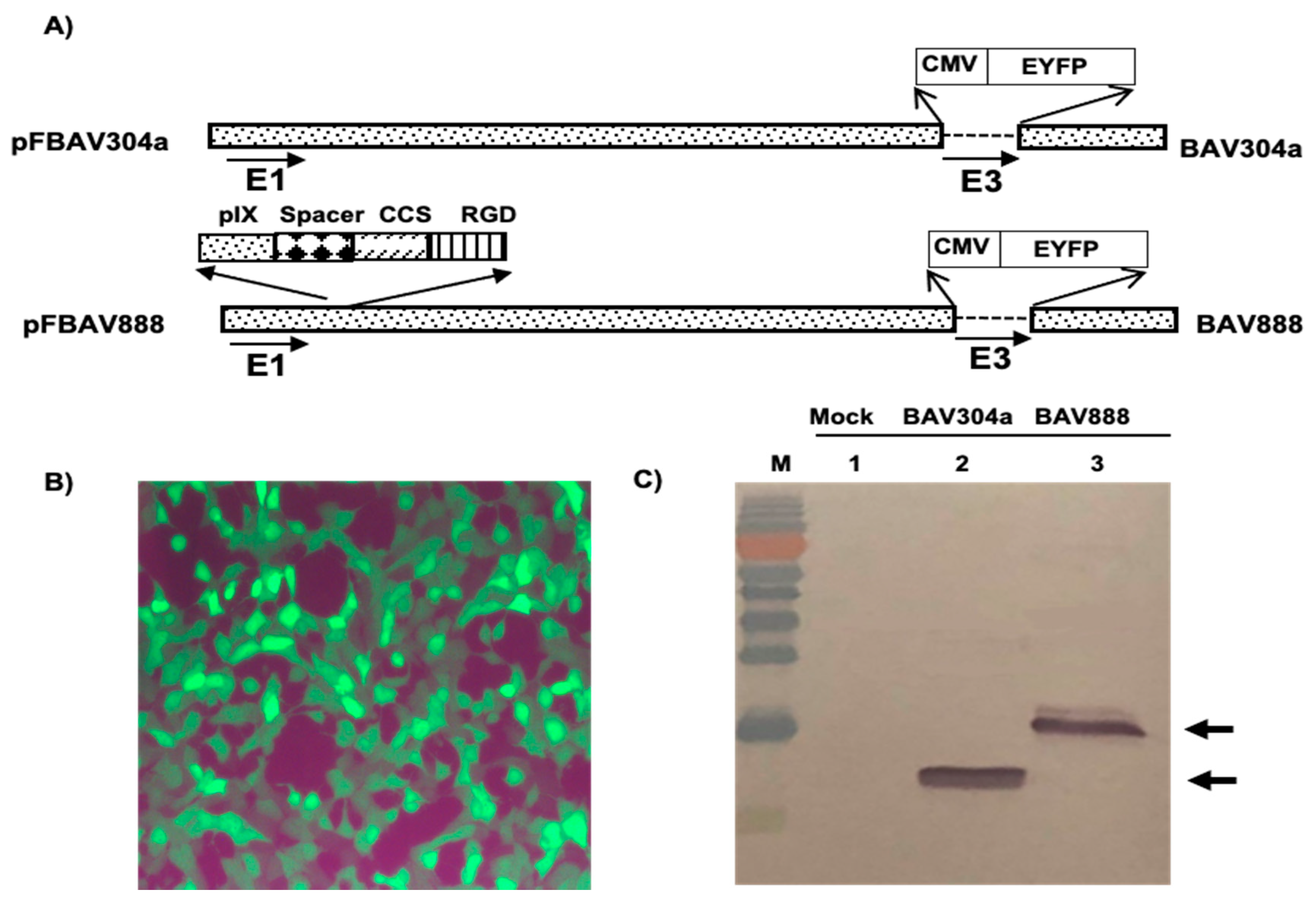
3.6. Isolation of Recombinant BAV888
3.7. Transduction of PBMC Subpopulations by BAV888
3.8. Viral Genome and Transgene Expression in Transduced Bovine Monocytes (CD14+)
3.9. Viral Gene Transcription in Transduced Monocytes (CD14+)
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tatsis, N.; Ertl, H.C. Adenoviruses as vaccine vectors. Mol. Ther. 2004, 10, 616–629. [Google Scholar] [CrossRef]
- Crystal, R.G. Transfer of genes to humans: Early lessons and obstacles to success. Science 1995, 270, 404–410. [Google Scholar] [CrossRef]
- Ayalew, L.E.; Pankaj Kumar, P.; Gaba, A.; Makadiya, N.; Tikoo, S.K. Bovine adenovirus-3 as a vaccine delivery vehicle. Vaccine 2015, 33, 493–499. [Google Scholar] [CrossRef]
- Lehmkuhl, H.D.; Smith, M.H.; Dierks, R.E. A bovine adenovirus type 3: Isolation, characterization and experimental infection in calves. Arch. Virol. 1975, 48, 39–46. [Google Scholar] [CrossRef]
- Baxi, M.K.; Deregt, D.; Robertson, J.; Babiuk, L.A.; Schlapp, T.; Tikoo, S.K. Recombinant bovine adenovirus type 3 expressing bovine viral diarrhea virus glycoprotein E2 induces an immune response in cotton rats. Virology 2000, 278, 234–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brownlie, R.; Kumar, P.; Babiuk, L.A.; Tikoo, S.K. Recombinant bovine adenovirus-3 co-expressing bovine respiratory syncytial virus glycoprotein G and truncated glycoprotein gD of bovine herpesvirus-1 induce immune responses in cotton rats. Mol. Biotechnol. 2015, 57, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.S.; Idamakanti, N.; Chen, Y.; Whale, T.; Babiuk, L.A.; Mehtali, M.; Tikoo, S.K. Replication-Defective Bovine Adenovirus Type 3 as an Expression Vector. J. Virol. 1999, 73, 9137–9144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakhartchouk, A.N.; Reddy, P.S.; Baxi, M.; Baca-Estrada, M.E.; Mehtali, M.; Babiuk, L.A.; Tikoo, S.K. Construction and characterization of E3-deleted bovine adenovirus type 3 expressing full-length and truncated form of bovine herpesvirus type 1 glycoprotein gD. Virology 1998, 250, 220–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakhartchouk, A.N.; Pyne, C.; Mutwiri, G.K.; Papp, Z.; Baca-Estrada, M.E.; Griebel, P.; Babiuk, L.A.; Tikoo, S.K. Mucosal Immunization of calves with recombinant bovine adenovirus-3 induction of protective immunity to bovine herpesvirus-1. J. Gen. Virol. 1999, 80, 1263–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, P.S.; Idamakanti, N.; Pyne, C.; Zakhartchouk, A.N.; Godson, D.L.; Papp, Z.; Baca-Estrada, M.E.; Babiuk, L.A.; Mutwiri, G.K.; Tikoo, S.K. The immunogenicity and efficacy of replication-defective and replication-competent bovine adenovirus-3 expressing bovine herpes-1 glycoprotein gD in cattle. Vet. Immunol. Immunopathol. 2000, 76, 257–268. [Google Scholar] [CrossRef]
- Du, E.; Tikoo, S.K. Efficient Replication and Generation of Recombinant Bovine Adenovirus-3 in Non-bovine Cotton Rat Lung Cells Expressing I-SceI Endonuclease. J. Gene Med. 2010, 12, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Tollefson, A.E.; Kuppuswamy, M.; Shashkova, E.V.; Doronin, K.; Wold, W.S. Preparation and titration of CsCl-banded adenovirus stocks. In Adenovirus Methods and Protocols; Humana Press: Totowa, NJ, USA, 2007; Volume 1, pp. 223–235. [Google Scholar]
- Kulshreshtha, V.; Babiuk, L.A.; Tikoo, S.K. Role of bovine adenovirus-3 33 K protein in viral replication. Virology 2004, 323, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Reddy, P.S.; Babiuk, L.A.; Tikoo, S.K. Bovine adenovirus type 3 E1B small protein is essential for growth in bovine fibroblast cells. Virology 2001, 288, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Pyne, C.; Tikoo, S.K. Determination of bovine adenovirus-3 titer based on immunohistochemical detection of DNA binding protein in infected cells. J. Virol. Methods 2001, 94, 147–153. [Google Scholar] [CrossRef]
- Paterson, C.P.; Ayalew, L.E.; Tikoo, S.K. Mapping of nuclear import signal and importin α3 binding regions of 52K protein of bovine adenovirus 3. Virology 2012, 432, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Makadiya, N.; Gaba, A.; Tikoo, S.K. Cleavage of bovine adenovirus type 3 non-structural 100 K protein by protease is required for nuclear localization in infected cells but is not essential for virus replication. J. Gen. Virol. 2015, 96, 2749–2763. [Google Scholar] [CrossRef]
- Ayalew, L.E.; Gaba, A.; Kumar, P.; Tikoo, S.K. Conserved regions of bovine adenovirus-3 pVIII contain functional domains involved in nuclear localization and packaging in mature infectious virions. J. Gen. Virol. 2014, 95, 1743–1754. [Google Scholar] [CrossRef] [Green Version]
- Zakhartchouk, A.; Connors, W.; Van Kessel, A.; Tikoo, S.K. Bovine adenovirus type 3 containing heterologous protein in the C-terminus of minor capsid protein IX. Virology 2004, 320, 291–300. [Google Scholar] [CrossRef] [Green Version]
- de Vrij, J.; Uil, T.G.; Van Den Hengel, S.K.; Cramer, S.J.; Koppers-Lalic, D.; Verweij, M.C.; Hoeben, R.C. Adenovirus targeting to HLA-A1/MAGE-A1-positive tumor cells by fusing a single-chain T-cell receptor with minor capsid protein IX. Gene Ther. 2008, 15, 978. [Google Scholar] [CrossRef]
- Chartier, C.; Degryse, E.; Gantzer, M.; Dieterle, A.; Pavirani, A.; Mehtali, M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996, 70, 4805–4810. [Google Scholar] [CrossRef] [Green Version]
- Arsenault, R.J.; Jalal, S.; Babiuk, L.A.; Potter, A.; Griebel, P.J.; Napper, S. Kinome analysis of Toll-like receptor signaling in bovine monocytes. J. Recept. Signal Transduct. 2009, 29, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Whale, T.A.; Beskorwayne, T.K.; Babiuk, A.L.; Griebel, P.J. Bovine polymorphonuclear cells passively acquire membrane lipids and integral membrane proteins from apoptotic and necrotic cells. J. Leukoc. Biol. 2006, 79, 1226–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, W.C.; Davis, J.E.; Hamilton, M.J. Use of monoclonal antibodies and flow cytometry to cluster and analyze leukocyte differentiation molecules. In Monoclonal Antibody Protocols; Davis, W.C., Ed.; The Humana Press Inc.: Totowa, NJ, USA, 1995; pp. 149–167. [Google Scholar]
- Yamakawa, Y.; Pennelegion, C.; Willcocks, S.; Stalker, A.; Machugh, N.; Burt, D.; Coffey, T.J.; Werling, D. Identification and functional characterization of a bovine orthologue to DC-SIGN. J. Leukoc. Biol. 2008, 83, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- González-Cano, P.; Arsic, N.; Popowych, Y.I.; Griebel, P.J. Two functionally distinct myeloid dendritic cell subpopulations are present in bovine blood. Dev. Comp. Immunol. 2014, 44, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Gaba, A.; Ayalew, L.; Makadiya, N.; Tikoo, S. Proteolytic cleavage of bovine adenovirus 3-encoded pVIII. J. Virol. 2017, 91, e00211-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, E.S.; McIntyre, T.M.; Prescott, S.M.; Zimmerman, G.A. The leukocyte integrins. J. Biol. Chem. 2000, 275, 23409–23412. [Google Scholar] [CrossRef] [Green Version]
- Stewart, M.; Thiel, M.; Hogg, N. Leukocyte integrins. Curr. Opin. Cell Biol. 1995, 7, 690–696. [Google Scholar] [CrossRef]
- Wu, H.; Seki, T.; Dmitriev, I.; Uil, T.; Kashentseva, E.; Han, T.; Curiel, D.T. Double modification of adenovirus fiber with RGD and polylysine motifs improves coxsackievirus–adenovirus receptor-independent gene transfer efficiency. Hum. Gene Ther. 2002, 13, 1647–1653. [Google Scholar] [CrossRef]
- Gogev, S.; Georgin, J.P.; Schynts, F.; Vanderplasschen, A.; Thiry, E. Bovine herpesvirus 1 glycoprotein D expression in bovine upper respiratory tract mediated by a human adenovirus type 5. Vet. Res. 2004, 35, 715–721. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Tikoo, S.K. Altered tropism of recombinant bovine adenovirus type-3 expressing chimeric fiber. Virus Res. 2004, 99, 9–15. [Google Scholar] [CrossRef]
- Bergelson, J.M.; Cunningham, J.A.; Droguett, G.; Kurt-Jones, E.A.; Krithivas, A.; Hong, J.S.; Finberg, R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 1997, 275, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Wickham, T.J.; Mathias, P.; Cheresh, D.A.; Nemerow, G.R. Integrins α v β 3 and α v β 5 promote adenovirus internalization but not virus attachment. Cell 1993, 73, 309–319. [Google Scholar] [CrossRef]
- Li, X.; Bangari, D.S.; Sharma, A.; Mittal, S.K. Bovine adenovirus serotype 3 utilizes sialic acid as a cellular receptor for virus entry. Virology 2009, 392, 162–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segerman, A.; Lindman, K.; Mei, Y.F.; Allard, A.; Wadell, G. Adenovirus types 11p and 35 attach to and infect primary lymphocytes and monocytes, but hexon expression in T-cells requires prior activation. J. Virol. 2006, 349, 96–111. [Google Scholar] [CrossRef] [Green Version]
- Cotter, M.J.; Zaiss, A.K.; Muruve, D.A. Neutrophils Interact with Adenovirus Vectors via Fc Receptors and Complement Receptor 1. J. Virol. 2005, 79, 14622–14631. [Google Scholar] [CrossRef] [Green Version]
- Whale, T.A.; Wilson, H.L.; Tikoo, S.K.; Babiuk, L.K.; Griebel, P.J. Pivotal Advance: Passively acquired membrane proteins alter the functional capacity of bovine polymorphonuclear cells. J. Leukoc. Biol. 2005, 80, 481–491. [Google Scholar] [CrossRef]
- Adams, W.C.; Bond, E.; Havenga, M.J.; Holterman, L.; Goudsmit, J.; Karlsson, p.; Hedestam, G.B.; Koup, R.A.; Loré, K. Adenovirus serotype 5 infects human dendritic cells via a coxsackie virus-adenovirus receptor-independent receptor pathway mediated by lactoferrin and DC-SIGN. J. Gen. Virol. 2009, 90, 1600–1610. [Google Scholar] [CrossRef]
- Kessler, T.; Hamprecht, K.; Feuchtinger, T.; Jahn, G. Dendritic cells are susceptible to infection with wild-type adenovirus, inducing a differentiation arrest in precursor cells and inducing a strong T-cell stimulation. J. Gen. Virol. 2010, 91, 1150–1154. [Google Scholar] [CrossRef]
- Lv, Y.; Xiao, F.-J.; Wang, Y.; Zou, X.-H.; Wang, H.; Wang, H.-Y.; Wang, L.-S.; Lu, Z.-Z. Efficient gene transfer into T lymphocytes by fiber—Modified human adenovirus-5. BMC Biotechnol. 2019, 19, 23. [Google Scholar] [CrossRef]
- Reddy, P.S.; Idamakanti, N.; Zakhartchouk, A.N.; Baxi, M.K.; Lee, J.B.; Pyne, C.; Babiuk, L.A.; Tikoo, S.K. Nucleotide sequence genome organization, and transcription map of bovine adenovirus- ype 3. J. Virol. 1998, 72, 1394–1402. [Google Scholar] [CrossRef] [Green Version]
- Steinman, R.M.; Hemmi, H. Dendritic cells: Translating innate to adaptive immunity. In From Innate Immunity to Immunological Memory; Springer: Berlin/Heidelberg, Germany, 2006; pp. 17–58. [Google Scholar]
- Trombetta, E.S.; Mellman, I. Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 2005, 23, 975–1028. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Ginhoux, F.; Jakubzick, C.; van Rooijen, N.; Merad, M.; Randolph, G.J. Immature monocytes acquire antigens from other cells in the bone marrow and present them to T cells after maturing in the periphery. J. Exp. Med. 2006, 203, 583–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Cell Surface Marker | Isotype | Catalog No | Supplier |
|---|---|---|---|
| CD14 | IgG1 | BOV2109 | Washington State University |
| CD3 | IgG1 | BOV2009 | Washington State University |
| CD21 | IgG1 | BOV2031 | Washington State University |
| CD335 | IgG1 | BOV2147 | Washington State University |
| CD209 | IgG2a | BOV2133 | Washington State University |
| MHCI | IgG2a | BOV2120 | Washinton state University |
| MHCII | IgG2a | BOV2125 | Washington State University |
| CD21 | IgG1 | MCA1424GA | AbD Serotec |
| CD11c | IgM | BAQ153A | VMRD |
| CD209 | IgG2b | 551186 | BD Biosciences |
| CD335 | IgG1 | MCA2365 | AbD Serotech |
| CD3 | IgG1 | MM1A | VMRD |
| CD14 | IgG1 | MM61A | VMRD |
| RReactivity | Fluorophore | Target IgG class | Catalog No | Supplier |
|---|---|---|---|---|
| Rat anti-mouse | APC | IgG1 | 17-4015-80 | Thermo Fisher Scientific |
| Goat anti-mouse | APC | IgG2a | 17-4010-82 | Thermo Fisher Scientific |
| Rat anti-mouse | APC | IgM | 550676 | BD Biosciences |
| Goat anti-mouse | PE | IgG2b | P-21149 | Invitrogen |
| Rat anti-mouse | APC | IgG1 | 560089 | BD Pharmingen |
| Goat anti-mouse | APC | IgM | F0117 | R & D systems |
| Target Gene | Direction | Sequence (5′ to 3′) |
|---|---|---|
| EYFP | Forward | AAGCTGACCCTGAAGTTCATCTC |
| Reverse | CTTGTAGTTGCCGTCGTCCTTGAA | |
| pVIII | Forward | CAGGTGCCAGTCAAGATTAC |
| BAdV-3 | Reverse | ATGGCCGACTGAGTCATAAG |
| β-actin | Forward | GATCTGGCACCACACCTTCTAC |
| Reverse | AGGCATACAGGGACAGCACA | |
| Hexon | Forward | TGCTTCTTGCAAACACGACG |
| BAdV-3 | Reverse | CCAATCTGAACCCCCGACAA |
| 19 K | Forward | ATCGCACTGGAGTGTGGAAG |
| BAdV-3 | Reverse | GGCACCACAAACACGTCAAA |
| 52 K | Forward | ACCCTGGGTTTGATGCACTT |
| BAdV-3 | Reverse | AGCTTCCCCAAAAATGCCCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khosa, S.; Bravo Araya, M.; Griebel, P.; Arsic, N.; Tikoo, S.K. Bovine Adenovirus-3 Tropism for Bovine Leukocyte Sub-Populations. Viruses 2020, 12, 1431. https://doi.org/10.3390/v12121431
Khosa S, Bravo Araya M, Griebel P, Arsic N, Tikoo SK. Bovine Adenovirus-3 Tropism for Bovine Leukocyte Sub-Populations. Viruses. 2020; 12(12):1431. https://doi.org/10.3390/v12121431
Chicago/Turabian StyleKhosa, Sugandhika, Maria Bravo Araya, Philip Griebel, Natasa Arsic, and Suresh K. Tikoo. 2020. "Bovine Adenovirus-3 Tropism for Bovine Leukocyte Sub-Populations" Viruses 12, no. 12: 1431. https://doi.org/10.3390/v12121431
APA StyleKhosa, S., Bravo Araya, M., Griebel, P., Arsic, N., & Tikoo, S. K. (2020). Bovine Adenovirus-3 Tropism for Bovine Leukocyte Sub-Populations. Viruses, 12(12), 1431. https://doi.org/10.3390/v12121431






