Structure and Function of the T4 Spackle Protein Gp61.3
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of Expression Vectors
2.2. Protein Expression
2.3. Purification of Gp61.3
2.4. Purification of Gp5*
2.5. Purification of Gp51-372 and Gp5G322D1-372
2.6. Purification of T4L
2.7. Purification of Gp61.3-Gp5Lys Complex
2.8. Crystallization and Data Collection
2.9. Structure Determination
2.10. Lysozyme Halo Assay
2.11. Analytical Ultracentrifugation
2.12. Cell Lysis Assay
2.13. Molecular Dynamics
2.14. Sequence Alignments and Molecular Graphics Generation
3. Results
3.1. Gp61.3 is Translocated into the Periplasm and Its Signal Sequence Is Cleaved
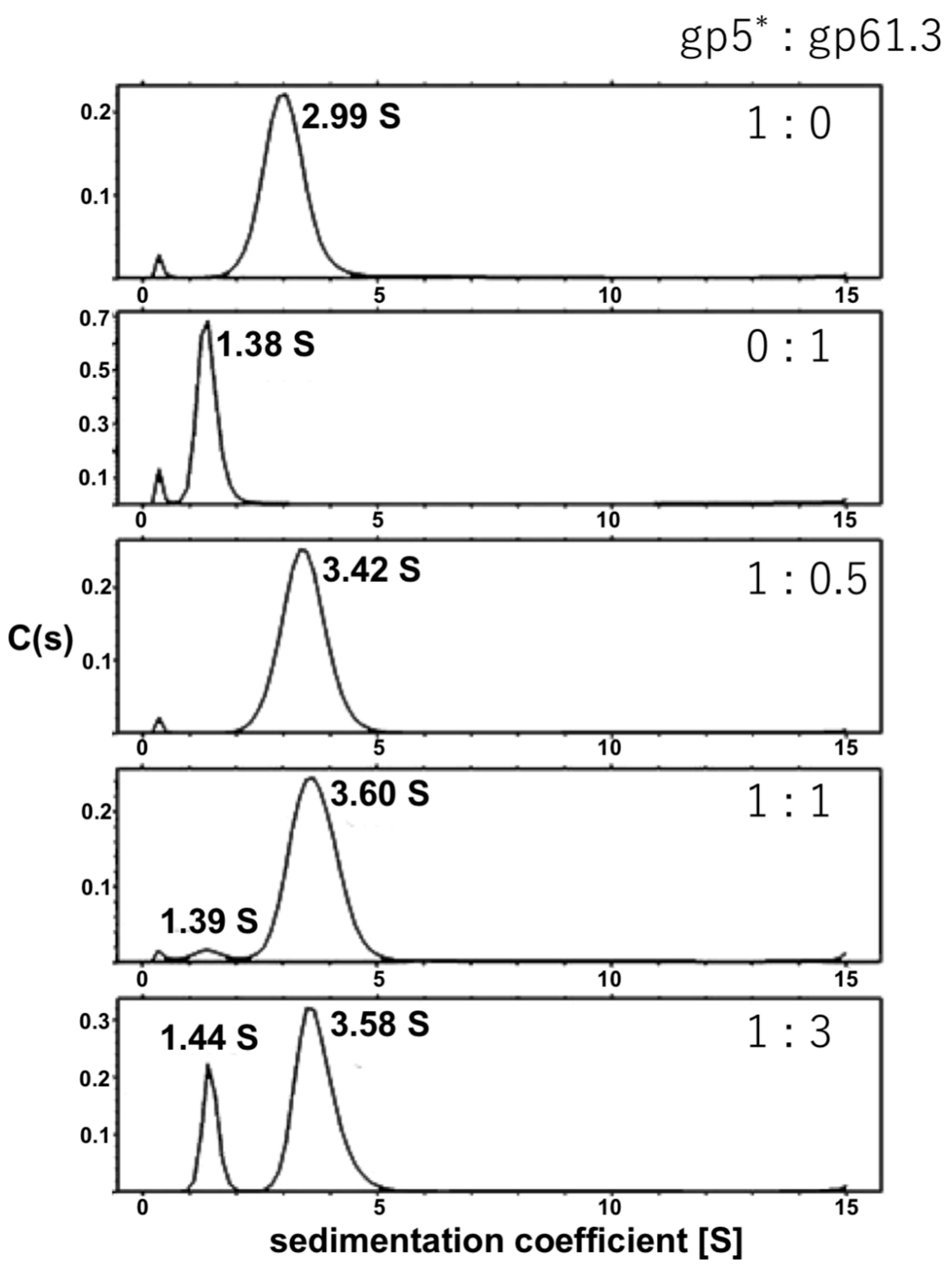
3.2. Gp61.3 Forms a 1:1 Complex with Gp5*
3.3. Gp61.3 Inhibits Lysozyme Activity of Gp5 but Does Not Affect the Activity of T4L In Vitro
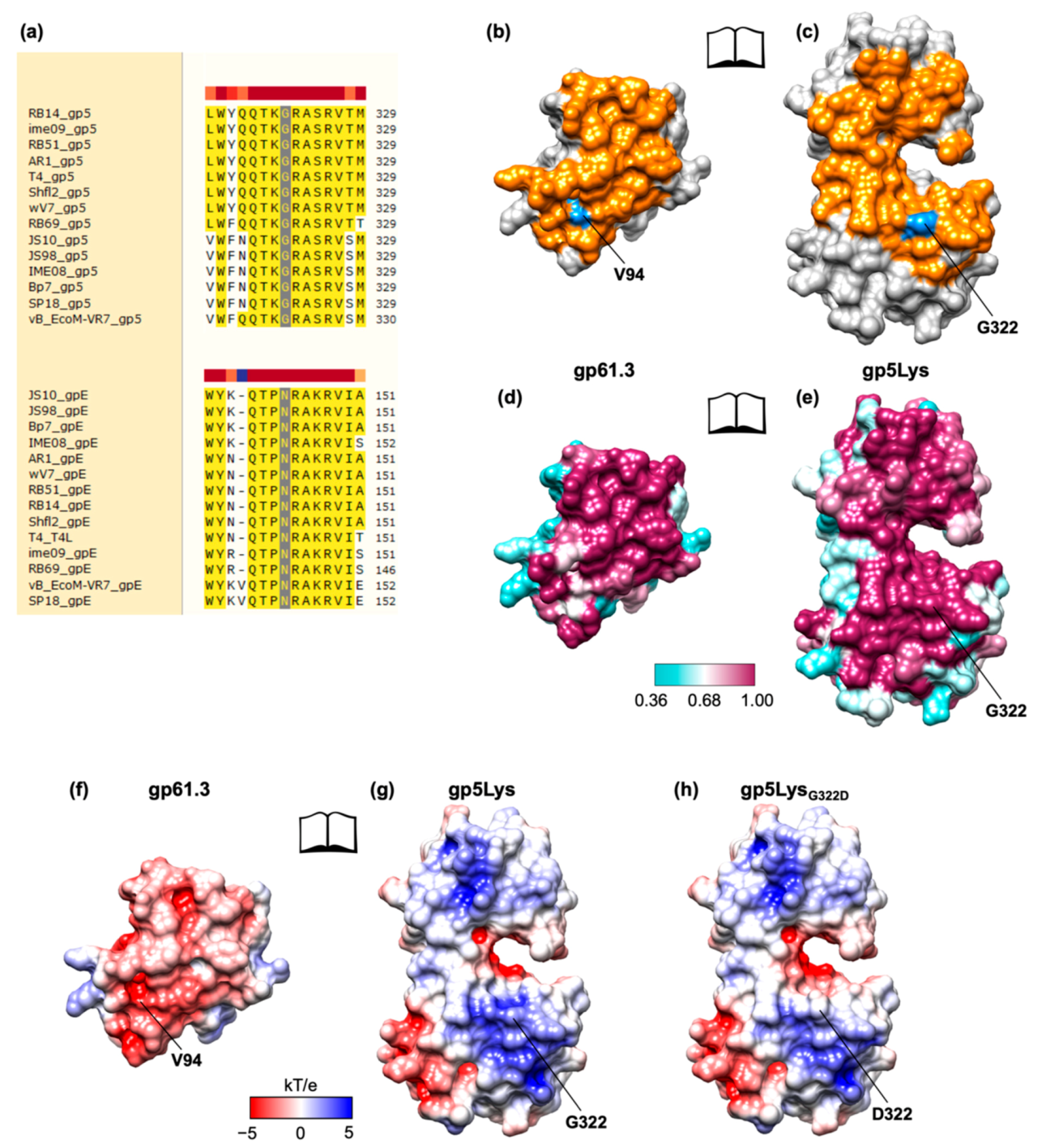
3.4. The Crystal Structure of Gp61.3 Shows That It Has a Novel Fold
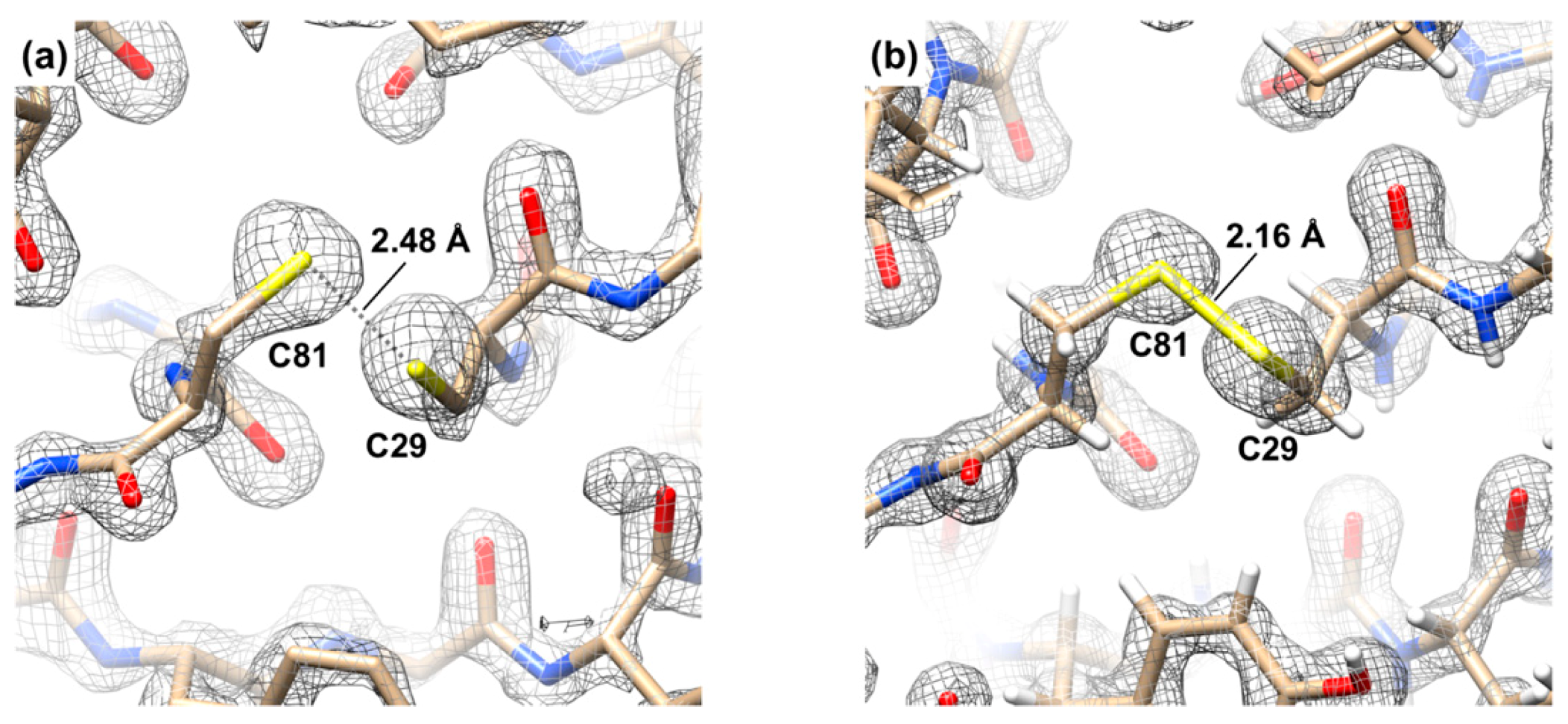
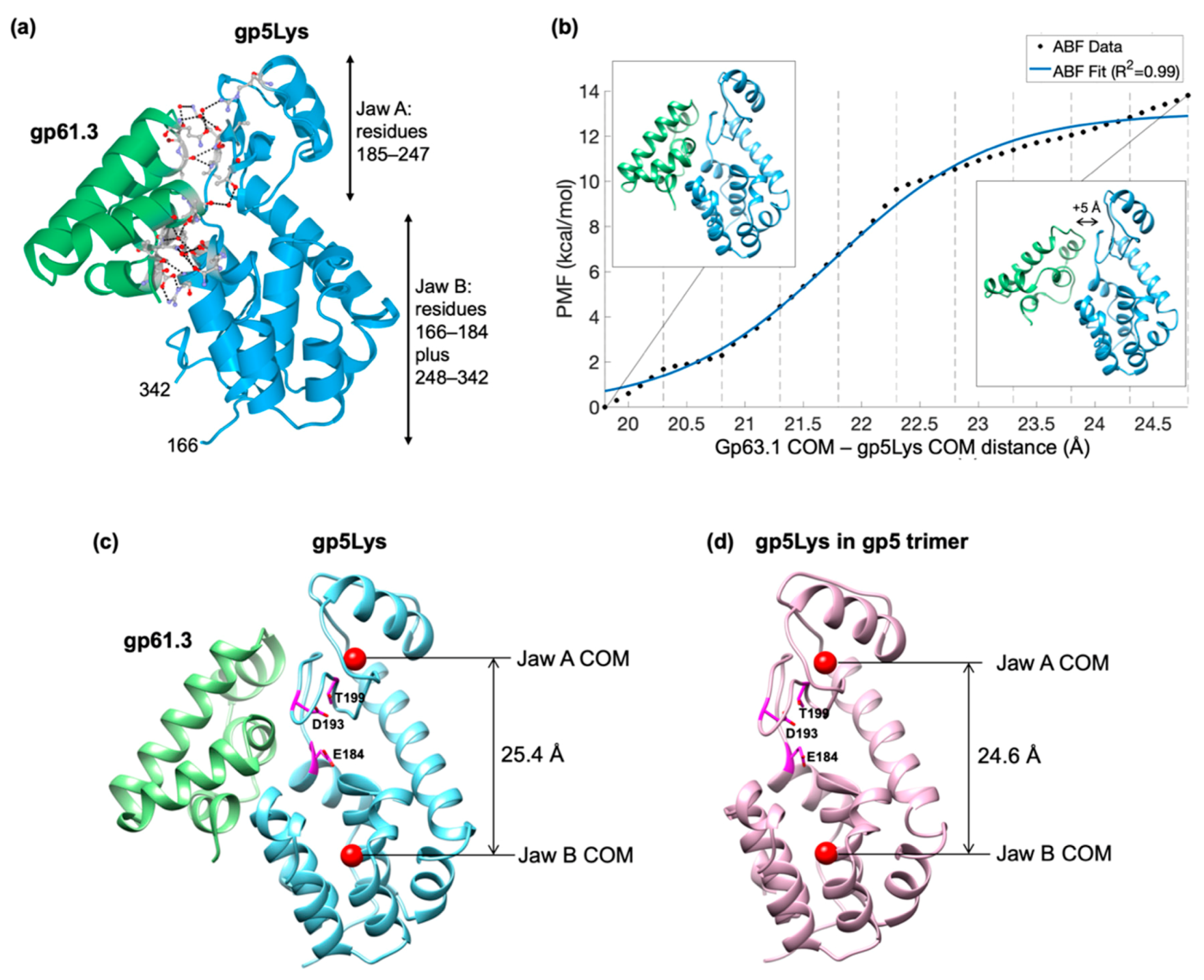
3.5. The Crystal Structure of the Gp61.3–Gp5Lys Complex
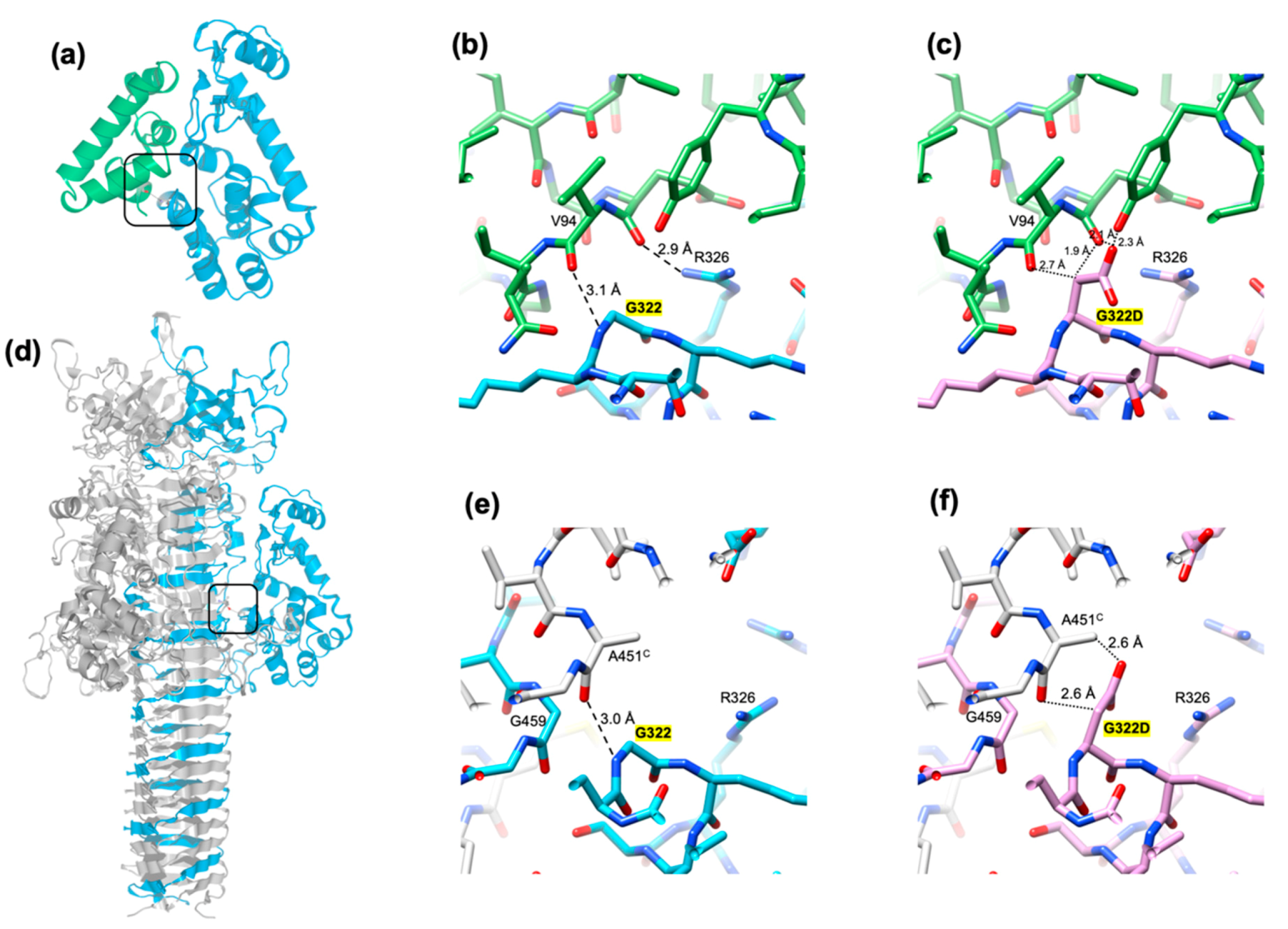
3.6. The Critical Location of G322D Mutation in Gp5Lys
3.7. Gp5 Can Penetrate the E. coli Inner Membrane from the Inside on Its Own
4. Discussion
Supplementary Materials
Note Added during the Proofreading
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuroki, R.; Weaver, L.H.; Matthews, B.W. A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme. Science 1993, 262, 2030–2033. [Google Scholar] [CrossRef] [PubMed]
- Mukai, F.; Streisinger, G.; Miller, B. Mechanism of lysis in phage T4-infected cells. Virology 1967, 33, 398–404. [Google Scholar] [CrossRef]
- Emrich, J. Lysis of T4-infected bacteria in absence of lysozyme. Virology 1968, 35, 158–165. [Google Scholar] [CrossRef]
- Kao, S.H.; McClain, W.H. Baseplate protein of bacteriophage-T4 with both structural and lytic functions. J. Virol. 1980, 34, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.H.; McClain, W.H. Roles of bacteriophage-T4 gene-5 and gene-s products in cell-lysis. J. Virol. 1980, 34, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Arisaka, F.; Ishii, S.I. Isolation and characterization of the bacteriophage-T4 tail-associated lysozyme. J. Virol. 1985, 54, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Hoshida, K.; Arisaka, F. Mapping of functional sites on the primary structure of the tail lysozyme of bacteriophage T4 by mutational analysis. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 1998, 1384, 243–252. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis and the interaction between free phages and infected cells. In The Molecular Biology of Bacteriophage T4; Karam, J.D., Kutter, E., Carlson, K., Guttman, B., Eds.; ASM Press: Washington, DC, USA, 1994; pp. 397–405. [Google Scholar]
- Abedon, S.T. Bacteriophage T4 resistance to lysis-inhibition collapse. Genet. Res. 1999, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Look Who’s Talking: T-Even Phage Lysis Inhibition, the Granddaddy of Virus-Virus Intercellular Communication Research. Viruses 2019, 11, 951. [Google Scholar] [CrossRef]
- Okamoto, K.; Yutsudo, M. Participation of the s gene product of phage T4 in the establishment of resistance to T4 ghosts. Virology 1974, 58, 369–376. [Google Scholar] [CrossRef]
- Kanamaru, S.; Gassner, N.C.; Ye, N.Z.; Takeda, S.; Arisaka, F. The C-terminal fragment of the precursor tail lysozyme of bacteriophage T4 stays as a structural component of the baseplate after cleavage. J. Bacteriol. 1999, 181, 2739–2744. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, S.; Leiman, P.G.; Kostyuchenko, V.A.; Chipman, P.R.; Mesyanzhinov, V.V.; Arisaka, F.; Rossmann, M.G. Structure of the cell-puncturing device of bacteriophage T4. Nature 2002, 415, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Kai, T.; Ueno, H.; Otsuka, Y.; Morimoto, W.; Yonesaki, T. Gene 61.3 of bacteriophage t4 is the spackle gene. Virology 1999, 260, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Buth, S.A.; Shneider, M.M.; Scholl, D.; Leiman, P.G. Structure and Analysis of R1 and R2 Pyocin Receptor-Binding Fibers. Viruses 2018, 10, 427. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. Sect. A 2008, 64, 112–122. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 1997, 276, 307–326. [Google Scholar]
- Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2010, 66, 470–478. [Google Scholar] [CrossRef]
- Langer, G.; Cohen, S.X.; Lamzin, V.S.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D-Struct. Biol. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D-Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Battye, T.G.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G.W. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2011, 67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D-Struct. Biol. 2006, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.H.; Schuck, P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 2006, 90, 4651–4661. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. Model 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Henin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Loncharich, R.J.; Brooks, B.R.; Pastor, R.W. Langevin Dynamics of Peptides—The Frictional Dependence of Isomerization Rates of N-Acetylalanyl-N′-Methylamide. Biopolymers 1992, 32, 523–535. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant-Pressure Molecular-Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.H.; Pastor, R.W.; Brooks, B.R. Constant-Pressure Molecular-Dynamics Simulation—The Langevin Piston Method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Lu, H.; Schulten, K. Steered molecular dynamics simulations of force-induced protein domain unfolding. Proteins 1999, 35, 453–463. [Google Scholar] [CrossRef]
- Darve, E.; Rodriguez-Gomez, D.; Pohorille, A. Adaptive biasing force method for scalar and vector free energy calculations. J. Chem. Phys. 2008, 128, 144120. [Google Scholar] [CrossRef] [PubMed]
- Henin, J.; Fiorin, G.; Chipot, C.; Klein, M.L. Exploring Multidimensional Free Energy Landscapes Using Time-Dependent Biases on Collective Variables. J. Chem. Theory Comput. 2010, 6, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone phi, psi and Side-Chain chi(1) and chi(2) Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Moussa, S.H.; Kuznetsov, V.; Tran, T.A.T.; Sacchettini, J.C.; Young, R. Protein determinants of phage T4 lysis inhibition. Protein Sci. 2012, 21, 571–582. [Google Scholar] [CrossRef]
- Lall, S.D.; Eriho, B.E.; Jay, J.M. Comparison for four methods for extracting periplasmic proteins. J. Microbiol. Methods 1989, 9, 195–199. [Google Scholar] [CrossRef]
- Malherbe, G.; Humphreys, D.P.; Dave, E. A robust fractionation method for protein subcellular localization studies in Escherichia coli. Biotechniques 2019, 66, 171–178. [Google Scholar] [CrossRef]
- Holm, L.; Rosenstrom, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Cranford-Smith, T.; Huber, D. The way is the goal: How SecA transports proteins across the cytoplasmic membrane in bacteria. Fems Microbiol. Lett. 2018, 365, fny093. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Amoros, D.; Garcia de la Torre, J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011, 101, 892–898. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Comer, J.; Gumbart, J.C.; Henin, J.; Lelievre, T.; Pohorille, A.; Chipot, C. The Adaptive Biasing Force Method: Everything You Always Wanted To Know but Were Afraid To Ask. J. Phys. Chem. B 2015, 119, 1129–1151. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.P.; Zaccai, N.R.; Fleming, P.J.; Gessmann, D.; Fleming, K.G. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc. Natl. Acad. Sci. USA 2013, 110, 4285–4290. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Kurniawan, F.; Banerjee, S.; Moeller, N.H.; Aihara, H. Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing. Acta Crystallogr. Sect. D Struct. Biol. 2020, 76, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; King, J. Genetic control of bacteriophage T4 baseplate morphogenesis: III. Formation of the central plug and overall assembly pathway. J. Mol. Biol. 1975, 99, 695–716. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; Mccammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Ramanculov, E.; Young, R. Genetic analysis of the T4 holin: Timing and topology. Gene 2001, 265, 25–36. [Google Scholar] [CrossRef]
- Sanghamitra, N.J.M.; Inaba, H.; Arisaka, F.; Wang, D.O.; Kanamaru, S.; Kitagawa, S.; Ueno, T. Plasma membrane translocation of a protein needle based on a triple-stranded beta-helix motif. Mol. Biosyst. 2014, 10, 2677–2683. [Google Scholar] [CrossRef] [PubMed]
- Baase, W.A.; Liu, L.; Tronrud, D.E.; Matthews, B.W. Lessons from the lysozyme of phage T4. Protein Sci. 2010, 19, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Weaver, L.H.; Matthews, B.W. Structure of bacteriophage T4 lysozyme refined at 1.7 Å resolution. J. Mol. Biol. 1987, 193, 189–199. [Google Scholar] [CrossRef]
- Yirdaw, R.B.; McHaourab, H.S. Direct Observation of T4 Lysozyme Hinge-Bending Motion by Fluorescence Correlation Spectroscopy. Biophys. J. 2012, 103, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Wozniak, J.A.; Matthews, B.W. Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme. J. Mol. Biol. 1995, 250, 527–552. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, S.; Ishiwata, Y.; Suzuki, T.; Rossmann, M.G.; Arisaka, F. Control of bacteriophage T4 tall lysozyme activity during the infection process. J. Mol. Biol. 2005, 346, 1013–1020. [Google Scholar] [CrossRef]
- Callewaert, L.; Aertsen, A.; Deckers, D.; Vanoirbeek, K.G.A.; Vanderkelen, L.; Van Herreweghe, J.M.; Masschalck, B.; Nakimbugwe, D.; Robben, J.; Michiels, C.W. A new family of lysozyme inhibitors contributing to lysozyme tolerance in gram-negative bacteria. PLoS Pathog. 2008, 4, e1000019. [Google Scholar] [CrossRef]
- Leysen, S.; Vanderkelen, L.; Van Asten, K.; Vanheuverzwijn, S.; Theuwis, V.; Michiels, C.W.; Strelkov, S.V. Structural characterization of the PliG lysozyme inhibitor family. J. Struct. Biol. 2012, 180, 235–242. [Google Scholar] [CrossRef]
- Whitney, J.C.; Chou, S.; Russell, A.B.; Biboy, J.; Gardiner, T.E.; Ferrin, M.A.; Brittnacher, M.; Vollmer, W.; Mougous, J.D. Identification, Structure, and Function of a Novel Type VI Secretion Peptidoglycan Glycoside Hydrolase Effector-Immunity Pair. J. Biol. Chem. 2013, 288, 26616–26624. [Google Scholar] [CrossRef]
- Yum, S.; Kim, M.J.; Xu, Y.; Jin, X.L.; Yoo, H.Y.; Park, J.W.; Gong, J.H.; Choe, K.M.; Lee, B.L.; Ha, N.C. Structural basis for the recognition of lysozyme by MliC, a periplasmic lysozyme inhibitor in Gram-negative bacteria. Biochem. Biophys. Res. Commun. 2009, 378, 244–248. [Google Scholar] [CrossRef]






| Crystal | Gp61.3 SeMet | Gp61.3-gp5Lys |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.97941 | 1.0000 |
| Number of frames | 325 | 360 |
| Frame width (°) | 1.0 | 1.0 |
| Space group | P21 | P212121 |
| Cell dimensions a, b, c (Å) | 30.376, 46.921, 75.277 | 46.72, 69.18, 83.82 |
| Resolution (Å) | 39.8–1.60 (1.64–1.60) * | 17.92–1.15 (1.21–1.15) |
| Rmeas | 0.064 (0.158) | 0.109 (1.885) |
| <I>/<σI> | 16.0 (5.8) | 12.2 (1.8) |
| Completeness (%) | 94.2 (73.7) | 99.7 (99.8) |
| Multiplicity | 3.3 (3.1) | 13.5 (12.6) |
| Anomalous signal # | 4.91 (2.42) | N.A. $ |
| Refinement | ||
| Resolution | 19.90–1.60 | 17.92–1.15 |
| No. reflections used in refinement | 51,366 | 97,671 |
| No. atoms (non-H) | 2272 | 2630 |
| Protein | 1870 | 2195 |
| Ligand/ion | 3 | 53 |
| Water | 399 | 382 |
| Rwork/Rfree | 0.179/0.228 | 0.107/0.122 |
| Average B factor (Å2) | 18.0 | 16.3 |
| Protein | 15.6 | 13.5 |
| Ligand/Ion | 33.1 | 32.7 |
| Water | 29.1 | 30.1 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.008 |
| Bond angles (°) | 0.493 | 1.066 |
| Ramachandran plot statistics | ||
| Favored (%) | 99.01 | 97.39 |
| Allowed (%) | 0.91 | 1.61 |
| Outliers (%) | 0.00 | 0.00 |
| PDB accession code | 7CN6 | 7CN7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanamaru, S.; Uchida, K.; Nemoto, M.; Fraser, A.; Arisaka, F.; Leiman, P.G. Structure and Function of the T4 Spackle Protein Gp61.3. Viruses 2020, 12, 1070. https://doi.org/10.3390/v12101070
Kanamaru S, Uchida K, Nemoto M, Fraser A, Arisaka F, Leiman PG. Structure and Function of the T4 Spackle Protein Gp61.3. Viruses. 2020; 12(10):1070. https://doi.org/10.3390/v12101070
Chicago/Turabian StyleKanamaru, Shuji, Kazuya Uchida, Mai Nemoto, Alec Fraser, Fumio Arisaka, and Petr G. Leiman. 2020. "Structure and Function of the T4 Spackle Protein Gp61.3" Viruses 12, no. 10: 1070. https://doi.org/10.3390/v12101070
APA StyleKanamaru, S., Uchida, K., Nemoto, M., Fraser, A., Arisaka, F., & Leiman, P. G. (2020). Structure and Function of the T4 Spackle Protein Gp61.3. Viruses, 12(10), 1070. https://doi.org/10.3390/v12101070





