Isolation of a Divergent Strain of Bovine Parainfluenza Virus Type 3 (BPIV3) Infecting Cattle in China
Abstract
:1. Introduction
2. Materials and Methods
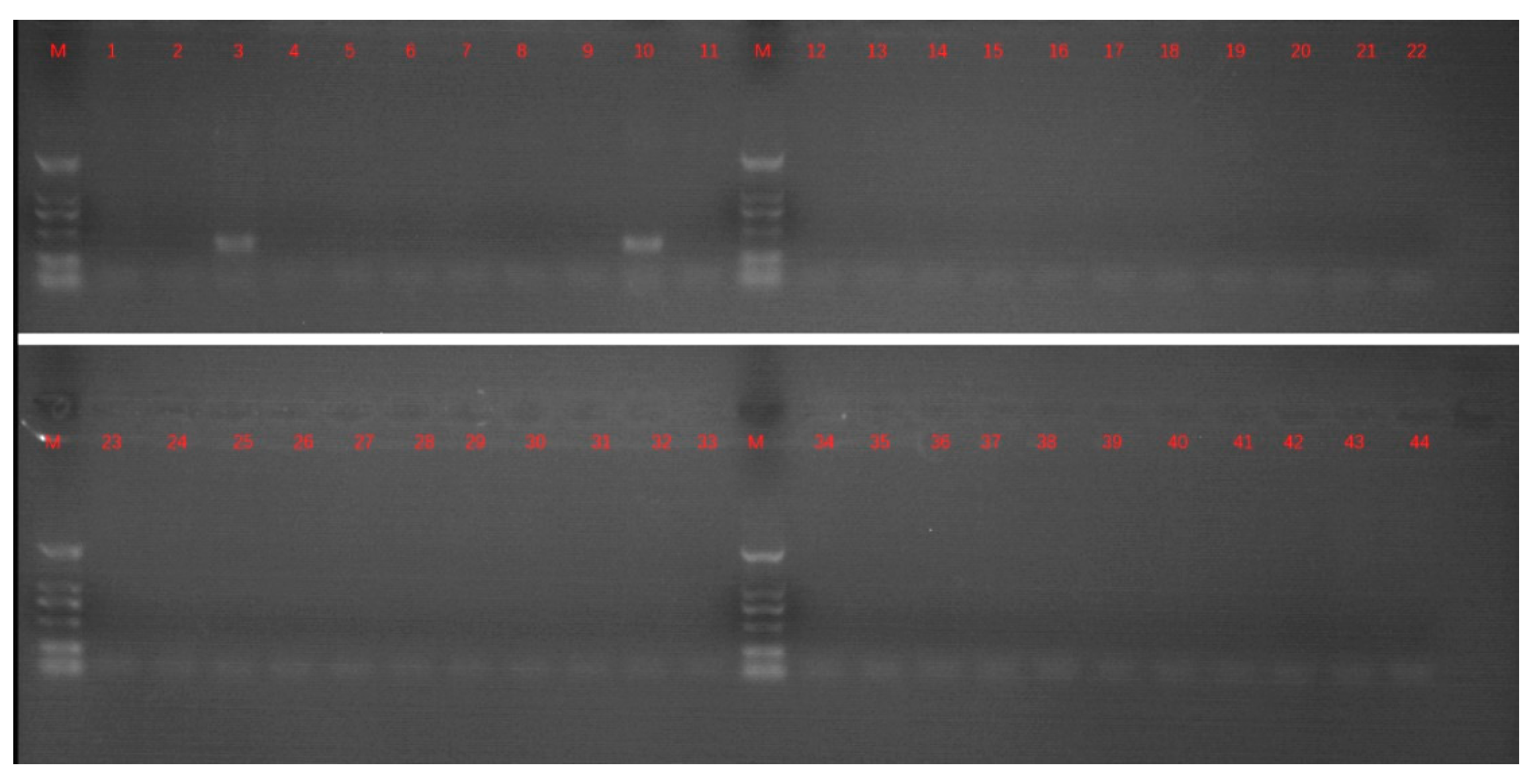
2.1. Sample Treatment and PCR Detection
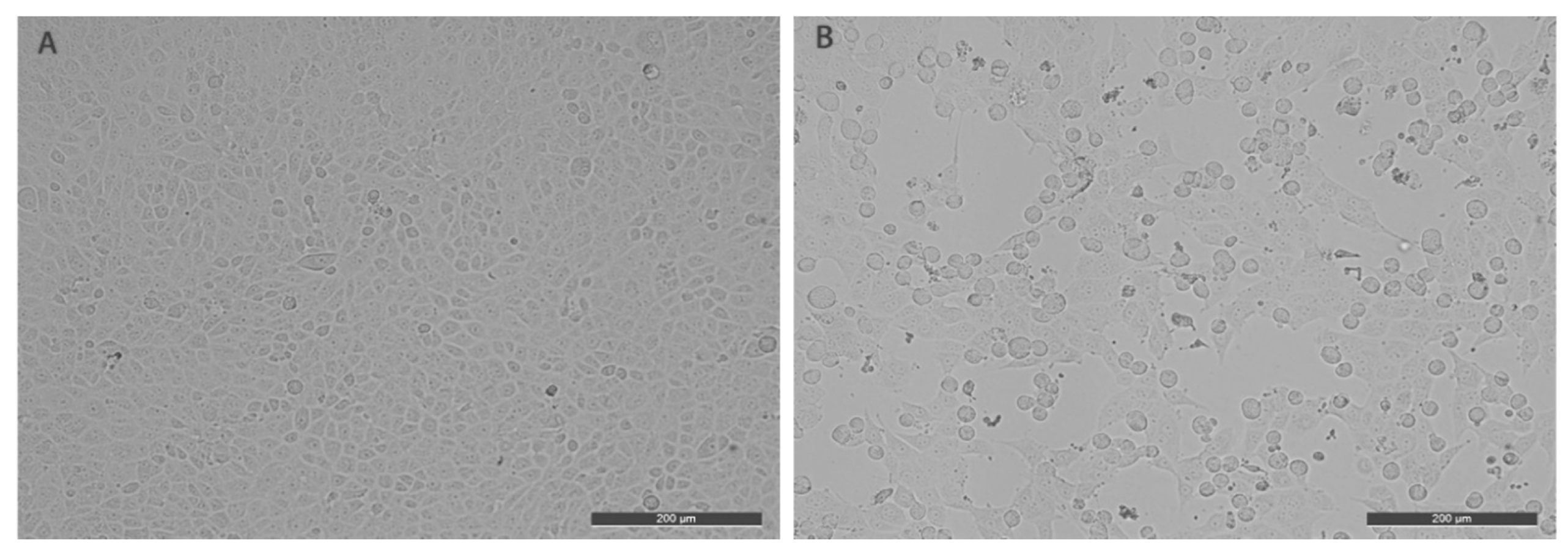
2.2. Cell Cultivation and Virus Isolation
2.3. Cloning and Whole Genome Sequencing
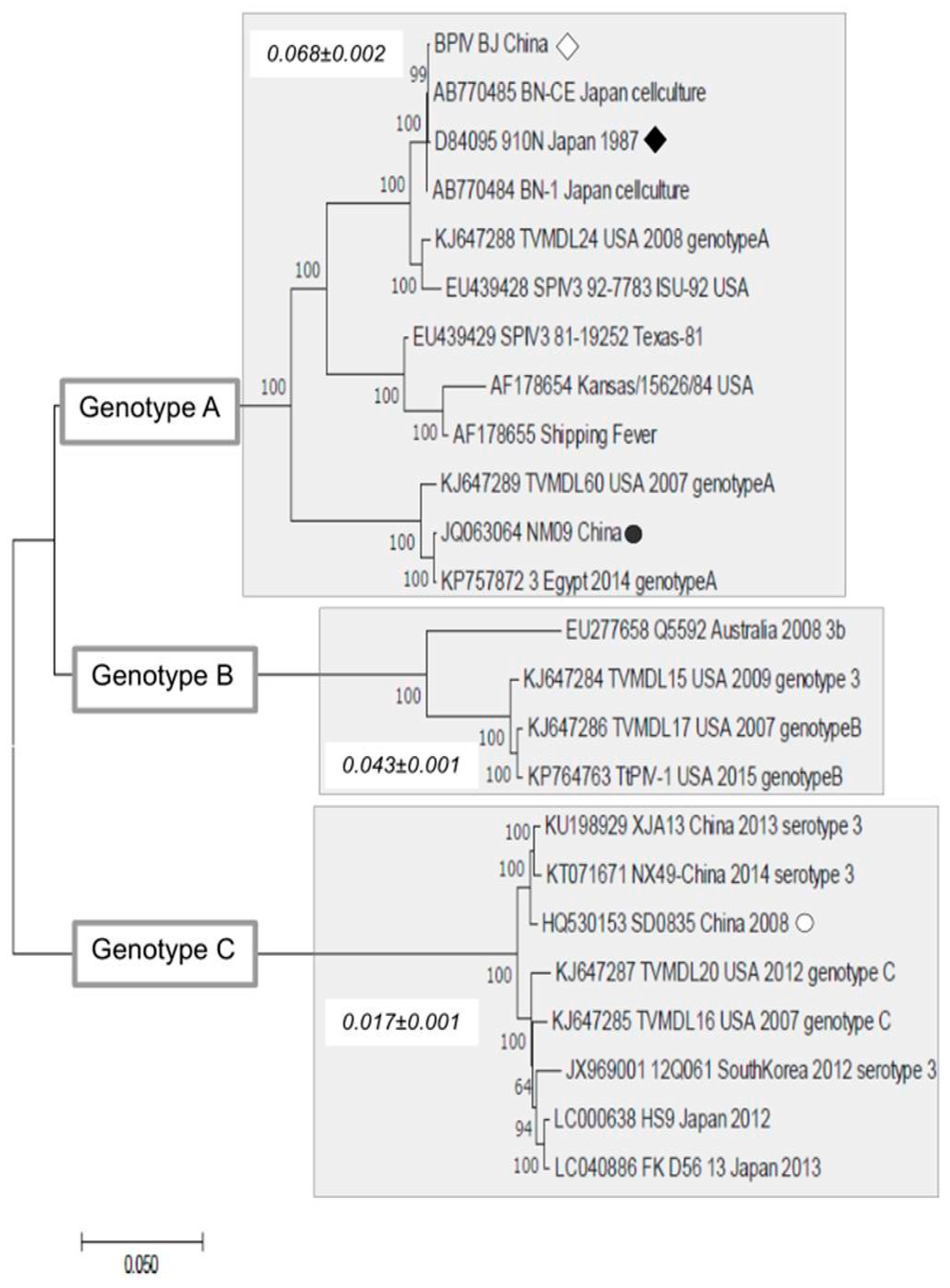
2.4. Sequence Analysis
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Reisinger, R.C.; Heddleston, K.L.; Manthei, C.A. A myxovirus (sf-4) associated with shipping fever of cattle. J. Am. Vet. Med. Assoc. 1959, 135, 147–152. [Google Scholar] [PubMed]
- Fulton, R.W.; Blood, K.S.; Panciera, R.J.; Payton, M.E.; Ridpath, J.F.; Confer, A.W.; Saliki, J.T.; Burge, L.T.; Welsh, R.D.; Johnson, B.J.; et al. Lung pathology and infectious agents in fatal feedlot pneumonias and relationship with mortality, disease onset, and treatments. J. Vet. Diagn. Investig. 2009, 21, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Hoerlein, A.B.; Mansfield, M.E.; Abinanti, F.R.; Huebner, R.J. Studies of shipping fever of cattle. I. Para-influenza 3 virus antibodies in feeder calves. J. Am. Vet. Med. Assoc. 1959, 135, 153–160. [Google Scholar] [PubMed]
- Murray, G.M.; More, S.J.; Sammin, D.; Casey, M.J.; McElroy, M.C.; O’Neill, R.G.; Byrne, W.J.; Earley, B.; Clegg, T.A.; Ball, H.; et al. Pathogens, patterns of pneumonia, and epidemiologic risk factors associated with respiratory disease in recently weaned cattle in ireland. J. Vet. Diagn. Investig. 2017, 29, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.A. Bovine parainfluenza-3 virus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 575–593. [Google Scholar] [CrossRef] [PubMed]
- Bailly, J.E.; McAuliffe, J.M.; Skiadopoulos, M.H.; Collins, P.L.; Murphy, B.R. Sequence determination and molecular analysis of two strains of bovine parainfluenza virus type 3 that are attenuated for primates. Virus Genes 2000, 20, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.F.; Zhu, Y.M.; Dong, X.M.; Cai, H.; Ma, L.; Wang, S.; Yan, H.; Wang, X.Z.; Xue, F. Pathogenesis of a genotype c strain of bovine parainfluenza virus type 3 infection in albino guinea pigs. Virus Res. 2014, 188, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Horwood, P.F.; Gravel, J.L.; Mahony, T.J. Identification of two distinct bovine parainfluenza virus type 3 genotypes. J. Gen. Virol. 2008, 89, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Ohkura, T.; Shimizu, M.; Akiyama, M.; Kameyama, K.; Takeuchi, K. Complete genome sequence of the first isolate of genotype c bovine parainfluenza virus type 3 in japan. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Maidana, S.S.; Lomonaco, P.M.; Combessies, G.; Craig, M.I.; Diodati, J.; Rodriguez, D.; Parreno, V.; Zabal, O.; Konrad, J.L.; Crudelli, G.; et al. Isolation and characterization of bovine parainfluenza virus type 3 from water buffaloes (bubalus bubalis) in argentina. BMC Vet. Res. 2012, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Neill, J.D.; Ridpath, J.F.; Valayudhan, B.T. Identification and genome characterization of genotype b and genotype c bovine parainfluenza type 3 viruses isolated in the united states. BMC Vet. Res. 2015, 11, 112. [Google Scholar] [CrossRef] [PubMed]
- Oem, J.K.; Lee, E.Y.; Lee, K.K.; Kim, S.H.; Lee, M.H.; Hyun, B.H. Molecular characterization of a korean bovine parainfluenza virus type 3 isolate. Vet. Microbiol. 2013, 162, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, T.; Kokuho, T.; Konishi, M.; Kameyama, K.; Takeuchi, K. Complete genome sequences of bovine parainfluenza virus type 3 strain bn-1 and vaccine strain bn-ce. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.L.; Zhu, Y.M.; Zhou, Y.H.; Lv, C.; Yan, H.; Ma, L.; Shi, H.F.; Xue, F. Identification of three antigen epitopes on the nucleocapsid protein of the genotype c of bovine parainfluenza virus type 3. Vet. Microbiol. 2015, 178, 61–69. [Google Scholar] [CrossRef]
- Wen, Y.J.; Shi, X.C.; Wang, F.X.; Wang, W.; Zhang, S.Q.; Li, G.; Song, N.; Chen, L.Z.; Cheng, S.P.; Wu, H. Phylogenetic analysis of the bovine parainfluenza virus type 3 from cattle herds revealing the existence of a genotype a strain in china. Virus Genes 2012, 45, 542–547. [Google Scholar] [CrossRef]
- Zhu, Y.M.; Shi, H.F.; Gao, Y.R.; Xin, J.Q.; Liu, N.H.; Xiang, W.H.; Ren, X.G.; Feng, J.K.; Zhao, L.P.; Xue, F. Isolation and genetic characterization of bovine parainfluenza virus type 3 from cattle in china. Vet. Microbiol. 2011, 149, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.A. How efficacious are vaccines against bovine respiratory syncytial virus in cattle? Vet. Microbiol. 2017, 206, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Grissett, G.P.; White, B.J.; Larson, R.L. Structured literature review of responses of cattle to viral and bacterial pathogens causing bovine respiratory disease complex. J. Vet. Intern. Med. 2015, 29, 770–780. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
| Protein Name (Position) | BPIV-BJ and D84095 (Position) | BPIV-BJ and JQ063064 (Position) | BPIV-BJ and HQ530153 (Position) |
|---|---|---|---|
| NP (111-1658) | none | R(556)G; V(628)I; N(1420)D; Q(1429)R; S(1471)N; S(1489)L; E(1504)D; S(1575)N; P(1582)S; K(1585)R; S(1588)P; N(1594)D; D(1606)N; | L(157)I; S(385)N; S(431)P; S(448)G; S(602)A; I(629)V; Q(692)R; A(769)S; I(1066)V; S(1237)N; E(1249)D; D(1252)E; R(556)G; V(1264)I; N(1306)K; R(556)G; S(1316)I; H(1333)Y; R(556)G; S(1351)T; A(1381)I; G(1417)T; N(1420)D; E(1423)D; I(1442)V; T(1465)V; R(1468)S; S(1471)N; D(1475)K; T(1483)A; E(1486)G; V(1492)T; E(1498)D; I(1501)A; E(1504)N; I(1510)L; K(1513)G; T(1516)V; K(1570)R; S(1576)N; D(1579)E; P(1583)S; N(1494)D; A(1600)T; D(1606)N; T(1609)A; N(1612)D; N(1541)S; |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leal, É.; Liu, C.; Zhao, Z.; Deng, Y.; Villanova, F.; Liang, L.; Li, J.; Cui, S. Isolation of a Divergent Strain of Bovine Parainfluenza Virus Type 3 (BPIV3) Infecting Cattle in China. Viruses 2019, 11, 489. https://doi.org/10.3390/v11060489
Leal É, Liu C, Zhao Z, Deng Y, Villanova F, Liang L, Li J, Cui S. Isolation of a Divergent Strain of Bovine Parainfluenza Virus Type 3 (BPIV3) Infecting Cattle in China. Viruses. 2019; 11(6):489. https://doi.org/10.3390/v11060489
Chicago/Turabian StyleLeal, Élcio, Cun Liu, Zhanzhong Zhao, Yong Deng, Fabiola Villanova, Lin Liang, Jinxiang Li, and Shangjin Cui. 2019. "Isolation of a Divergent Strain of Bovine Parainfluenza Virus Type 3 (BPIV3) Infecting Cattle in China" Viruses 11, no. 6: 489. https://doi.org/10.3390/v11060489
APA StyleLeal, É., Liu, C., Zhao, Z., Deng, Y., Villanova, F., Liang, L., Li, J., & Cui, S. (2019). Isolation of a Divergent Strain of Bovine Parainfluenza Virus Type 3 (BPIV3) Infecting Cattle in China. Viruses, 11(6), 489. https://doi.org/10.3390/v11060489






