Correlation in Expression between LTR Retrotransposons and Potential Host Cis-Targets during Infection of Antherea pernyi with ApNPV Baculovirus
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of LTR-Retrotransposons
2.2. Classification and Phylogenetic Analysis of LTR-Retrotransposons
2.3. Analysis of the Env Genes from AyLTRs
2.4. Identification of Genes Located in the Neighborhood of AyLTRs in the A. yamamai Genome
2.5. Analysis of RNA-Seq Data
3. Results
3.1. De novo Detection of AyLTRs in the Antheraea yamamai Genome
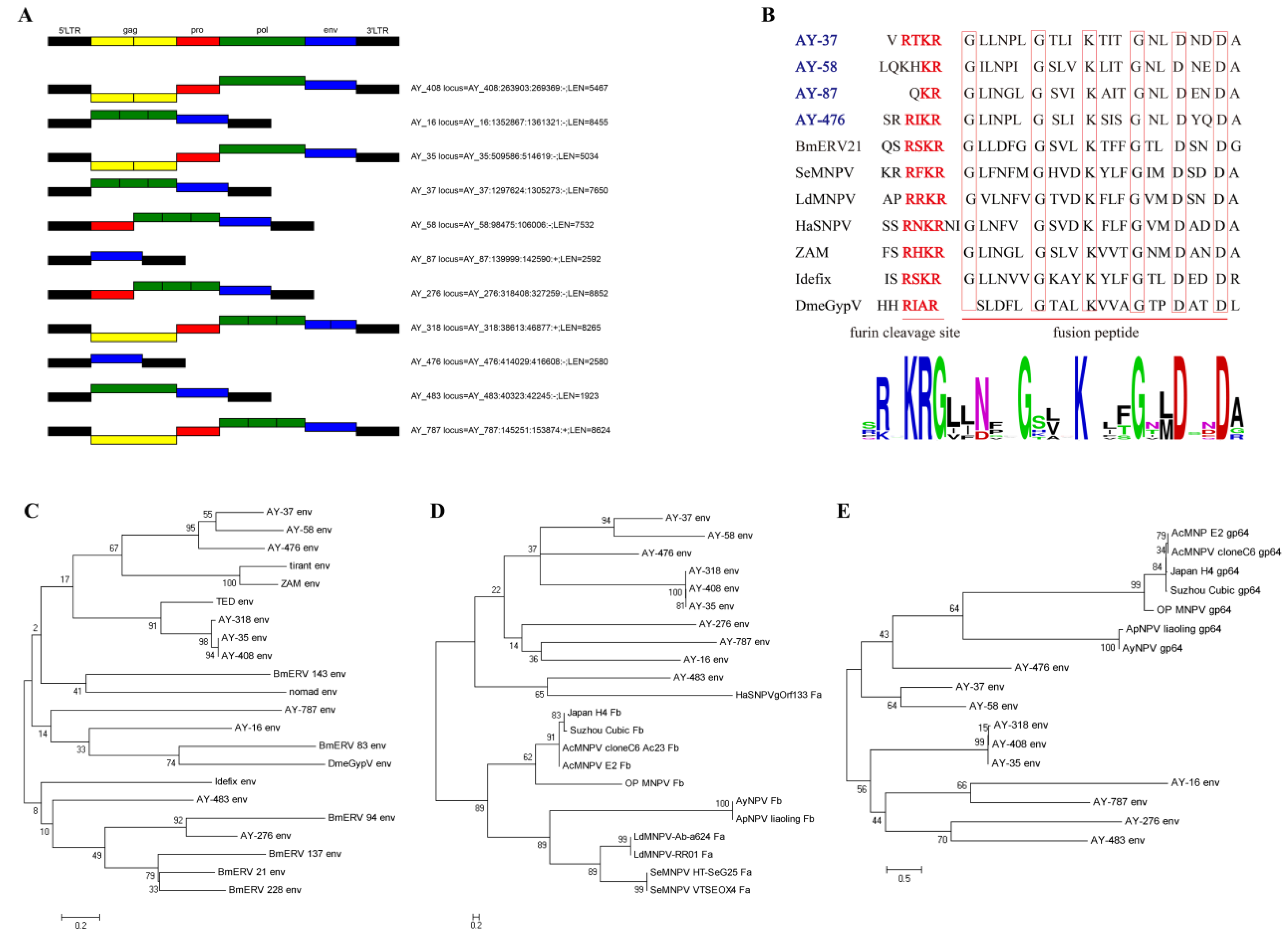
3.2. Phylogenetic Analysis and Classification of Full-Length AyLTRs
3.3. Analysis of Env Genes from Full-Length AyLTRs
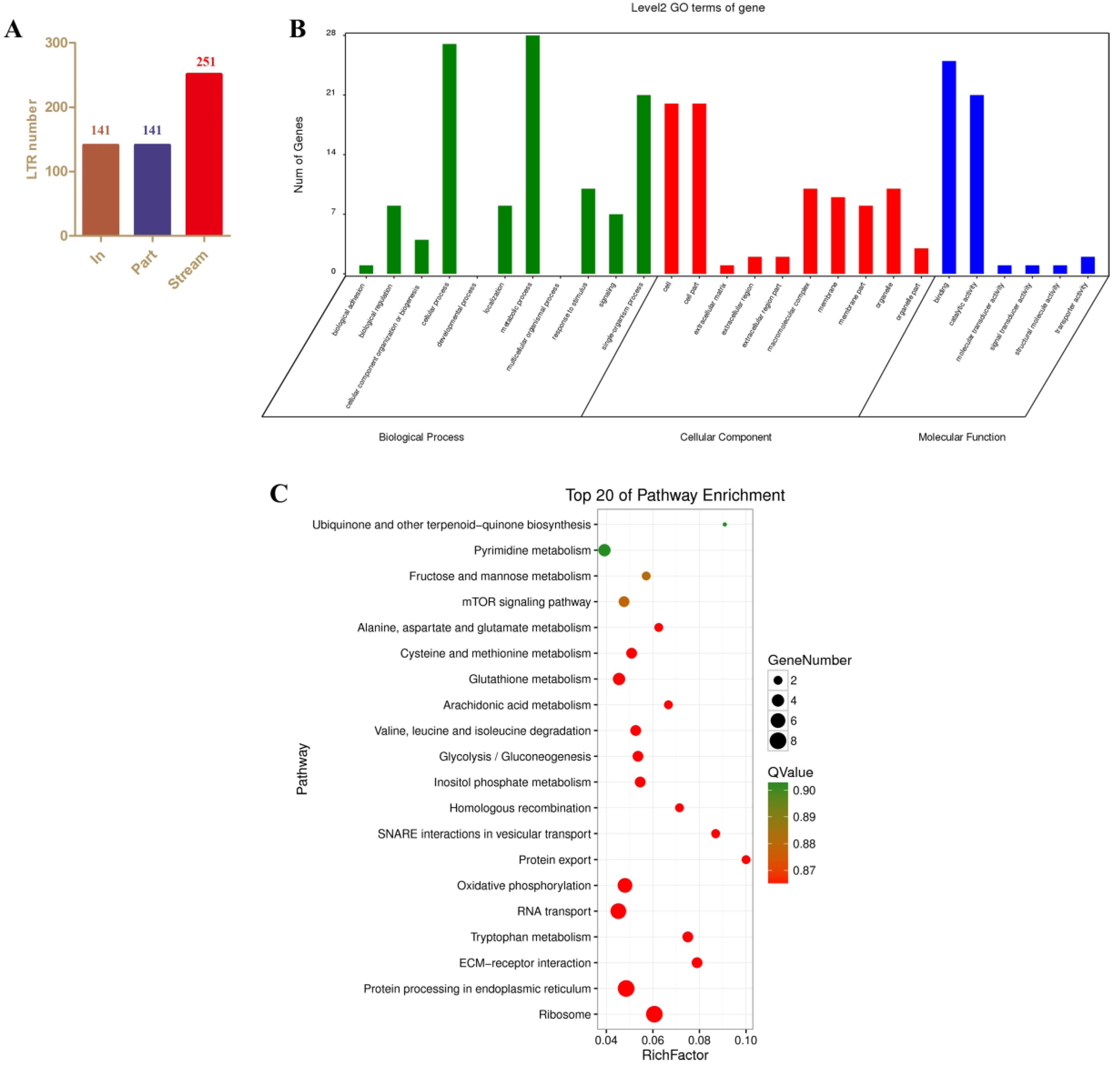
3.4. Identification and Analysis of (Potentially Cis-Target) Genes that Occur in the Neighborhood of AyLTRs
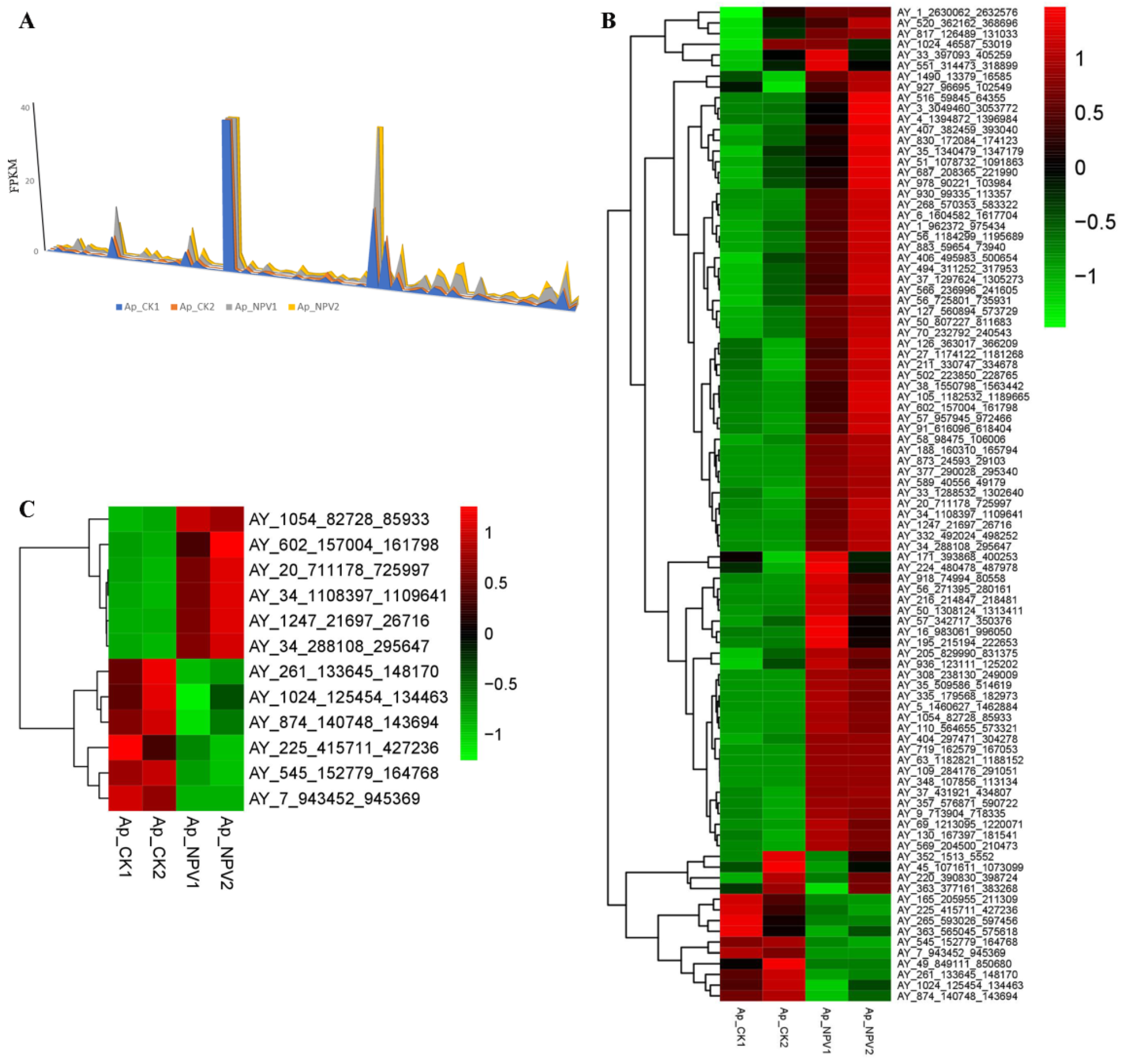
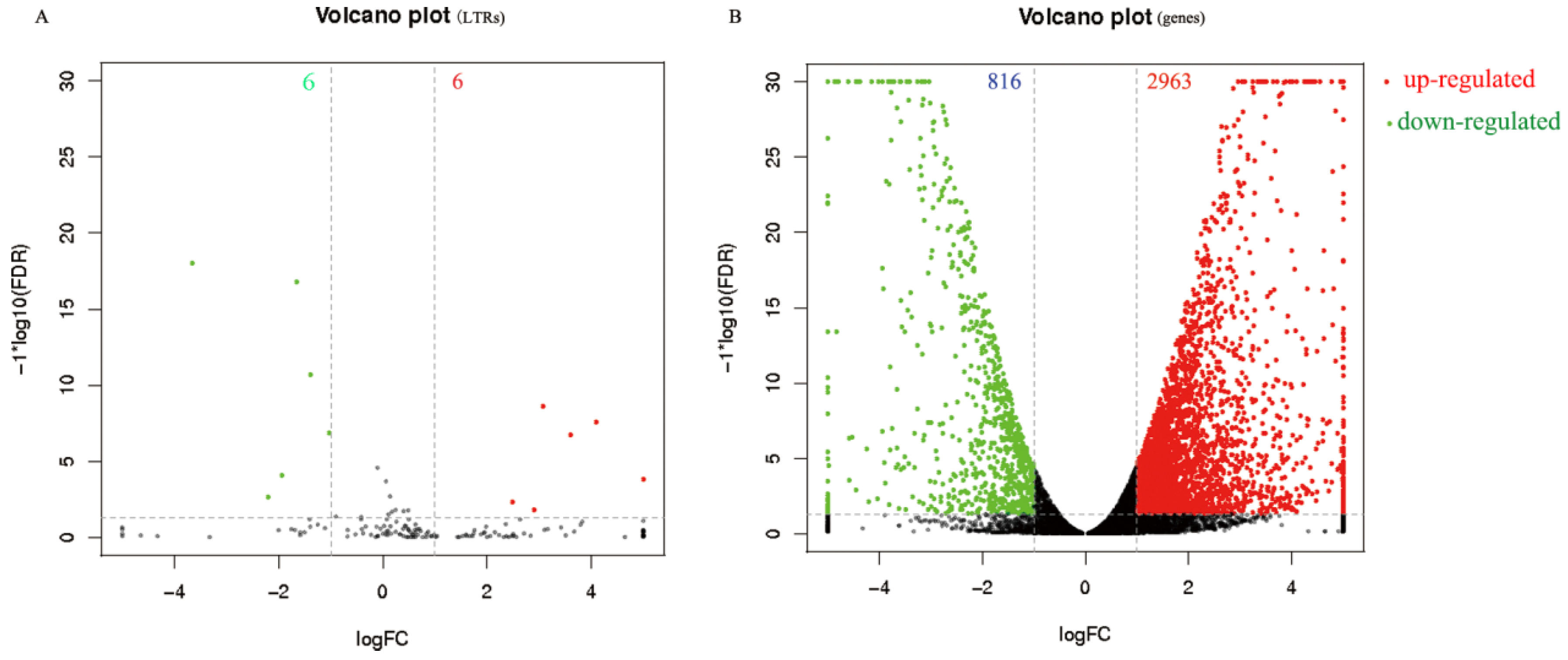
3.5. Transcriptome Analysis of LTR-Retrotransposons in A. pernyi Larval Midgut Samples
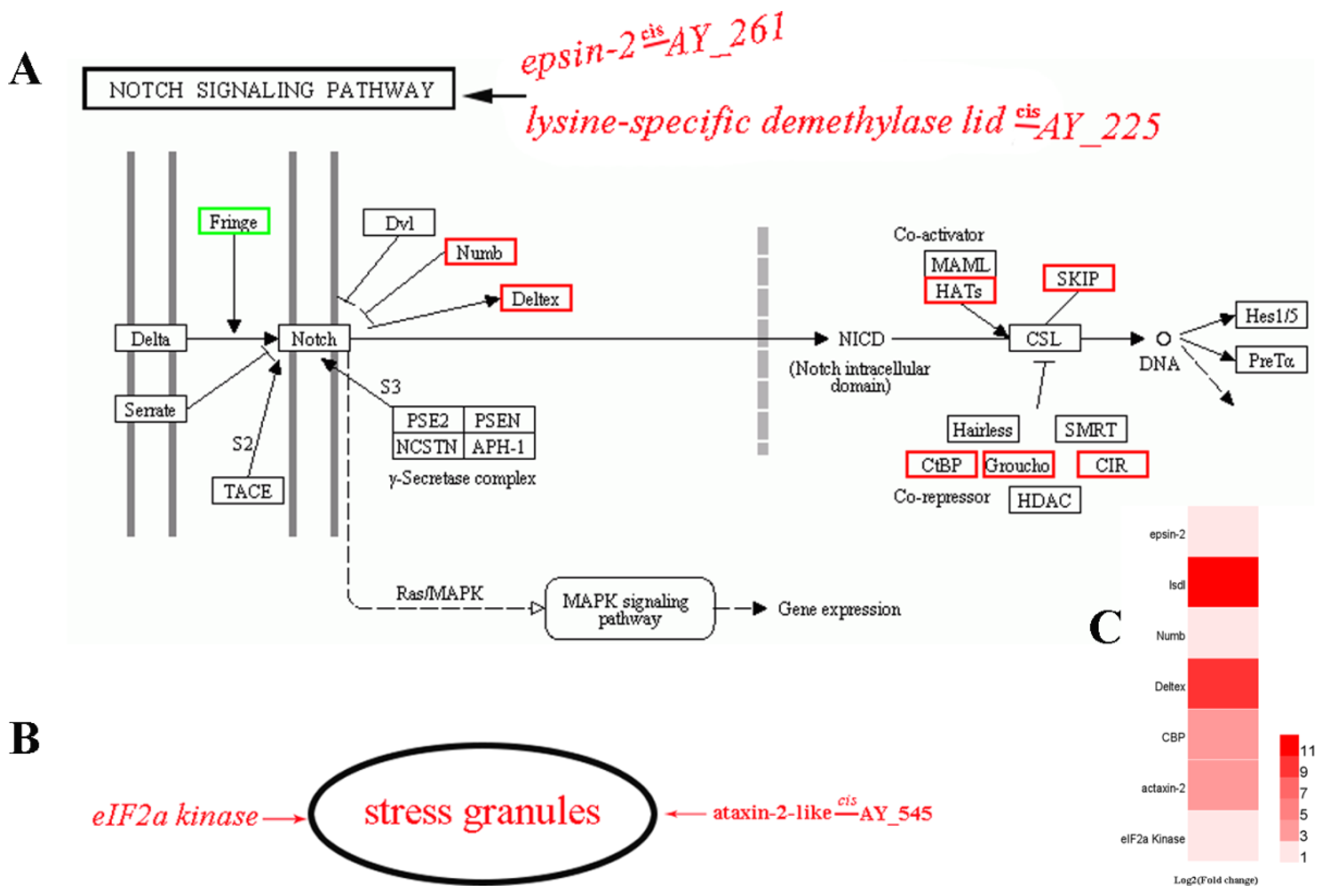
3.6. Bioinformatics Analysis of Notch Signaling and Stress Granule (SG) Regulation Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, J.; Han, D. Safety evaluation of protein of silkworm (Antheraea pernyi) pupae. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Boil. Res. Assoc. 2006, 44, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, Z.; Lin, L.; Terenius, O. Antheraea pernyi (Lepidoptera: Saturniidae) and Its Importance in Sericulture, Food Consumption, and Traditional Chinese Medicine. J. Econ. Èntomol. 2017, 110, 1404–1411. [Google Scholar] [CrossRef] [PubMed]
- Li, X.S.; Wang, G.B.; Sun, Y.; Liu, W.; He, Y.Z.; Wang, F.C.; Jiang, Y.R.; Qin, L. Transcriptome Analysis of the Midgut of the Chinese Oak Silkworm Antheraea pernyi Infected with Antheraea pernyi Nucleopolyhedrovirus. PLoS ONE 2016, 11, e0165959. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Chen, K.P.; Zhang, Z.; Yao, Q.; Gao, G.T.; Zhao, Y. Phylogenetic analysis of Nosema antheraeae (Microsporidia) isolated from Chinese oak silkworm, Antheraea pernyi. J. Eukaryot. Microbiol. 2006, 53, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, Y.; Wang, Y.; Li, X.; Yang, R.; Yu, Z.; Qin, L. The Toll Signaling Pathway in the Chinese Oak Silkworm, Antheraea pernyi: Innate Immune Responses to Different Microorganisms. PLoS ONE 2016, 11, e0160200. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Y.; Liu, W.; Zhou, J.L.; Zeng, J.; Wang, X.H.; Jiang, Y.R.; Li, D.H.; Qin, L. Upregulation of a Trypsin-Like Serine Protease Gene in Antheraea pernyi (Lepidoptera: Saturniidae) Strains Exposed to Different Pathogens. J. Econ. Èntomol. 2017, 110, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhu, S.; Wang, L.; Yu, F.; Shen, W. Cloning and sequence analysis of the Antheraea pernyi nucleopolyhedrovirus gp64 gene. J. Biosci. 2005, 30, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Federici, B.A.; Hice, R.H. Organization and molecular characterization of genes in the polyhedrin region of the Anagrapha falcifera multinucleocapsid NPV. Arch. Virol. 1997, 142, 333–348. [Google Scholar] [CrossRef]
- Havecker, E.R.; Gao, X.; Voytas, D.F. The diversity of LTR retrotransposons. Genome Boil. 2004, 5, 225. [Google Scholar] [CrossRef][Green Version]
- Nefedova, L.; Kim, A. Mechanisms of LTR-Retroelement Transposition: Lessons from Drosophila melanogaster. Viruses 2017, 9. [Google Scholar] [CrossRef]
- Bolisetty, M.; Blomberg, J.; Benachenhou, F.; Sperber, G.; Beemon, K. Unexpected diversity and expression of avian endogenous retroviruses. mBio 2012, 3, e00344-12. [Google Scholar] [CrossRef] [PubMed]
- Mommert, M.; Tabone, O.; Oriol, G.; Cerrato, E.; Guichard, A.; Naville, M.; Fournier, P.; Volff, J.N.; Pachot, A.; Monneret, G.; et al. LTR-retrotransposon transcriptome modulation in response to endotoxin-induced stress in PBMCs. BMC Genom. 2018, 19, 522. [Google Scholar] [CrossRef]
- Hurst, T.P.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Balada, E.; Vilardell-Tarres, M.; Ordi-Ros, J. Implication of human endogenous retroviruses in the development of autoimmune diseases. Int. Rev. Immunol. 2010, 29, 351–370. [Google Scholar] [CrossRef]
- Kim, S.R.; Kwak, W.; Kim, H.; Caetano-Anolles, K.; Kim, K.Y.; Kim, S.B.; Choi, K.H.; Kim, S.W.; Hwang, J.S.; Kim, M.; et al. Genome sequence of the Japanese oak silk moth, Antheraea yamamai: The first draft genome in the family Saturniidae. GigaScience 2018, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yukuhiro, K.; Kanda, T.; Tamura, T. Preferential codon usage and two types of repetitive motifs in the fibroin gene of the Chinese oak silkworm, Antheraea pernyi. Insect Mol. Boil. 1997, 6, 89–95. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Kurtz, S.; Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Steinbiss, S.; Willhoeft, U.; Gremme, G.; Kurtz, S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009, 37, 7002–7013. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: the protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Wang, X.; Ren, F.; Zhang, N.; Zhou, Y.; Sun, J. Genome-Wide Characterization of Endogenous Retroviruses in Bombyx mori Reveals the Relatives and Activity of env Genes. Front. Microbiol. 2018, 9, 1732. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Boil. 2013, 14, R36. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Griffiths, D.J. Endogenous retroviruses in the human genome sequence. Genome Boil. 2001, 2, REVIEWS1017. [Google Scholar]
- De Souza, F.S.; Franchini, L.F.; Rubinstein, M. Exaptation of transposable elements into novel cis-regulatory elements: Is the evidence always strong? Mol. Boil. Evol. 2013, 30, 1239–1251. [Google Scholar] [CrossRef]
- Li, Z.; Ouyang, H.; Zheng, M.; Cai, B.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. Integrated Analysis of Long Non-coding RNAs (LncRNAs) and mRNA Expression Profiles Reveals the Potential Role of LncRNAs in Skeletal Muscle Development of the Chicken. Front. Physiol. 2016, 7, 687. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Li, X.; Qin, L. The origin and dispersal of the domesticated Chinese oak silkworm, Antheraea pernyi, in China: a reconstruction based on ancient texts. J. Insect Sci. 2010, 10, 180. [Google Scholar] [CrossRef]
- Bowen, N.J.; McDonald, J.F. Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res. 2001, 11, 1527–1540. [Google Scholar] [CrossRef]
- Teysset, L.; Burns, J.C.; Shike, H.; Sullivan, B.L.; Bucheton, A.; Terzian, C. A Moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J. Virol. 1998, 72, 853–856. [Google Scholar]
- Llorens, J.V.; Clark, J.B.; Martinez-Garay, I.; Soriano, S.; de Frutos, R.; Martinez-Sebastian, M.J. Gypsy endogenous retrovirus maintains potential infectivity in several species of Drosophilids. BMC Evol. Boil. 2008, 8, 302. [Google Scholar] [CrossRef]
- Leblanc, P.; Desset, S.; Giorgi, F.; Taddei, A.R.; Fausto, A.M.; Mazzini, M.; Dastugue, B.; Vaury, C. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J. Virol. 2000, 74, 10658–10669. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Henikoff, S.; Eickbush, T.H. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000, 10, 1307–1318. [Google Scholar] [CrossRef]
- Rohrmann, G.F.; Karplus, P.A. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol. Boil. 2001, 1, 1. [Google Scholar]
- Cohen, C.J.; Lock, W.M.; Mager, D.L. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 2009, 448, 105–114. [Google Scholar] [CrossRef]
- Mangeney, M.; de Parseval, N.; Thomas, G.; Heidmann, T. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J. Gen. Virol. 2001, 82, 2515–2518. [Google Scholar] [CrossRef]
- Santoni, F.A.; Guerra, J.; Luban, J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 2012, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Aswad, A.; Katzourakis, A. Paleovirology and virally derived immunity. Trends Ecol. Evol. 2012, 27, 627–636. [Google Scholar] [CrossRef]
- Katzourakis, A.; Aswad, A. Evolution: Endogenous Viruses Provide Shortcuts in Antiviral Immunity. Curr. Boil. CB 2016, 26, R427–R429. [Google Scholar] [CrossRef]
- Hurst, T.P.; Magiorkinis, G. Activation of the innate immune response by endogenous retroviruses. J. Gen. Virol. 2015, 96, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Lekha, G.; Gupta, T.; Awasthi, A.K.; Murthy, G.N.; Trivedy, K.; Ponnuvel, K.M. Genome wide microarray based expression profiles associated with BmNPV resistance and susceptibility in Indian silkworm races of Bombyx mori. Genomics 2015, 106, 393–403. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]
- Gao, J.; Xiong, Y.; Wang, Y.; Wang, Y.; Zheng, G.; Xu, H. Hepatitis B virus X protein activates Notch signaling by its effects on Notch1 and Notch4 in human hepatocellular carcinoma. Int. J. Oncol. 2016, 48, 329–337. [Google Scholar] [CrossRef]
- Fan, Y.; Gao, X.; Chen, J.; Liu, Y.; He, J.J. HIV Tat Impairs Neurogenesis through Functioning As a Notch Ligand and Activation of Notch Signaling Pathway. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 11362–11373. [Google Scholar] [CrossRef]
- Hsiao, H.W.; Liu, W.H.; Wang, C.J.; Lo, Y.H.; Wu, Y.H.; Jiang, S.T.; Lai, M.Z. Deltex1 is a target of the transcription factor NFAT that promotes T cell anergy. Immunity 2009, 31, 72–83. [Google Scholar] [CrossRef]
- Dragan, A.I.; Hargreaves, V.V.; Makeyeva, E.N.; Privalov, P.L. Mechanisms of activation of interferon regulator factor 3: The role of C-terminal domain phosphorylation in IRF-3 dimerization and DNA binding. Nucleic Acids Res. 2007, 35, 3525–3534. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, K.; Yoo, D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 2016, 489, 252–268. [Google Scholar] [CrossRef]
- Armbruester, V.; Sauter, M.; Roemer, K.; Best, B.; Hahn, S.; Nty, A.; Schmid, A.; Philipp, S.; Mueller, A.; Mueller-Lantzsch, N. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J. Virol. 2004, 78, 10310–10319. [Google Scholar] [CrossRef]
- Chen, H.; Ko, G.; Zatti, A.; Di Giacomo, G.; Liu, L.; Raiteri, E.; Perucco, E.; Collesi, C.; Min, W.; Zeiss, C.; et al. Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both epsin 1 and epsin 2 in mice. Proc. Natl. Acad. Sci. USA 2009, 106, 13838–13843. [Google Scholar] [CrossRef]
- Di Stefano, L.; Walker, J.A.; Burgio, G.; Corona, D.F.; Mulligan, P.; Naar, A.M.; Dyson, N.J. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011, 25, 17–28. [Google Scholar] [CrossRef]
- Kaehler, C.; Isensee, J.; Nonhoff, U.; Terrey, M.; Hucho, T.; Lehrach, H.; Krobitsch, S. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS ONE 2012, 7, e50134. [Google Scholar] [CrossRef]
- Malinowska, M.; Niedzwiedzka-Rystwej, P.; Tokarz-Deptula, B.; Deptula, W. Stress granules (SG) and processing bodies (PB) in viral infections. Acta Biochim. Pol. 2016, 63, 183–188. [Google Scholar] [CrossRef]
- McCormick, C.; Khaperskyy, D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017, 17, 647–660. [Google Scholar] [CrossRef]


| Sample Name | Raw Reads | Clean Reads | Total Mapped | All Gene Num |
|---|---|---|---|---|
| Ap_CK1 | 74,427,158 | 71,599,652 | 66.90% | 10,535 |
| Ap_CK2 | 59,939,546 | 57,723,648 | 67.39% | 10,334 |
| Ap_NPV1 | 73,741,696 | 70,342,118 | 64.60% | 11,041 |
| Ap_NPV2 | 64,399,928 | 61,942,502 | 63.31% | 11,152 |
| ID | Ap_CK_fpkm | Ap_NPV_fpkm | log2(FC) | FDR |
|---|---|---|---|---|
| AY_1247_21697_26716 | 0.001 | 0.835 | 9.705632 | 0.000158 |
| AY_602_157004_161798 | 0.355 | 6.095 | 4.101735 | 2.88 × 10−8 |
| AY_20_711178_725997 | 0.17 | 2.07 | 3.606024 | 1.85 × 10−7 |
| AY_34_1108397_1109641 | 110.745 | 934.43 | 3.076845 | 2.57 × 10−9 |
| AY_1054_82728_85933 | 0.43 | 3.23 | 2.909126 | 0.016659 |
| AY_34_288108_295647 | 0.625 | 3.52 | 2.493647 | 0.005137 |
| AY_1024_125454_134463 | 1.165 | 0.575 | −1.0187 | 1.49 × 10−7 |
| AY_874_140748_143694 | 0.9 | 0.37 | −1.2824 | 0.001709 |
| AY_261_133645_148170 | 1 | 0.38 | −1.39593 | 1.94 × 10−11 |
| AY_545_152779_164768 | 12.995 | 4.155 | −1.64504 | 1.65 × 10−17 |
| AY_225_415711_427236 | 0.305 | 0.08 | −1.93074 | 8.78 × 10−5 |
| AY_7_943452_945369 | 0.71 | 0.001 | −9.47168 | 0.001699 |
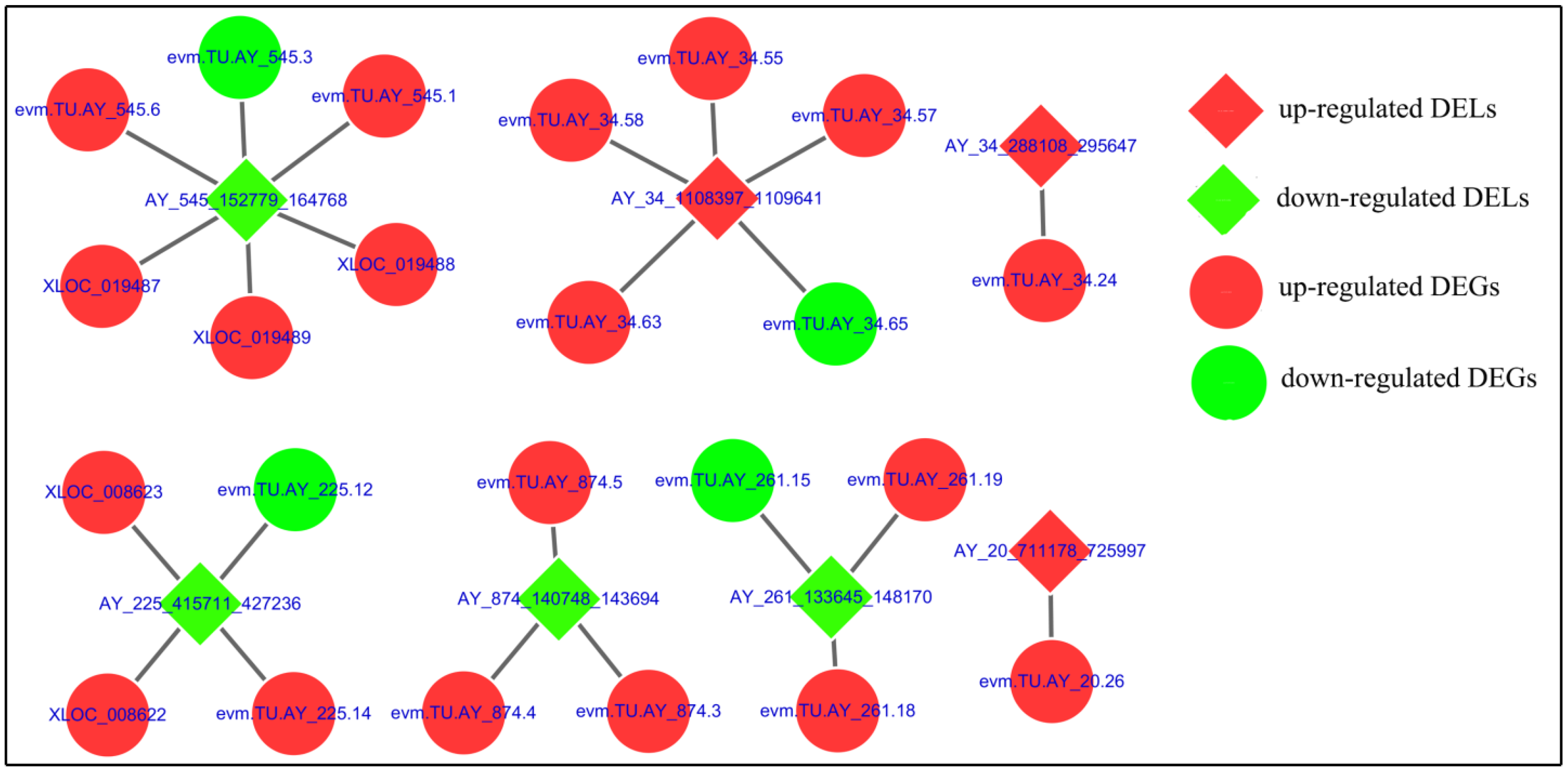
| LTR ID | Gene ID | Gene Name | log2(FC) |
|---|---|---|---|
| AY_20_711178_725997 | evm.TU.AY_20.26 | probable E3 ubiquitin-protein ligase sinah [Papilio xuthus] | 2.269461 |
| AY_225_415711_427236 | evm.TU.AY_225.12 | facilitated trehalose transporter Tret1-like [Bombyx mori] | −2.0158 |
| AY_225_415711_427236 | XLOC_008622 | lysine-specific demethylase lid [Papilio xuthus] | 11.34707 |
| AY_225_415711_427236 | evm.TU.AY_225.14 | lysine-specific demethylase lid isoform X2 [Bombyx mori] | 3.464566 |
| AY_225_415711_427236 | XLOC_008623 | -- | 11.94398 |
| AY_261_133645_148170 | evm.TU.AY_261.15 | glucose dehydrogenase [FAD, quinone]-like [Bombyx mori] | −1.71236 |
| AY_261_133645_148170 | evm.TU.AY_261.19 | protein singed [Bombyx mori] | 1.400687 |
| AY_261_133645_148170 | evm.TU.AY_261.18 | epsin-2 [Amyelois transitella] | 1.159083 |
| AY_34_288108_295647 | evm.TU.AY_34.24 | endonuclease and reverse transcriptase-like protein [Bombyx mori] | 1.318608 |
| AY_34_1108397_1109641 | evm.TU.AY_34.65 | protein transport protein Sec61 subunit alpha isoform 2 [Papilio machaon] | −1.35623 |
| AY_34_1108397_1109641 | evm.TU.AY_34.55 | tyrosine-protein kinase CSK isoform X1 [Bombyx mori] | 2.137612 |
| AY_34_1108397_1109641 | evm.TU.AY_34.57 | DET1 homolog [Amyelois transitella] | 1.614551 |
| AY_34_1108397_1109641 | evm.TU.AY_34.58 | polyubiquitin-B [Drosophila bipectinata] | 2.66362 |
| AY_34_1108397_1109641 | evm.TU.AY_34.63 | midnolin [Papilio machaon] | 1.787524 |
| AY_545_152779_164768 | XLOC_019487 | protein split ends isoform X1 [Amyelois transitella] | 5.019194 |
| AY_545_152779_164768 | XLOC_019488 | -- | 3.989946 |
| AY_545_152779_164768 | XLOC_019489 | protein split ends isoform X1 [Amyelois transitella] | 4.9572955 |
| AY_545_152779_164768 | evm.TU.AY_545.6 | ataxin-2-like protein [Bombyx mori] | 3.027068 |
| AY_545_152779_164768 | evm.TU.AY_545.3 | malate dehydrogenase, mitochondrial [Amyelois transitella] | −1.42245 |
| AY_545_152779_164768 | evm.TU.AY_545.1 | nuclear receptor corepressor 1 [Bombyx mori] | 4.495695 |
| AY_874_140748_143694 | evm.TU.AY_874.5 | reverse transcriptase [Danaus plexippus] | 9.748193 |
| AY_874_140748_143694 | evm.TU.AY_874.3 | zinc finger protein OZF-like [Papilio xuthus] | 2.395929 |
| AY_874_140748_143694 | evm.TU.AY_874.4 | zinc finger protein 431-like [Bombyx mori] | 2.895531 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, M.; Ren, F.; Zhou, Y.; Zhang, N.; Lu, Q.; Swevers, L.; Sun, J. Correlation in Expression between LTR Retrotransposons and Potential Host Cis-Targets during Infection of Antherea pernyi with ApNPV Baculovirus. Viruses 2019, 11, 421. https://doi.org/10.3390/v11050421
Feng M, Ren F, Zhou Y, Zhang N, Lu Q, Swevers L, Sun J. Correlation in Expression between LTR Retrotransposons and Potential Host Cis-Targets during Infection of Antherea pernyi with ApNPV Baculovirus. Viruses. 2019; 11(5):421. https://doi.org/10.3390/v11050421
Chicago/Turabian StyleFeng, Min, Feifei Ren, Yaohong Zhou, Nan Zhang, Qiuyuan Lu, Luc Swevers, and Jingchen Sun. 2019. "Correlation in Expression between LTR Retrotransposons and Potential Host Cis-Targets during Infection of Antherea pernyi with ApNPV Baculovirus" Viruses 11, no. 5: 421. https://doi.org/10.3390/v11050421
APA StyleFeng, M., Ren, F., Zhou, Y., Zhang, N., Lu, Q., Swevers, L., & Sun, J. (2019). Correlation in Expression between LTR Retrotransposons and Potential Host Cis-Targets during Infection of Antherea pernyi with ApNPV Baculovirus. Viruses, 11(5), 421. https://doi.org/10.3390/v11050421





