New Method for Differentiation of Granuloviruses (Betabaculoviruses) Based on Real-Time Polymerase Chain Reaction (Real-Time PCR)
Abstract
1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis of Granuloviruses
2.2. Determination of Betabaculovirus Representative Group
2.3. Virus Purification and DNA Isolation
2.4. Granulin, Late Expression Factor-9 and Late Expression Factor-8 Nucleotide Sequence Alignments and Degenerate Primer Design; Amino Acid Alignment of the PCR Products.
2.5. Real-Time PCR Reaction
2.6. Agarose Gel Electrophoresis
3. Results
3.1. Phylogenetic Analysis and the Representative group of Betabaculovirus
3.2. Granulin, Late Expression Factor-9, and Late Expression Factor-8 Nucleotide and Amino Acid Sequence Alignment
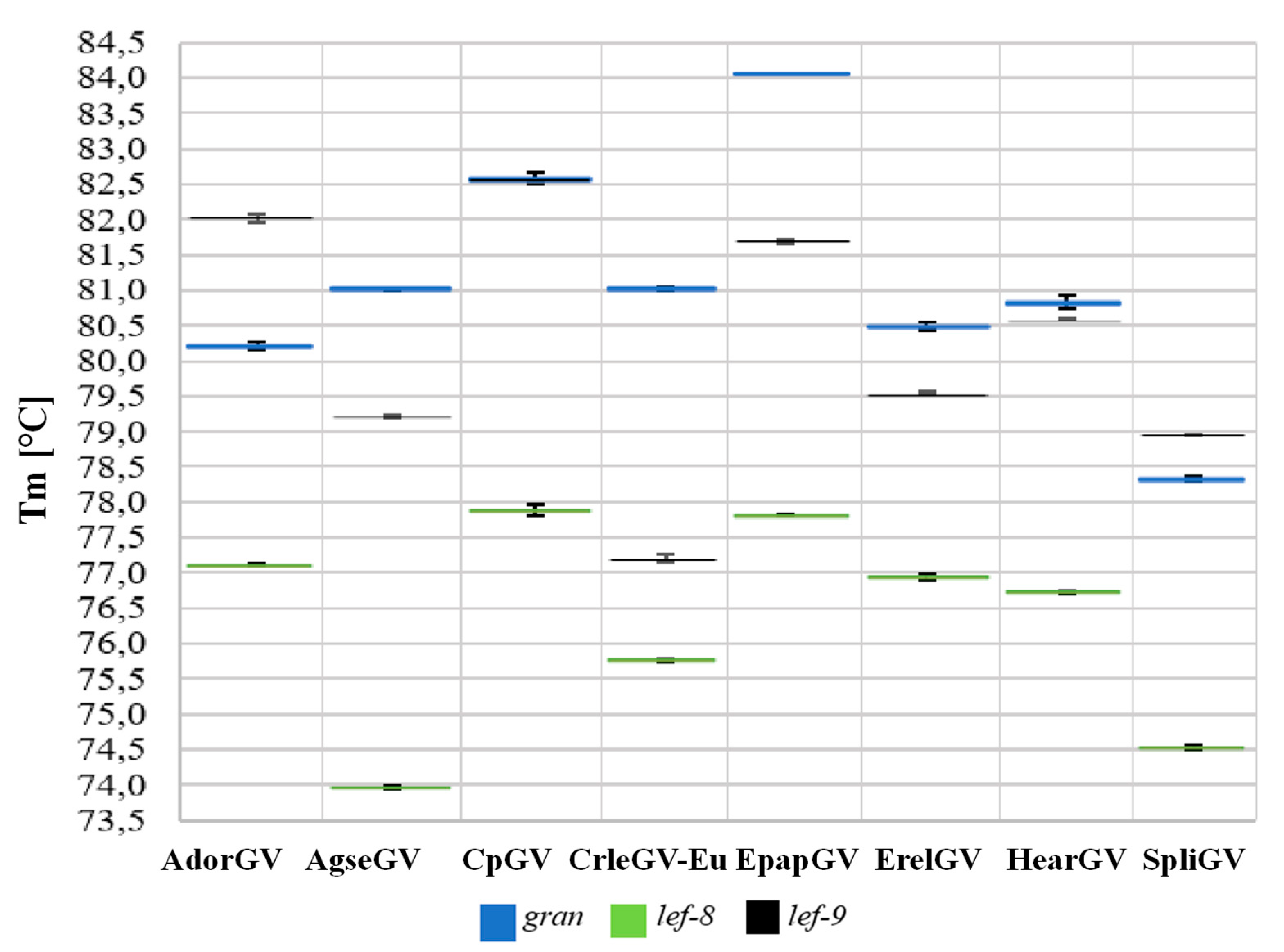
3.3. Real-Time PCR Assay
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Martignioni, M.; Iwai, P.J. A catalogue of viral diseases of insects, mites and thicks. In Microbial Control of Pests and Plant Diseases; Burges, H.D., Ed.; Arcade: London, UK, 1987; pp. 897–911. [Google Scholar]
- Szewczyk, B.; Souza, M.L.; Castro, M.E.B.; Moscardi, M.L.; Moscardi, F. Baculovirus Biopesticides. In Pesticides–Formulation, Effects, Fate; Stoytcheva, M., Ed.; Intech Open: London, UK, 2011; pp. 25–36. [Google Scholar]
- Haase, S.; Sciocco-Cap, A.; Romanowski, V. Baculovirus insecticides in Latin America: Historical overview, current status and future perspectives. Viruses 2015, 7, 2230–2267. [Google Scholar] [CrossRef] [PubMed]
- Blissard, G.W.; Rohrmann, G.F. Baculovirus Diversity and Molecular Biology. Annu. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef]
- Lauzon, H.A.M.; Lucarotti, C.J.; Krell, P.J.; Feng, Q.; Retnakaran, A.; Arif, B.M. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 2004, 78, 7023–7035. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, T.; Ko, R.; Okano, K.; Seong, S.I.; Goto, C.; Maeda, S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 1999, 262, 277–297. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.A.; Blissard, G.W.; Bonning, B.C.; Cory, J.S.; Herniou, E.A.; Rohrmann, G.F.; Theilmann, D.A.; Thiem, S.M.; Vlak, J.M. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006, 151, 1257–1266. [Google Scholar] [CrossRef]
- Herniou, E.A.; Jehle, J.A. Baculovirus phylogeny and evolution. Curr. Drug Targets 2007, 8, 1043–1050. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Polyhedrin structure. J. Gen. Virol. 1986, 67, 1499–1513. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus Molecular Biology, 3rd ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013. [Google Scholar]
- Federici, B.A. Baculovirus pathogenesis. In The Baculoviruses; Miller, L.K., Ed.; Springer Science & Business Media: New York, NY, USA, 1997; pp. 33–59. [Google Scholar]
- Harrison, R.L.; Popham, H.J.R. Genomic sequence analysis of a granulovirus isolated from the Old World bollworm, Helicoverpa armigera. Virus Genes 2008, 36, 565–581. [Google Scholar] [CrossRef]
- Luque, T.; Finch, R.; Crook, N.; O’Reilly, D.R.; Winstanley, D. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 2001, 82, 2531–2547. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.; Kirkman, W.; Richards, G.; Stephen, P.; Moore, S.D.; Kirkman, W.; Richards, G.I.; Stephen, P.R. The Cryptophlebia Leucotreta Granulovirus—10 Years of Commercial Field Use. Viruses 2015, 7, 1284–1312. [Google Scholar] [CrossRef]
- Tanada, Y.; Kaya, H. Insect Pathology, 1st ed.; Academic Press: Cambridge, MA, USA, 1993; ISBN 978-0-08-092625-4. [Google Scholar]
- Ferrelli, M.L.; Salvador, R.; Biedma, M.E.; Berretta, M.F.; Haase, S.; Sciocco-Cap, A.; Ghiringhelli, P.D.; Romanowski, V. Genome of Epinotia aporema granulovirus (EpapGV), a polyorganotropic fast killing betabaculovirus with a novel thymidylate kinase gene. BMC Genom. 2012, 13, 548. [Google Scholar] [CrossRef]
- Federici, B.A.; Stern, V.M. Replication and occlusion of a granulosis virus in larval and adult midgut epithelium of the western grapeleaf skeletonizer, Harrisina brillians. J. Invertebr. Pathol. 1990, 56, 401–414. [Google Scholar] [CrossRef]
- Bideshi, D.K.; Bigot, Y.; Federici, B.A. Molecular characterization and phylogenetic analysis of the Harrisina brillians granulovirus granulin gene. Arch. Virol. 2000, 145, 1933–1945. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Zhu, Z.; Liu, X.; Hou, D.; Wang, J.; Zhang, L.; Wang, M.; Kou, Z.; Wang, H.; Deng, F.; et al. The Complete Genome of a New Betabaculovirus from Clostera anastomosis. PLoS ONE 2015, 10, e0132792. [Google Scholar] [CrossRef] [PubMed]
- Miele, S.A.B.; Garavaglia, M.J.; Belaich, M.N.; Ghiringhelli, P.D. Baculovirus: Molecular Insights on Their Diversity and Conservation. Int. J. Evol. Biol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Garavaglia, M.J.; Miele, S.A.B.; Iserte, J.A.; Belaich, M.N.; Ghiringhelli, P.D. The ac53, ac78, ac101, and ac103 Genes Are Newly Discovered Core Genes in the Family Baculoviridae. J. Virol. 2012, 86, 12069–12079. [Google Scholar] [CrossRef]
- Javed, M.A.; Biswas, S.; Willis, L.G.; Harris, S.; Pritchard, C.; Oers, M.M.; van Donly, B.C.; Erlandson, M.A.; Hegedus, D.D.; Theilmann, D.A. Autographa californica Multiple Nucleopolyhedrovirus AC83 is a per os infectivity factor (PIF) protein required for occlusion-derived virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the PIF complex in ODV envelopes. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Guarino, L.A.; Xu, B.; Jin, J.; Dong, W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 1998, 72, 7985–7991. [Google Scholar]
- Jehle, J.A.; Lange, M.; Wang, H.; Hu, Z.; Wang, Y.; Hauschild, R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 2006, 346, 180–193. [Google Scholar] [CrossRef]
- Liu, H.; Niu, Y.D.; Li, J.; Stanford, K.; McAllister, T.A. Rapid and accurate detection of bacteriophage activity against Escherichia coli O157:H7 by propidium monoazide real-time PCR. Biomed. Res. Int. 2014, 2014, 319351. [Google Scholar] [CrossRef] [PubMed]
- Groner, A. Specificity and safety of baculoviruses. In The Biology of Baculoviruses; Granados, R.R., Federici, B.A., Eds.; CRC Press: Boca Raton, FL, USA, 1986; Volume I, pp. 177–201. [Google Scholar]
- Harrison, R.L.; Mowery, J.D.; Rowley, D.L.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The complete genome sequence of a third distinct baculovirus isolated from the true armyworm, Mythimna unipuncta, contains two copies of the lef-7 gene. Virus Genes 2018, 54, 297–310. [Google Scholar] [PubMed]
- Harrison, R.L.; Bonning, B.C. The nucleopolyhedroviruses of Rachiplusia ou and Anagrapha falcifera are isolates of the same virus. J. Gen. Virol. 1999, 80, 2793–2798. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, R.R.; Maruniak, J.E. Detection and identification of multiple baculoviruses using the polymerase chain reaction (PCR) and restriction endonuclease analysis. J. Virol. Methods 1997, 63, 209–217. [Google Scholar] [CrossRef]
- Lange, M.; Wang, H.; Zhihong, H.; Jehle, J.A. Towards a molecular identification and classification system of lepidopteran-specific baculoviruses. Virology 2004, 325, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, B.; Barski, P.; Sihler, W.; Rabalski, L.; Skrzecz, I.; Hoyos-Carvajal, L.; de Souza, M.L. Detection and identification of baculovirus pesticides by multitemperature single-strand conformational polymorphism. J. Environ. Sci. Health B 2008, 43, 539–545. [Google Scholar] [CrossRef]
- Krejmer-Rabalska, M.; Rabalski, L.; Lobo de Souza, M.; Moore, S.D.; Szewczyk, B. New method for differentiation of granuloviruses (Betabaculoviruses) based on multitemperature single stranded conformational polymorphism. Int. J. Mol. Sci. 2017, 19. [Google Scholar] [CrossRef]
- Kaczanowski, R.; Trzeciak, L.; Kucharczyk, K. Multitemperature single-strand conformation polymorphism. Electrophoresis 2001, 22, 3539–3545. [Google Scholar] [CrossRef]
- Moreau, F.; Fetouchi, R.; Micalessi, I.; Brejeon, V.; Bacon, N.; Jannes, G.; Le Pendeven, C.; Lekbaby, B.; Kremsdorf, D.; Lacau Saint Guily, J.; et al. Detection and genotyping of human papillomavirus by real-time PCR assay. J. Clin. Virol. 2013, 56, 244–249. [Google Scholar] [CrossRef]
- Günes, A.; Marek, A.; Grafl, B.; Berger, E.; Hess, M. Real-time PCR assay for universal detection and quantitation of all five species of fowl adenoviruses (FAdV-A to FAdV-E). J. Virol. Methods 2012, 183, 147–153. [Google Scholar] [CrossRef]
- Arneodo, J.D.; König, G.A.; Berretta, M.F.; Rienzo, J.A.D.; Taboga, O.; Sciocco-Cap, A. Detection and kinetic analysis of Epinotia aporema granulovirus in its lepidopteran host by real-time PCR. Arch. Virol. 2012, 157, 1149–1153. [Google Scholar] [CrossRef]
- Afonso, C.L.; Tulman, E.R.; Lu, Z.; Balinsky, C.A.; Moser, B.A.; Becnel, J.J.; Rock, D.L.; Kutish, G.F. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 2001, 75, 11157–11165. [Google Scholar] [CrossRef] [PubMed]
- Herniou, E.A.; Olszewski, J.A.; O’Reilly, D.R.; Cory, J.S. Ancient coevolution of baculoviruses and their insect hosts. J. Virol. 2004, 78, 3244–3251. [Google Scholar] [CrossRef] [PubMed]
- Wennmann, J.T.; Keilwagen, J.; Jehle, J.A. Baculovirus Kimura two-parameter species demarcation criterion is confirmed by the distances of 38 core gene nucleotide sequences. J. Gen. Virol. 2018, 99, 1307–1320. [Google Scholar] [CrossRef]
- Brito, A.F.; de Braconi, C.T.; Weidmann, M.; Dilcher, M.; Alves, J.M.P.; Gruber, A.; de Zanotto, P.M.A. The Pangenome of the Anticarsia gemmatalis Multiple Nucleopolyhedrovirus (AgMNPV). Genome Biol. Evol. 2015, 8, 94–108. [Google Scholar] [CrossRef]
- Bonsall, D.; Black, S.; Howe, A.Y.; Chase, R.; Ingravallo, P.; Pak, I.; Brown, A.; Smith, D.A.; Bowden, R.; Barnes, E. Characterization of hepatitis C virus resistance to grazoprevir reveals complex patterns of mutations following on-treatment breakthrough that are not observed at relapse. Infect. Drug Resist. 2018, 11, 1119–1135. [Google Scholar] [CrossRef] [PubMed]
- Cory, J.S.; Green, B.M.; Paul, R.K.; Hunter-Fujita, F. Genotypic and phenotypic diversity of a baculovirus population within an individual insect host. J. Invertebr. Pathol. 2005, 89, 101–111. [Google Scholar] [CrossRef]
- Chateigner, A.; Bézier, A.; Labrousse, C.; Jiolle, D.; Barbe, V.; Herniou, E.A. Ultra Deep Sequencing of a Baculovirus Population Reveals Widespread Genomic Variations. Viruses 2015, 7, 3625–3646. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Radtke, P.; Eberle, K.E.; Gueli Alletti, G.; Jehle, J.A. Deciphering Single Nucleotide Polymorphisms and Evolutionary Trends in Isolates of the Cydia pomonella granulovirus. Viruses 2017, 9. [Google Scholar] [CrossRef]
- Van der Merwe, M.; Jukes, M.D.; Rabalski, L.; Knox, C.; Opoku-Debrah, J.K.; Moore, S.D.; Krejmer-Rabalska, M.; Szewczyk, B.; Hill, M.P. Genome Analysis and Genetic Stability of the Cryptophlebia leucotreta Granulovirus (CrleGV-SA) after 15 Years of Commercial Use as a Biopesticide. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Harrison, R.L.; Rowley, D.L.; Mowery, J.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The Complete Genome Sequence of a Second Distinct Betabaculovirus from the True Armyworm, Mythimna unipuncta. PLoS ONE 2017, 12, e0170510. [Google Scholar] [CrossRef] [PubMed]
- Asser-Kaiser, S.; Fritsch, E.; Undorf-Spahn, K.; Kienzle, J.; Eberle, K.E.; Gund, N.A.; Reineke, A.; Zebitz, C.P.W.; Heckel, D.G.; Huber, J.; et al. Rapid emergence of baculovirus resistance in codling moth due to dominant, sex-linked inheritance. Science 2007, 317, 1916–1918. [Google Scholar] [CrossRef] [PubMed]
- Alletti, G.G.; Sauer, A.J.; Weihrauch, B.; Fritsch, E.; Undorf-Spahn, K.; Wennmann, J.T.; Jehle, J.A. Using next generation sequencing to identify and quantify the genetic composition of resistance-breaking commercial isolates of Cydia pomonella granulovirus. Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Asser-Kaiser, S.; Heckel, D.G.; Jehle, J.A. Sex linkage of CpGV resistance in a heterogeneous field strain of the codling moth Cydia pomonella (L.). J. Invertebr. Pathol. 2010, 103, 59–64. [Google Scholar] [CrossRef]
- Asser-Kaiser, S.; Radtke, P.; El-Salamouny, S.; Winstanley, D.; Jehle, J.A. Baculovirus resistance in codling moth (Cydia pomonella L.) caused by early block of virus replication. Virology 2011, 410, 360–367. [Google Scholar] [CrossRef]
- Jehle, J.A.; Schulze-Bopp, S.; Undorf-Spahn, K.; Fritsch, E. Evidence for a second type of resistance against Cydia pomonella granulovirus in field populations of codling moths. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Sauer, A.J.; Schulze-Bopp, S.; Fritsch, E.; Undorf-Spahn, K.; Jehle, J.A. A third type of resistance to Cydia pomonella granulovirus in codling moths shows a mixed Z-linked and autosomal inheritance pattern. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
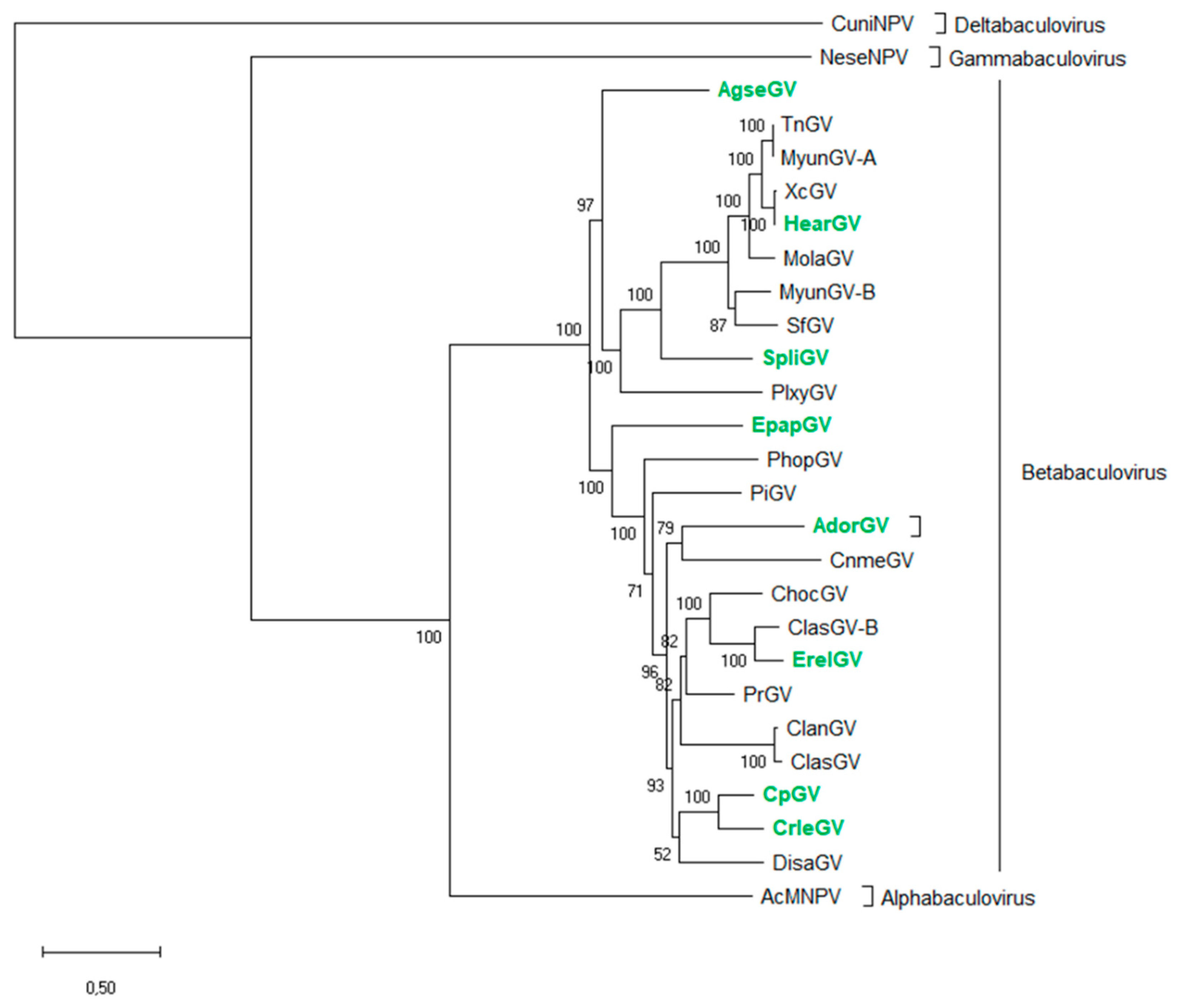
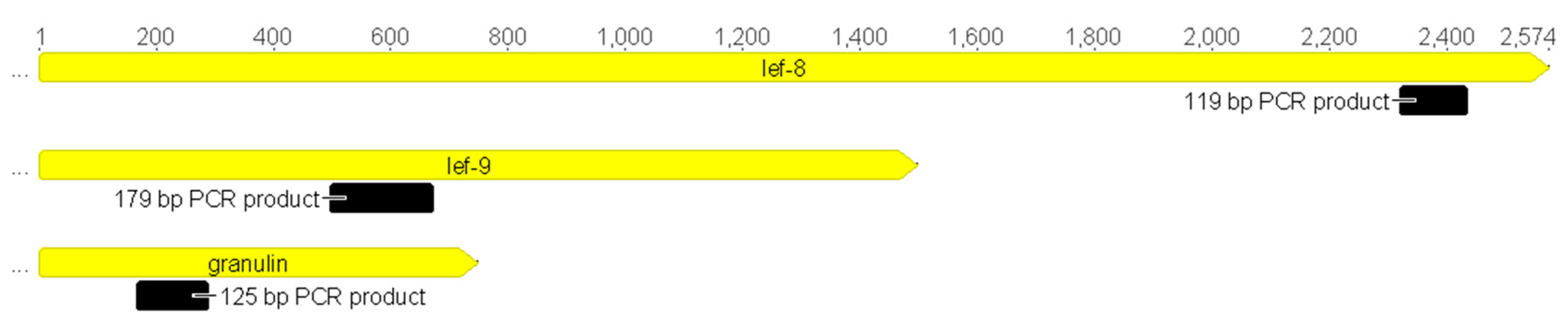

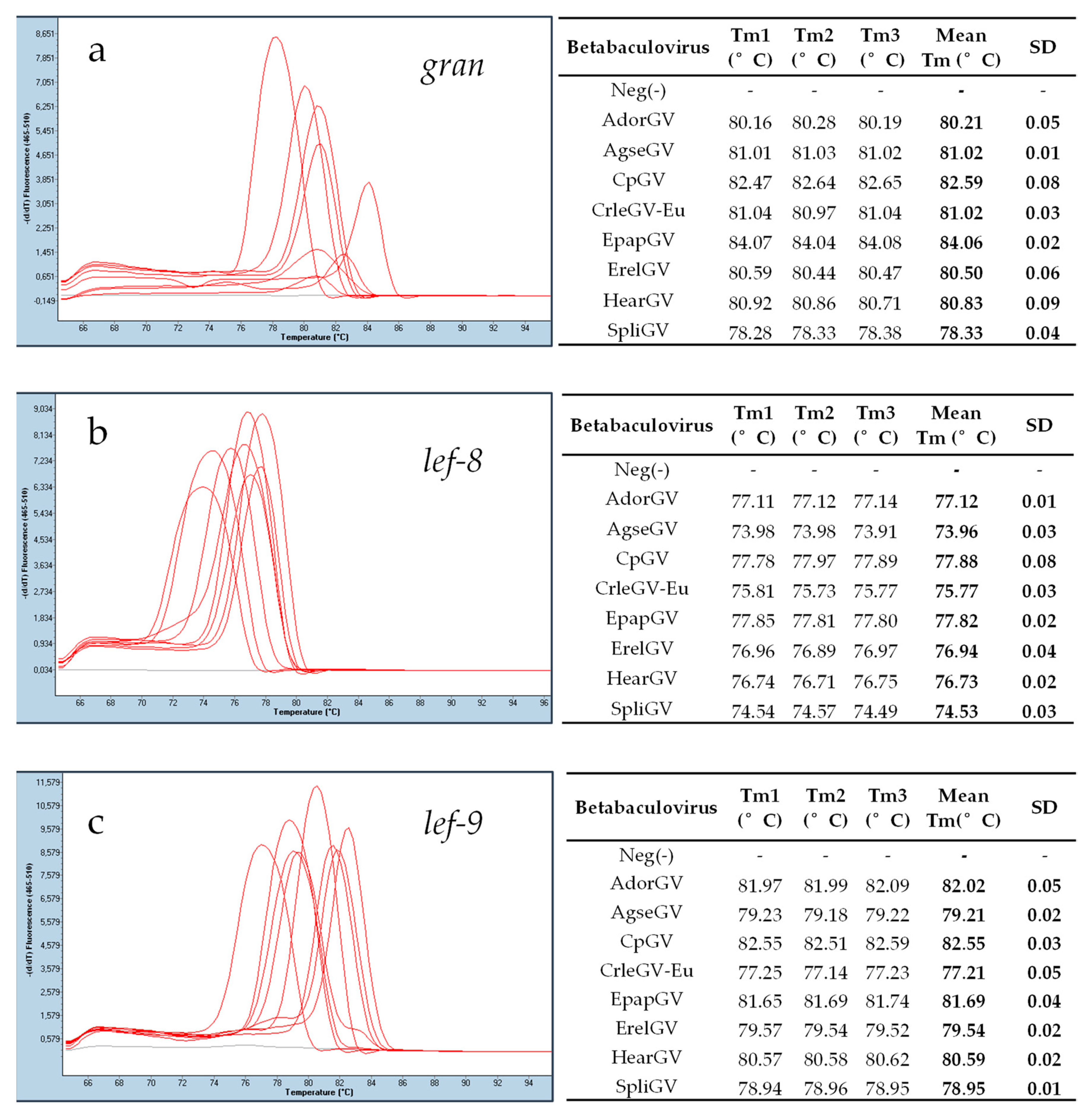
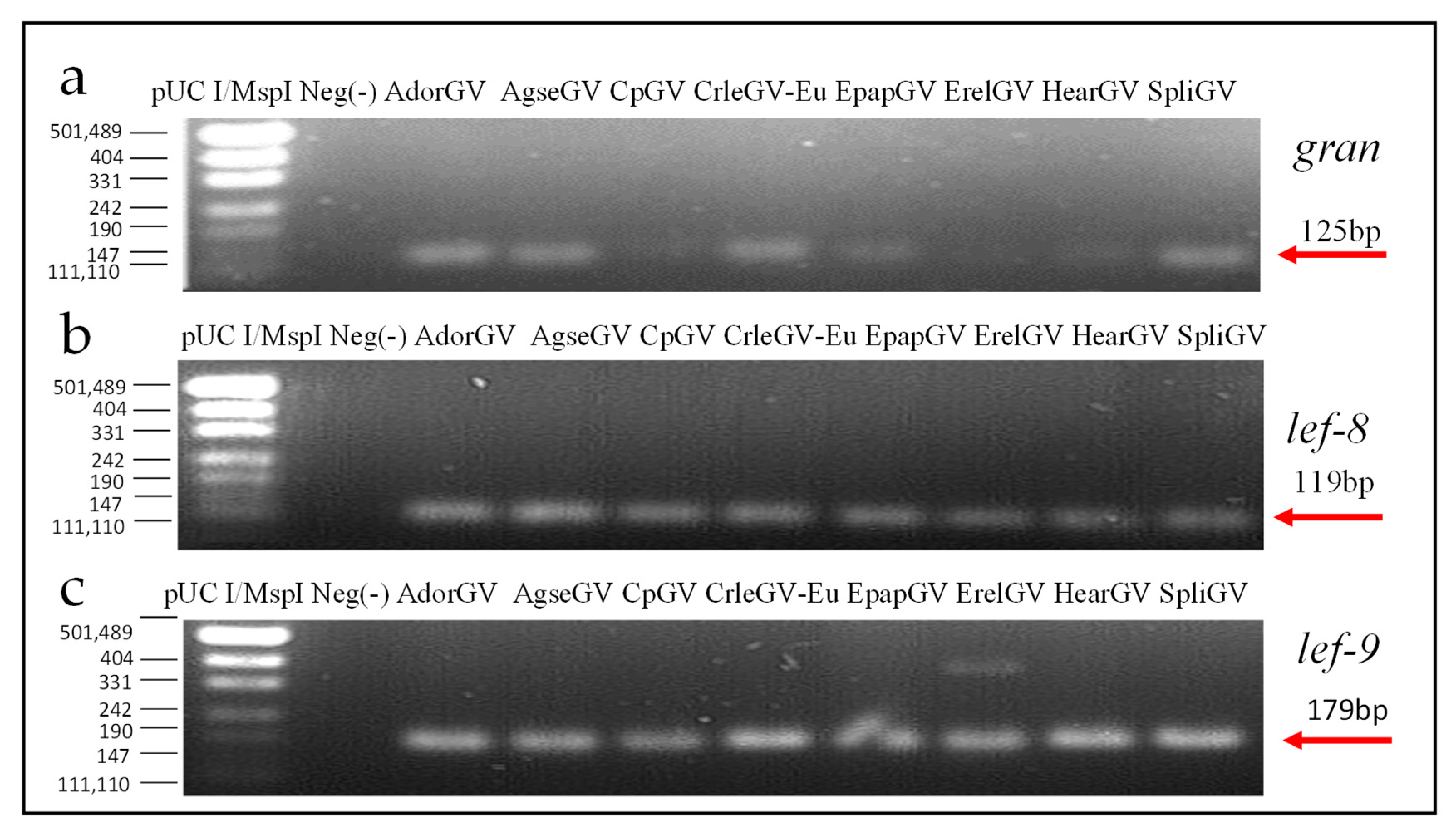
| Abrreviation | Betabaculovirus Name | Host Family | GenBank Accession Number | Genome Size (bp) |
|---|---|---|---|---|
| AdorGV | Adoxophyes orana granulovirus | Tortricidae | AF547984 | 99,657 |
| AgseGV | Agrotis segetum granulovirus | Noctuidae | AY522332 | 131,680 |
| ChocGV | Choristoneura occidentalis granulovirus | Tortricidae | DQ333351 | 104,710 |
| ClanGV | Clostera anachoreta granulovirus | Notodontidae | HQ116624 | 101,487 |
| ClasGV | Clostera anastomosis granulovirus | Notodontidae | KC179784 | 101,818 |
| ClasGV-B | Clostera anastomosis granulovirus B | Notodontidae | KR091910 | 107,439 |
| CnmeGV | Cnaphalocrocis medinalis granulovirus | Crambidae | KU593505 | 111,246 |
| CpGV | Cydia pomonella granulovirus | Tortricidae | U53466 | 123,500 |
| CrleGV | Cryptophlebia leucotreta granulovirus | Tortricidae | AY229987 | 110,907 |
| DisaGV | Diatraea saccharalis granulovirus | Crambidae | KP296186 | 98,392 |
| EpapGV | Epinotia aporema granulovirus | Tortricidae | JN408834 | 119,082 |
| ErelGV | Erinnyis ello granulovirus | Sphingidae | KJ406702 | 102,759 |
| HearGV | Helicoverpa armigera granulovirus | Noctuidae | EU255577 | 169,794 |
| MolaGV | Mocis latipes granulovírus | Noctuidae | KR011718 | 134,272 |
| MyunGV-A | Mythimna unipuncta granulovirus A | Noctuidae | EU678671 | 176,677 |
| MyunGV-B | Mythimna unipuncta granulovírus B | Noctuidae | KX855660 | 144,673 |
| PhopGV | Phthorimaea operculella granulovirus | Gelechiidae | AF499596 | 119,217 |
| PiGV | Plodia interpunctella granulovirus | Pyralidae | KX151395 | 112,536 |
| PlxyGV | Plutella xylostella granulovirus | Plutellidae | AF270937 | 100,999 |
| PrGV | Pieris rapae granulovirus isolate | Pieridae | GQ884143 | 108,592 |
| SfGV | Spodoptera frugiperda granulovirus | Noctuidae | KM371112 | 140,913 |
| SpliGV | Spodoptera litura granulovirus | Noctuidae | DQ288858 | 124,121 |
| TnGV | Trichoplusia ni granulovirus | Noctuidae | KU752557 | 175,360 |
| XcGV | Xestia c-nigrum granulovirus | Noctuidae | AF162221 | 178,733 |
| Isolate | Tm1 (°C) | Tm2 (°C) | Tm3 (°C) | Mean Tm (°C) | SD | |ΔMean Tm| (°C) | SNPs | |
|---|---|---|---|---|---|---|---|---|
| gran | Neg(-) | - | - | - | - | - | - | |
| CrleGV-Eu | 80.97 | 81.04 | 81.04 | 81.02 | 0.03 | 0.95 | 1 | |
| CrleGV-SA | 82.05 | 81.92 | 81.95 | 81.97 | 0.06 | |||
| lef-8 | Neg(-) | - | - | - | - | - | - | |
| CrleGV-Eu | 75.73 | 75.77 | 75.81 | 75.77 | 0.03 | 1.09 | 2 | |
| CrleGV-SA | 76.89 | 76.85 | 76.84 | 76.86 | 0.02 | |||
| lef-9 | Neg(-) | - | - | - | - | - | - | |
| CrleGV-Eu | 77.14 | 77.23 | 77.25 | 77.21 | 0.05 | 0.29 | 3 | |
| CrleGV-SA | 77.53 | 77.45 | 77.53 | 77.50 | 0.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krejmer-Rabalska, M.; Rabalski, L.; Jukes, M.D.; Lobo de Souza, M.; Moore, S.D.; Szewczyk, B. New Method for Differentiation of Granuloviruses (Betabaculoviruses) Based on Real-Time Polymerase Chain Reaction (Real-Time PCR). Viruses 2019, 11, 115. https://doi.org/10.3390/v11020115
Krejmer-Rabalska M, Rabalski L, Jukes MD, Lobo de Souza M, Moore SD, Szewczyk B. New Method for Differentiation of Granuloviruses (Betabaculoviruses) Based on Real-Time Polymerase Chain Reaction (Real-Time PCR). Viruses. 2019; 11(2):115. https://doi.org/10.3390/v11020115
Chicago/Turabian StyleKrejmer-Rabalska, Martyna, Lukasz Rabalski, Michael D. Jukes, Marlinda Lobo de Souza, Sean D. Moore, and Boguslaw Szewczyk. 2019. "New Method for Differentiation of Granuloviruses (Betabaculoviruses) Based on Real-Time Polymerase Chain Reaction (Real-Time PCR)" Viruses 11, no. 2: 115. https://doi.org/10.3390/v11020115
APA StyleKrejmer-Rabalska, M., Rabalski, L., Jukes, M. D., Lobo de Souza, M., Moore, S. D., & Szewczyk, B. (2019). New Method for Differentiation of Granuloviruses (Betabaculoviruses) Based on Real-Time Polymerase Chain Reaction (Real-Time PCR). Viruses, 11(2), 115. https://doi.org/10.3390/v11020115








