Evolution of a Landscape Phage Library in a Mouse Xenograft Model of Human Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
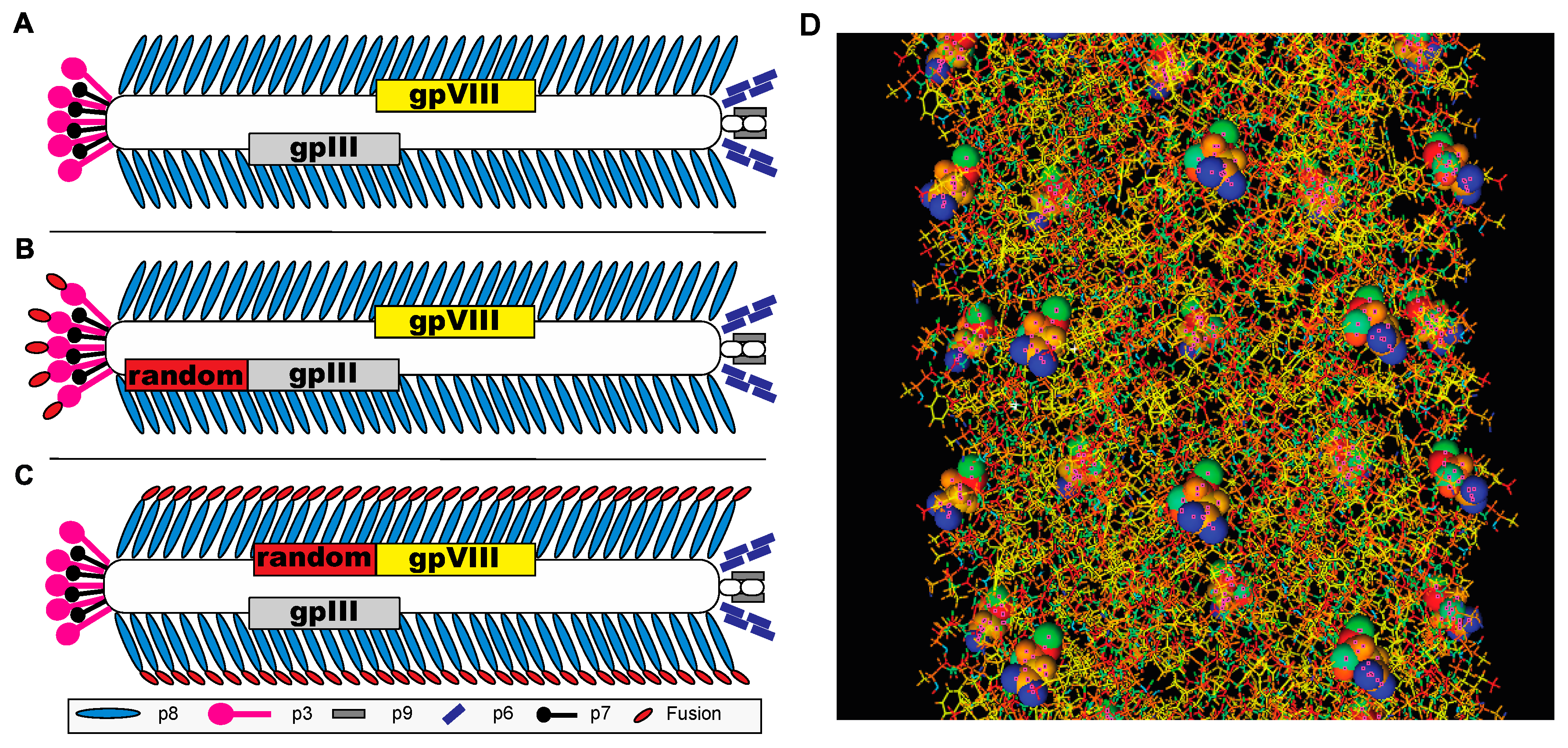
2.1. General Landscape Phage Procedures
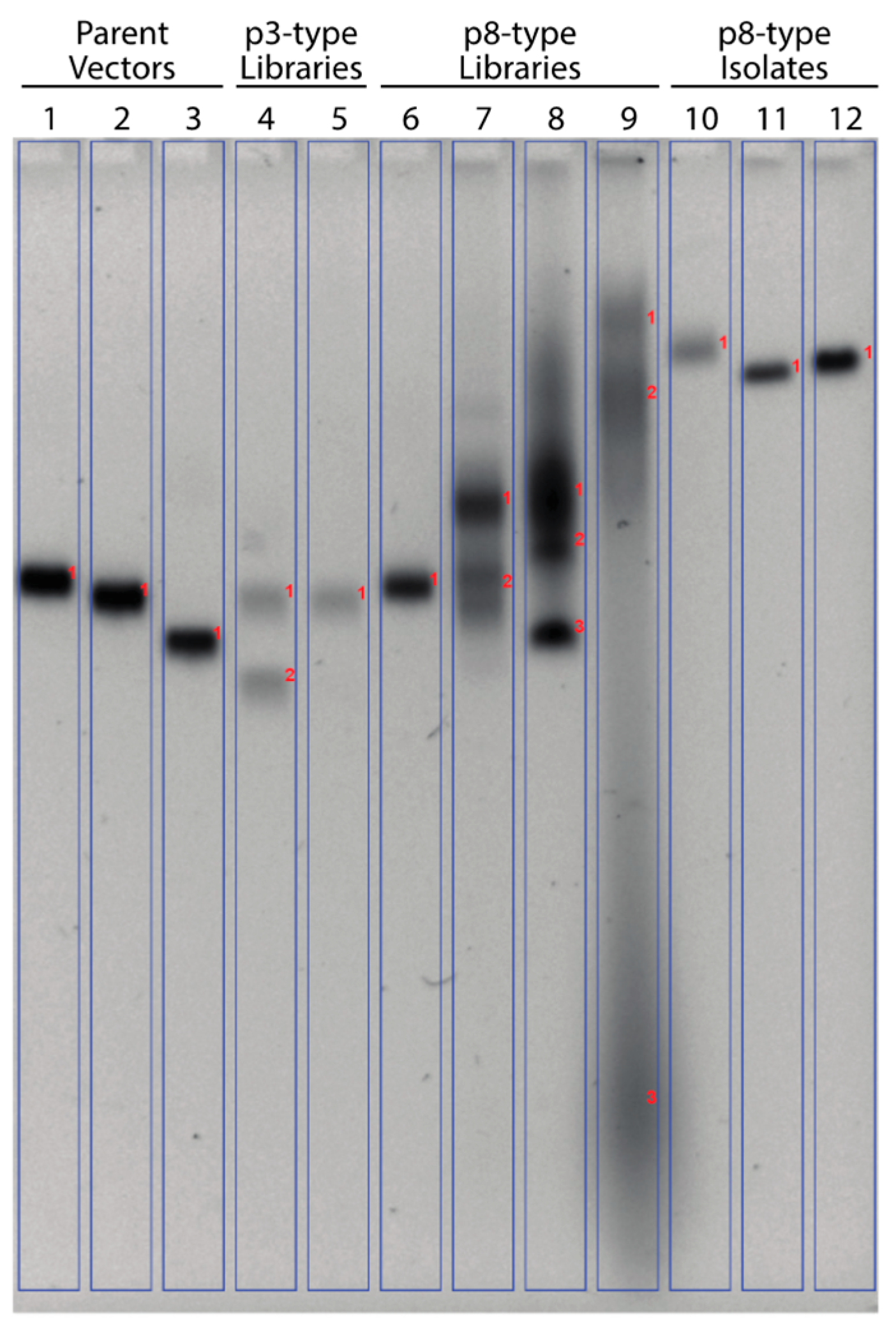
2.2. Agarose Gel Electrophoresis of Phage Libraries
2.3. Animals
2.4. Library Distribution and Tissue Collection
2.5. Tissue Processing and Archiving
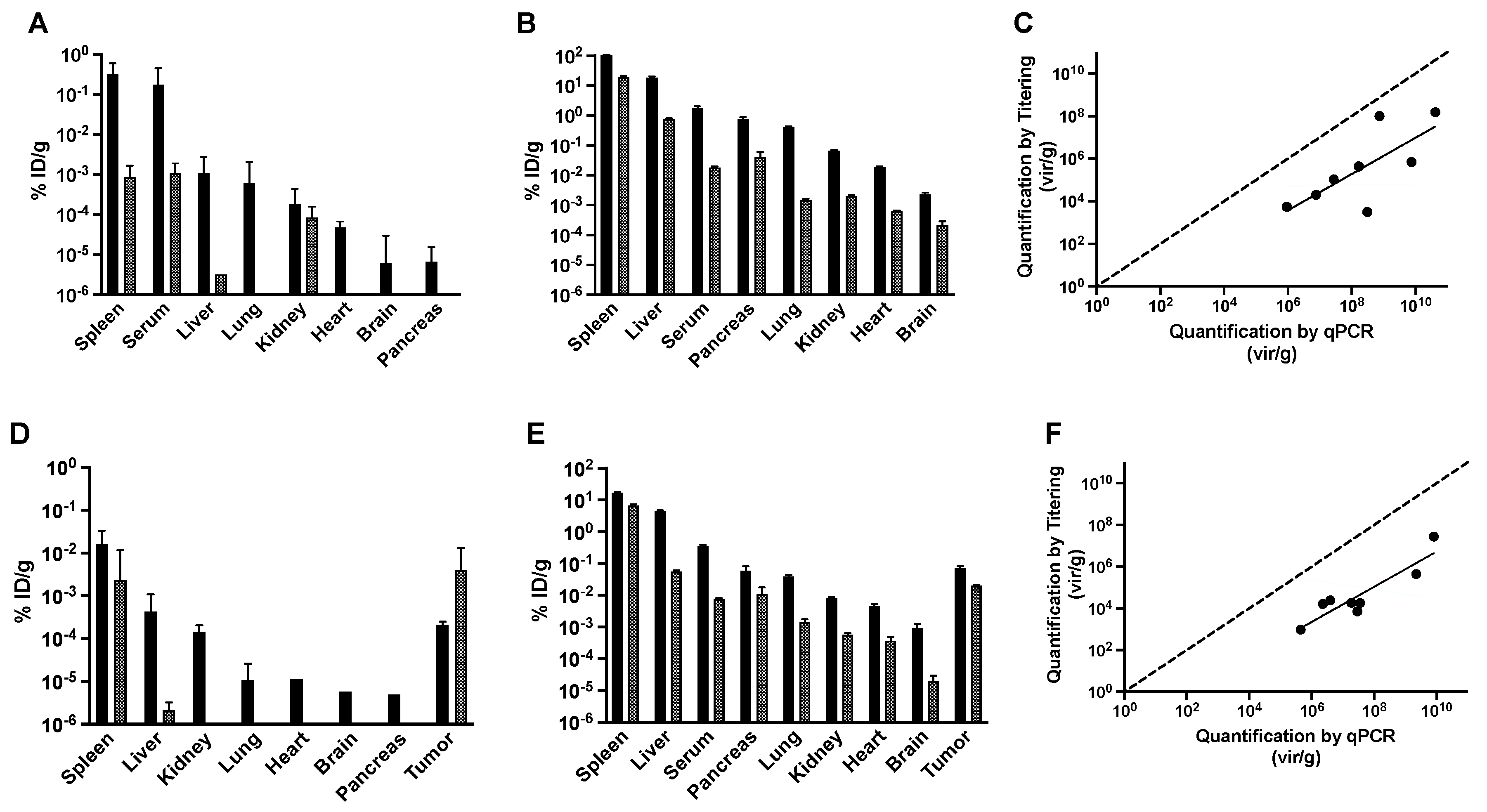
2.6. Quantification of Infectious Phages in Tissues by Titering
2.7. Quantification of Total Phage Particles in Tissues by qPCR
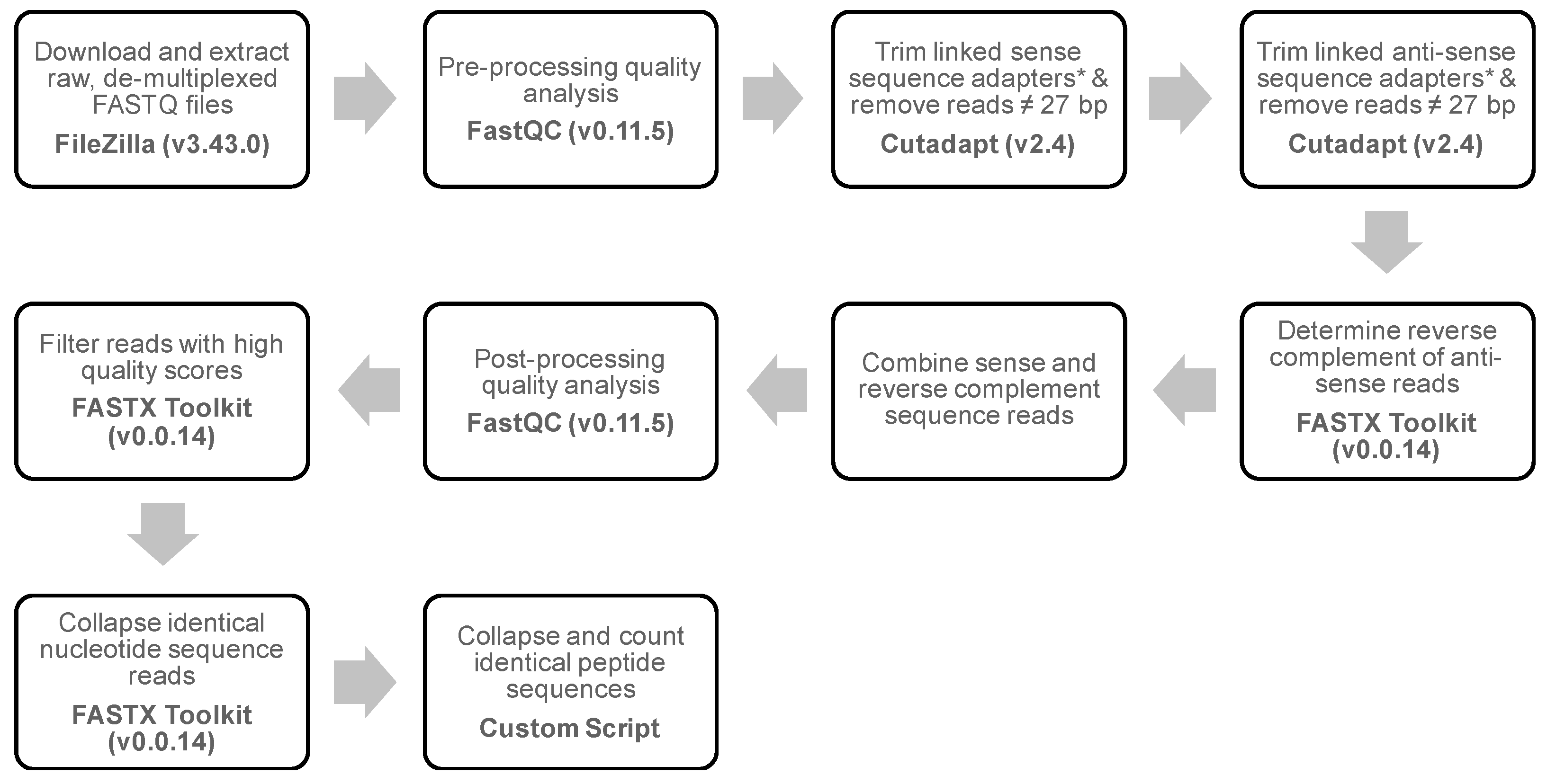
2.8. Next-Generation Sequencing of Phage Populations Enriched in Tissues
2.9. Bioinformatics Analysis of Tissue-Selective Phages
3. Results
3.1. Migration of Phage Display Libraries through a Simple Media
3.2. Tissue Distribution of Landscape Phage Display Library
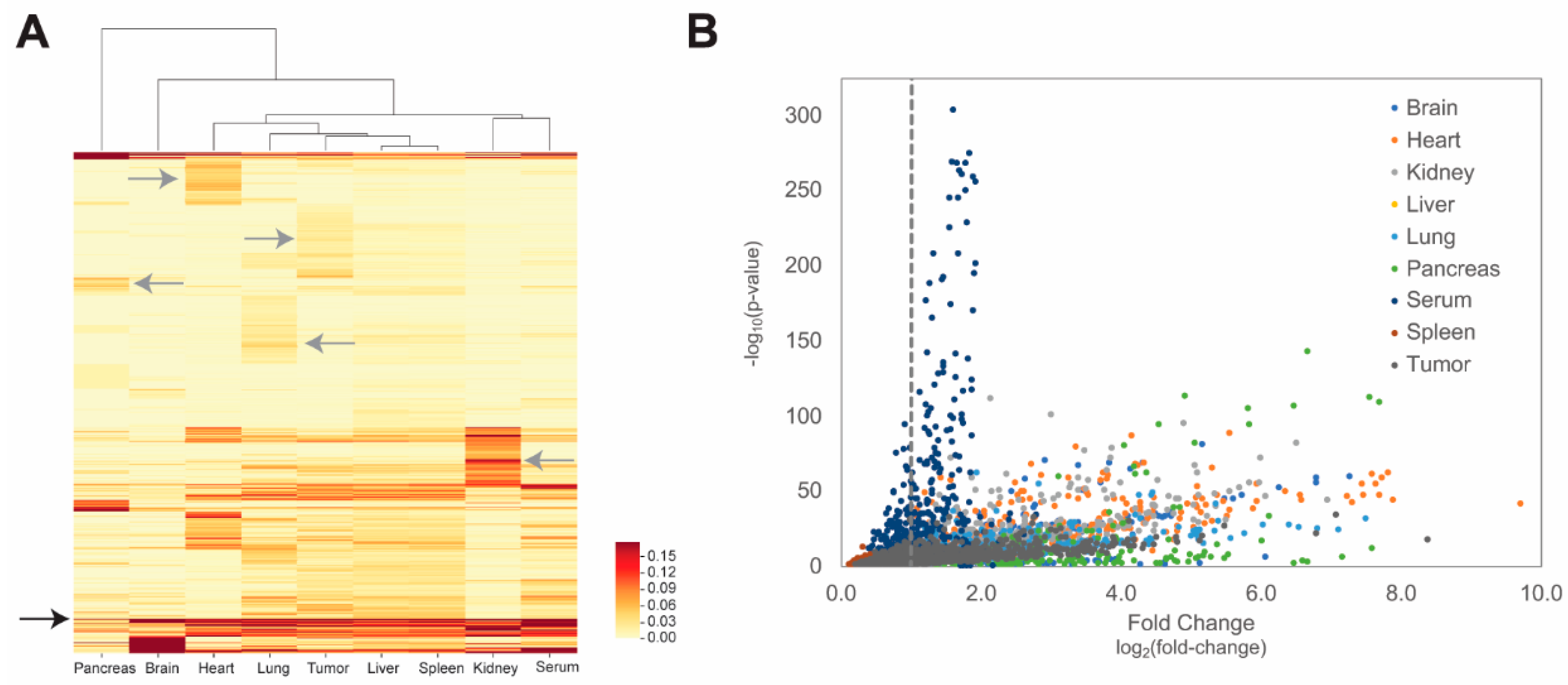
3.3. Molecular Evolution of Tissue-Selective Peptides Displayed on Landscape Phages
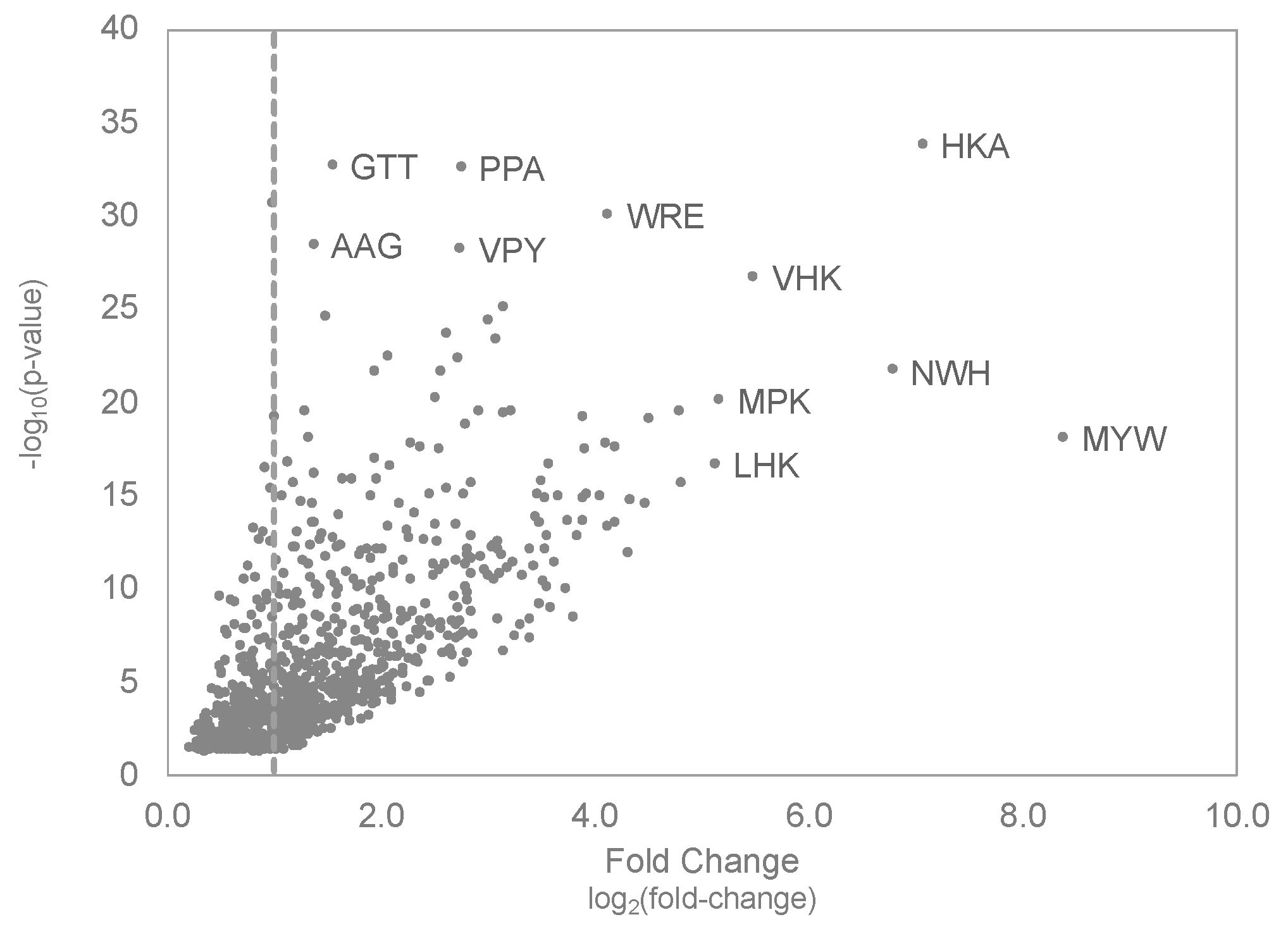
3.4. Molecular Evolution of Tumor-Selective Peptides Displayed on Landscape Phages
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scott, J.K.; Smith, G.P. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, R.; Ruoslahti, E. Organ targeting in vivo using phage display peptide libraries. Nature 1996, 380, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, R.; Koivunen, E.; Ruoslahti, E. A peptide isolated from phage display libraries is a structural and functional mimic of an rgd-binding site on integrins. J. Cell Biol. 1995, 130, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Parmley, S.F.; Smith, G.P. Antibody-selectable filamentous fd phage vectors: Affinity purification of target genes. Gene 1988, 73, 305–318. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.G.; Salamon, J.; Knopp, T.; Ittrich, H.; Adam, G.; Weller, H.; Jung, C. Magnetic particle imaging for in vivo blood flow velocity measurements in mice. Phys. Med. Biol. 2018, 63, 064001. [Google Scholar] [CrossRef]
- Stott, W.T.; Dryzga, M.D.; Ramsey, J.C. Blood-flow distribution in the mouse. J. Appl. Toxicol. 1983, 3, 310–312. [Google Scholar] [CrossRef]
- Rajotte, D.; Arap, W.; Hagedorn, M.; Koivunen, E.; Pasqualini, R.; Ruoslahti, E. From peptides to drugs via phage display. Drug Discov. Today 1998, 3, 370–378. [Google Scholar]
- Pasqualini, R.; Koivunen, E.; Ruoslahti, E. Alpha-v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 1997, 15, 542–546. [Google Scholar] [CrossRef]
- Pasqualini, R.; Koivunen, E.; Kain, R.; Lahdenranta, J.; Sakamoto, M.; Stryhn, A.; Ashmun, R.A.; Shapiro, L.H.; Arap, W.; Ruoslahti, E. Aminopeptidase n is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000, 60, 722–727. [Google Scholar]
- Arap, W.; Pasqualini, R.; Ruoslahti, E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 1998, 279, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Rajotte, D.; Arap, W.; Hagedorn, M.; Koivunen, E.; Pasqualini, R.; Ruoslahti, E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J. Clin. Investig. 1998, 102, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Larimer, B.M.; Thomas, W.D.; Smith, G.P.; Deutscher, S.L. Affinity maturation of an erbb2-targeted spect imaging peptide by in vivo phage display. Mol. Imaging Biol. 2014, 16, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Mi, P.; Cabral, H.; Kataoka, K. Ligand-installed nanocarriers toward precision therapy. Adv. Mater. 2019, 1902604. [Google Scholar] [CrossRef]
- Koivunen, E.; Arap, W.; Rajotte, D.; Lahdenranta, J.; Pasqualini, R. Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med. 1999, 40, 883–888. [Google Scholar]
- Ruoslahti, E. Targeting tumor vasculature with homing peptides from phage display. Semin. Cancer Biol. 2000, 10, 435–442. [Google Scholar] [CrossRef]
- Kolonin, M.; Pasqualini, R.; Arap, W. Molecular addresses in blood vessels as targets for therapy. Curr. Opin. Chem. Biol. 2001, 5, 308–313. [Google Scholar] [CrossRef]
- Arap, W.; Kolonin, M.G.; Trepel, M.; Lahdenranta, J.; Cardo-Vila, M.; Giordano, R.J.; Mintz, P.J.; Ardelt, P.U.; Yao, V.J.; Vidal, C.I.; et al. Steps toward mapping the human vasculature by phage display. Nat. Med. 2002, 8, 121–127. [Google Scholar] [CrossRef]
- Andrieu, J.; Re, F.; Russo, L.; Nicotra, F. Phage-displayed peptides targeting specific tissues and organs. J. Drug Target. 2019, 27, 555–565. [Google Scholar] [CrossRef]
- Grodzinski, P.; Kircher, M.; Goldberg, M.; Gabizon, A. Integrating nanotechnology into cancer care. ACS Nano 2019, 13, 7370–7376. [Google Scholar] [CrossRef]
- Landon, L.A.; Deutscher, S.L. Combinatorial discovery of tumor targeting peptides using phage display. J. Cell Biochem. 2003, 90, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Scodeller, P.; Simon-Gracia, L.; Kopanchuk, S.; Tobi, A.; Kilk, K.; Saalik, P.; Kurm, K.; Squadrito, M.L.; Kotamraju, V.R.; Rinken, A.; et al. Precision targeting of tumor macrophages with a cd206 binding peptide. Sci. Rep. 2017, 7, 14655. [Google Scholar] [CrossRef] [PubMed]
- Nel, A.; Ruoslahti, E.; Meng, H. New insights into “permeability” as in the enhanced permeability and retention effect of cancer nanotherapeutics. ACS Nano 2017, 11, 9567–9569. [Google Scholar] [CrossRef] [PubMed]
- Saalik, P.; Lingasamy, P.; Toome, K.; Mastandrea, I.; Rousso-Noori, L.; Tobi, A.; Simon-Gracia, L.; Hunt, H.; Paiste, P.; Kotamraju, V.R.; et al. Peptide-guided nanoparticles for glioblastoma targeting. J. Control. Release 2019, 308, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Leong, W.; Leong, K.W.; Chen, C.; Zhao, Y. Walking the line: The fate of nanomaterials at biological barriers. Biomaterials 2018, 174, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Babickova, J.; Tothova, L.; Boor, P.; Celec, P. In vivo phage display—A discovery tool in molecular biomedicine. Biotechnol. Adv. 2013, 31, 1247–1259. [Google Scholar] [CrossRef] [PubMed]
- Ledsgaard, L.; Kilstrup, M.; Karatt-Vellatt, A.; McCafferty, J.; Laustsen, A.H. Basics of antibody phage display technology. Toxins (Basel) 2018, 10, 236. [Google Scholar] [CrossRef]
- Kim, E.S. Directed evolution: A historial exploration into an evolutionary experimental system of nanobiotechnology, 1965–2006. Minerva 2008, 46, 463–484. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Smith, G.P. Vectors and modes of display. In Phage Display in Biotechnology and Drug Discovery, 1st ed.; Sidhu, S.S., Geyer, C.R., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 43–74. [Google Scholar]
- Petrenko, V.A.; Smith, G.P.; Gong, X.; Quinn, T. A library of organic landscapes on filamentous phage. Protein Eng. 1996, 9, 797–801. [Google Scholar] [CrossRef]
- Petrenko, V.A. Landscape phage: Evolution from phage display to nanobiotechnology. Viruses 2018, 10. [Google Scholar] [CrossRef]
- Mammen, M.; Choi, S.K.; Whitesides, G.M. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 1998, 37, 2754–2794. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Gillespie, J.W.; Xu, H.; O’Dell, T.; De Plano, L.M. Combinatorial avidity selection of mosaic landscape phages targeted at breast cancer cells-an alternative mechanism of directed molecular evolution. Viruses 2019, 11, 785. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A. Autonomous self-navigating drug-delivery vehicles: From science fiction to reality. Ther. Deliv. 2017, 8, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef] [PubMed]
- Neduva, V.; Russell, R.B. Linear motifs: Evolutionary interaction switches. FEBS Lett. 2005, 579, 3342–3345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davey, N.E.; Van Roey, K.; Weatheritt, R.J.; Toedt, G.; Uyar, B.; Altenberg, B.; Budd, A.; Diella, F.; Dinkel, H.; Gibson, T.J. Attributes of short linear motifs. Mol. Biosyst. 2012, 8, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Ivarsson, Y.; Jemth, P. Affinity and specificity of motif-based protein-protein interactions. Curr. Opin. Struct. Biol. 2019, 54, 26–33. [Google Scholar] [CrossRef]
- Almeida, J.P.; Chen, A.L.; Foster, A.; Drezek, R. In vivo biodistribution of nanoparticles. Nanomedicine (Lond) 2011, 6, 815–835. [Google Scholar] [CrossRef]
- Wei, Y.; Quan, L.; Zhou, C.; Zhan, Q. Factors relating to the biodistribution & clearance of nanoparticles & their effects on in vivo application. Nanomedicine (Lond) 2018, 13, 1495–1512. [Google Scholar]
- Kuzmicheva, G.A.; Jayanna, P.K.; Sorokulova, I.B.; Petrenko, V.A. Diversity and censoring of landscape phage libraries. Protein Eng. Des. Sel. 2009, 22, 9–18. [Google Scholar] [CrossRef]
- Brigati, J.R.; Samoylova, T.I.; Jayanna, P.K.; Petrenko, V.A. Phage display for generating peptide reagents. Curr. Protoc. Protein Sci. 2008, 51, 18.9.1–18.9.27. [Google Scholar]
- Kuzmicheva, G.A.; Jayanna, P.K.; Eroshkin, A.M.; Grishina, M.A.; Pereyaslavskaya, E.S.; Potemkin, V.A.; Petrenko, V.A. Mutations in fd phage major coat protein modulate affinity of the displayed peptide. Protein Eng. Des. Sel. 2009, 22, 631–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.; Dickerson, M.T.; Owen, N.K.; Landon, L.A.; Deutscher, S.L. Biodistribution of filamentous phage peptide libraries in mice. Mol. Biol. Rep. 2004, 31, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The miqe guidelines: Minimum information for publication of quantitative real-time pcr experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. Fastqc: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010; Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 July 2019).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Hannon, G. Fastx-toolkit: Fastq/a Short-Reads Pre-Processing Tools, 0.0.14. 2009. Available online: http://hannonlab.cshl.edu/fastx_toolkit/index.html (accessed on 24 July 2019).
- Chapman, B.A.; Chang, J.T. Biopython: Python tools for computational biology. ACM SIGBIO Newsletter 2000, 20, 15–19. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization, 0.9.0. 2017. Available online: https://seaborn.pydata.org/index.html (accessed on 3 September 2019).
- Kolonin, M.G.; Sun, J.; Do, K.A.; Vidal, C.I.; Ji, Y.; Baggerly, K.A.; Pasqualini, R.; Arap, W. Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB J. 2006, 20, 979–981. [Google Scholar] [CrossRef]
- Jones, E.; Oliphant, T.; Peterson, P. Scipy: Open Source Scientific Tools for Python. 2001. Available online: http://www.scipy.org/ (accessed on 3 September 2019).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B (Methodological) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Specthrie, L.; Bullitt, E.; Horiuchi, K.; Model, P.; Russel, M.; Makowski, L. Construction of a microphage variant of filamentous bacteriophage. J. Mol. Biol. 1992, 228, 720–724. [Google Scholar] [CrossRef]
- Mount, J.D.; Samoylova, T.I.; Morrison, N.E.; Cox, N.R.; Baker, H.J.; Petrenko, V.A. Cell targeted phagemid rescued by preselected landscape phage. Gene 2004, 341, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J. Animal models for studying prevention and treatment of breast cancer. In Animal Models for the Study of Human Disease; Conn, P.M., Ed.; Academic Press: New York, NY, USA, 2013; pp. 997–1018. [Google Scholar]
- Milioli, H.H.; Tishchenko, I.; Riveros, C.; Berretta, R.; Moscato, P. Basal-like breast cancer: Molecular profiles, clinical features and survival outcomes. BMC Med. Genomics 2017, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Toft, D.J.; Cryns, V.L. Minireview: Basal-like breast cancer: From molecular profiles to targeted therapies. Mol. Endocrinol. 2011, 25, 199–211. [Google Scholar] [CrossRef]
- Shultz, L.D.; Sidman, C.L. Genetically determined murine models of immunodeficiency. Annu. Rev. Immunol. 1987, 5, 367–403. [Google Scholar] [CrossRef]
- Krag, D.N.; Shukla, G.S.; Shen, G.P.; Pero, S.; Ashikaga, T.; Fuller, S.; Weaver, D.L.; Burdette-Radoux, S.; Thomas, C. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006, 66, 7724–7733. [Google Scholar] [CrossRef]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef]
- Rahman, L.; Bliskovski, V.; Kaye, F.J.; Zajac-Kaye, M. Evolutionary conservation of a 2-kb intronic sequence flanking a tissue-specific alternative exon in the ptbp2 gene. Genomics 2004, 83, 76–84. [Google Scholar] [CrossRef]
- Shadeo, A.; Lam, W.L. Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res. 2006, 8, R9. [Google Scholar] [CrossRef]
- Dias-Neto, E.; Nunes, D.N.; Giordano, R.J.; Sun, J.; Botz, G.H.; Yang, K.; Setubal, J.C.; Pasqualini, R.; Arap, W. Next-generation phage display: Integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS ONE 2009, 4, e8338. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Paci, E.; Dobson, C.M.; Karplus, M. Three key residues form a critical contact network in a protein folding transition state. Nature 2001, 409, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Cyert, M.S.; Moses, A.M. Short linear motifs—Ex nihilo evolution of protein regulation. Cell Commun. Signal. 2015, 13, 43. [Google Scholar] [CrossRef]
- Kolonin, M.G.; Bover, L.; Sun, J.; Zurita, A.J.; Do, K.A.; Lahdenranta, J.; Cardo-Vila, M.; Giordano, R.J.; Jaalouk, D.E.; Ozawa, M.G.; et al. Ligand-directed surface profiling of human cancer cells with combinatorial peptide libraries. Cancer Res. 2006, 66, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Wu, W.; Xiang, J.; Lv, Y.; Zheng, X.; Chen, Q.; Wang, H.; Wang, B.; Liu, Z.; Ma, F. Hepatocyte culture in autologous decellularized spleen matrix. Organogenesis 2015, 11, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Onufriev, A.V.; Alexov, E. Protonation and pk changes in protein-ligand binding. Q Rev. Biophys. 2013, 46, 181–209. [Google Scholar] [CrossRef]
- Marczynski, M.; Kasdorf, B.T.; Altaner, B.; Wenzler, A.; Gerland, U.; Lieleg, O. Transient binding promotes molecule penetration into mucin hydrogels by enhancing molecular partitioning. Biomater. Sci. 2018, 6, 3373–3387. [Google Scholar] [CrossRef]
- Zamboni, W.C.; Torchilin, V.; Patri, A.K.; Hrkach, J.; Stern, S.; Lee, R.; Nel, A.; Panaro, N.J.; Grodzinski, P. Best practices in cancer nanotechnology: Perspective from nci nanotechnology alliance. Clin. Cancer Res. 2012, 18, 3229–3241. [Google Scholar] [CrossRef]
- Saha, A.; Kim, Y.; Gewirtz, A.D.H.; Jo, B.; Gao, C.; McDowell, I.C.; Consortium, G.T.; Engelhardt, B.E.; Battle, A. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 2017, 27, 1843–1858. [Google Scholar] [CrossRef] [Green Version]
- Mack, K.L.; Phifer-Rixey, M.; Harr, B.; Nachman, M.W. Gene expression networks across multiple tissues are associated with rates of molecular evolution in wild house mice. Genes (Basel) 2019, 10, 225. [Google Scholar] [CrossRef]
- Pierson, E.; Consortium, G.T.; Koller, D.; Battle, A.; Mostafavi, S.; Ardlie, K.G.; Getz, G.; Wright, F.A.; Kellis, M.; Volpi, S.; et al. Sharing and specificity of co-expression networks across 35 human tissues. PLoS Comput. Biol. 2015, 11, e1004220. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Platig, J.; Fagny, M.; Chen, C.Y.; Paulson, J.N.; Lopes-Ramos, C.M.; DeMeo, D.L.; Quackenbush, J.; Glass, K.; Kuijjer, M.L. Understanding tissue-specific gene regulation. Cell Rep. 2017, 21, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Mimmi, S.; Maisano, D.; Quinto, I.; Iaccino, E. Phage display: An overview in context to drug discovery. Trends Pharmacol. Sci. 2019, 40, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.R.; Benoit, D.S.W. In vivo translation of peptide-targeted drug delivery systems discovered by phage display. Bioconjug. Chem. 2018, 29, 2161–2169. [Google Scholar] [CrossRef] [PubMed]
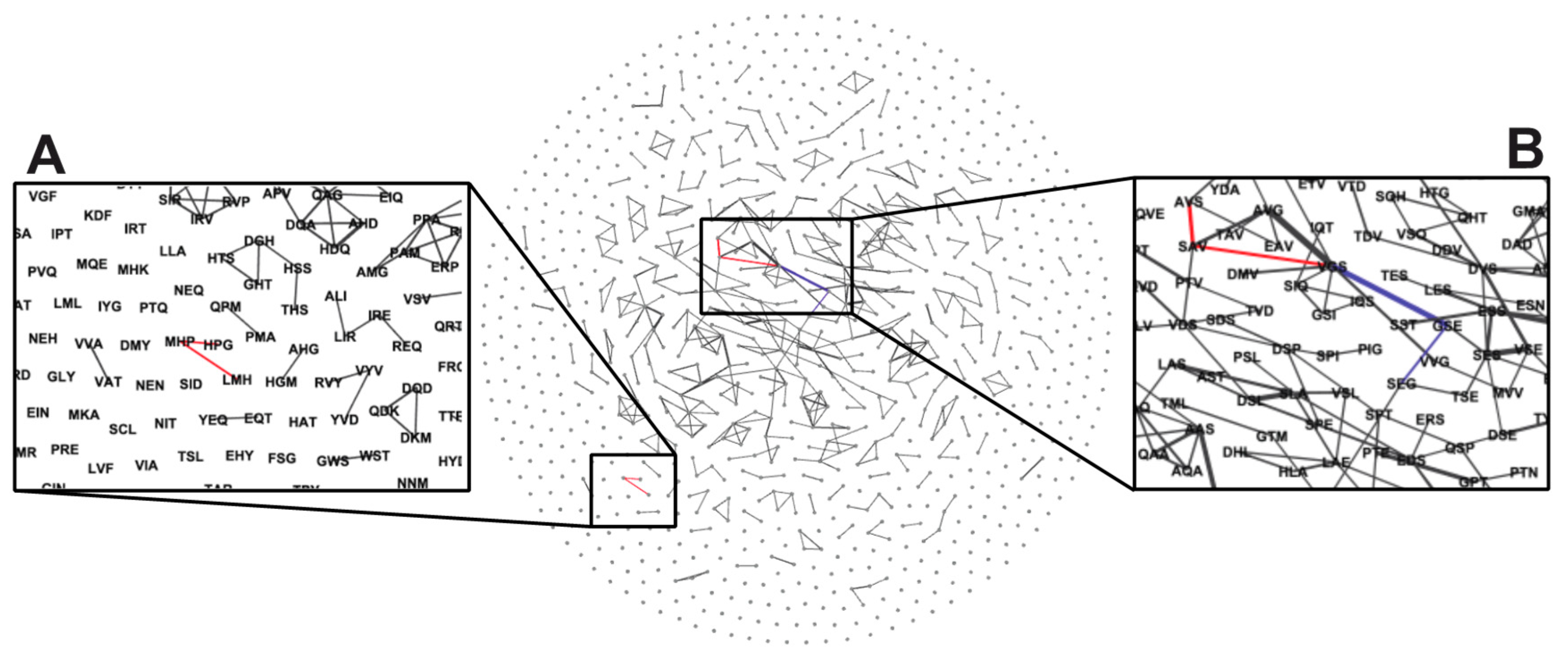
| MYW | LMH/MHP/HPG | VGS/AVS | VGS/SEG |
|---|---|---|---|
| AMYWDRASD | DLMHGPVMD | VGSAVSNEH | ASVGSEGDL |
| DMYWDGASD | DLMHPGAAD | VGSAVSSEH | ASVGSEGST |
| DMYWDKASD | DLMHPGAEG | VTDVGSAVS | DPSLVGSEG |
| DMYWDRADD | DLMHPGAID | DSSLVGSEG | |
| DMYWDRALA | DLMHPGAKD | VGSEGMVID | |
| DMYWDRALD | DLMHPGAMA | VGSEGSTTL | |
| DMYWDRAPD | DLMHPGAMD | VGSIQSEGT | |
| DMYWDRASA | DLMHPGAME | VGSTQSEGT | |
| DMYWDRASD | DLMHPGAMG | ||
| DMYWDRASG | DLMHPGAMH | ||
| DMYWDSASD | DLMHPGAMN | ||
| DMYWDSPSS | DLMHPGAMS | ||
| DMYWGRASD | DLMHPGAND | ||
| GMYWDRASD | DLMHPGASE | ||
| VMYWDRASD | DLMHPGATD | ||
| YMYWDRASD | GLMHPGAMD |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gillespie, J.W.; Yang, L.; De Plano, L.M.; Stackhouse, M.A.; Petrenko, V.A. Evolution of a Landscape Phage Library in a Mouse Xenograft Model of Human Breast Cancer. Viruses 2019, 11, 988. https://doi.org/10.3390/v11110988
Gillespie JW, Yang L, De Plano LM, Stackhouse MA, Petrenko VA. Evolution of a Landscape Phage Library in a Mouse Xenograft Model of Human Breast Cancer. Viruses. 2019; 11(11):988. https://doi.org/10.3390/v11110988
Chicago/Turabian StyleGillespie, James W., Liping Yang, Laura Maria De Plano, Murray A. Stackhouse, and Valery A. Petrenko. 2019. "Evolution of a Landscape Phage Library in a Mouse Xenograft Model of Human Breast Cancer" Viruses 11, no. 11: 988. https://doi.org/10.3390/v11110988
APA StyleGillespie, J. W., Yang, L., De Plano, L. M., Stackhouse, M. A., & Petrenko, V. A. (2019). Evolution of a Landscape Phage Library in a Mouse Xenograft Model of Human Breast Cancer. Viruses, 11(11), 988. https://doi.org/10.3390/v11110988







