Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism
Abstract
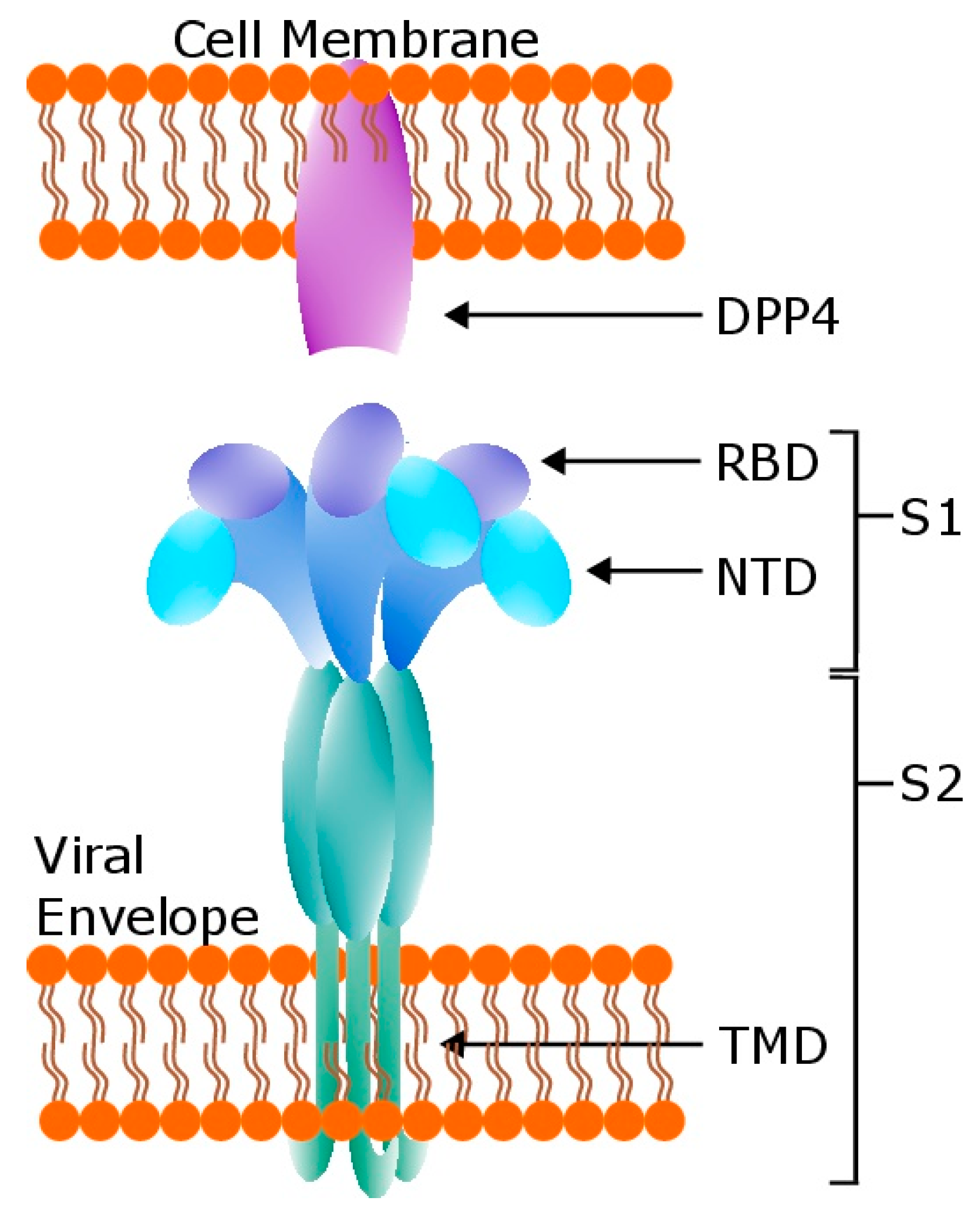
1. Introduction
2. Subunit Vaccines: Immunogenically Focused
2.1. Receptor Binding Domain
2.2. Full-Length S
2.3. N-Terminal Domain
3. DNA Vaccines: Efficient Protection
3.1. Full-Length S
3.2. S1 Domain
4. Viral Vector Vaccines: Optimized Delivery
5. Live Attenuated and Inactivated Vaccines: Situationally Useful
5.1. Inactivated
5.2. Live Attenuated
6. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Tsoi, H.W.; Wong, B.H.; Wong, S.S.; Leung, S.Y.; Chan, K.H.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 2005, 102, 14040–14045. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, V.; Khiabanian, H.; Rabadan, R. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N. Engl. J. Med. 2009, 361, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Gire, S.K.; Goba, A.; Andersen, K.G.; Sealfon, R.S.; Park, D.J.; Kanneh, L.; Jalloh, S.; Momoh, M.; Fullah, M.; Dudas, G.; et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 2014, 345, 1369–1372. [Google Scholar] [CrossRef] [PubMed]
- Baize, S.; Pannetier, D.; Oestereich, L.; Rieger, T.; Koivogui, L.; Magassouba, N.; Soropogui, B.; Sow, M.S.; Keita, S.; De Clerck, H.; et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014, 371, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [PubMed]
- Riski, H.; Hovi, T. Coronavirus infections of man associated with diseases other than the common cold. J. Med. Virol. 1980, 6, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Lau, S.K.; To, K.K.; Cheng, V.C.; Woo, P.C.; Yuen, K.Y. Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015, 28, 465–522. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Anthony, S.J.; Gilardi, K.; Menachery, V.D.; Goldstein, T.; Ssebide, B.; Mbabazi, R.; Navarrete-Macias, I.; Liang, E.; Wells, H.; Hicks, A.; et al. Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. mBio 2017, 8. [Google Scholar] [CrossRef]
- Menachery, V.D.; Yount, B.L., Jr.; Debbink, K.; Agnihothram, S.; Gralinski, L.E.; Plante, J.A.; Graham, R.L.; Scobey, T.; Ge, X.Y.; Donaldson, E.F.; et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015, 21, 1508–1513. [Google Scholar] [CrossRef]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- Muller, M.A.; Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.; Bosch, B.J.; Lattwein, E.; Hilali, M.; Musa, B.E.; et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014, 20, 2093–2095. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef] [PubMed]
- The Health Protection Agency (HPA) UK Novel Coronavirus Investigation Team. Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill. 2013, 18, 20427. [Google Scholar]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef] [PubMed]
- Public Health England/ISARIC. Treatment of MERS-CoV: Information for Clinicians, Clinical Decision-Making Support for Treatment of MERS-CoV Patients; Public Health England: London, UK, 2015. [Google Scholar]
- Vergara-Alert, J.; Vidal, E.; Bensaid, A.; Segales, J. Searching for animal models and potential target species for emerging pathogens: Experience gained from Middle East respiratory syndrome (MERS) coronavirus. One Health 2017, 3, 34–40. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Peck, K.M.; Cockrell, A.S.; Yount, B.L.; Scobey, T.; Baric, R.S.; Heise, M.T. Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection. J. Virol. 2015, 89, 4696–4699. [Google Scholar] [CrossRef]
- Cockrell, A.S.; Peck, K.M.; Yount, B.L.; Agnihothram, S.S.; Scobey, T.; Curnes, N.R.; Baric, R.S.; Heise, M.T. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J. Virol. 2014, 88, 5195–5199. [Google Scholar] [CrossRef]
- Coleman, C.M.; Matthews, K.L.; Goicochea, L.; Frieman, M.B. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J. Gen. Virol. 2014, 95 Pt 2, 408–412. [Google Scholar] [CrossRef]
- Cockrell, A.S.; Yount, B.L.; Scobey, T.; Jensen, K.; Douglas, M.; Beall, A.; Tang, X.C.; Marasco, W.A.; Heise, M.T.; Baric, R.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016, 2, 16226. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, K.; Wohlford-Lenane, C.; Agnihothram, S.S.; Fett, C.; Gale, M.J., Jr.; Baric, R.S.; Enjuanes, L.; Gallagher, T.; McCray, P.B., Jr.; et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA 2014, 111, 4970–4975. [Google Scholar] [CrossRef] [PubMed]
- Pascal, K.E.; Coleman, C.M.; Mujica, A.O.; Kamat, V.; Badithe, A.; Fairhurst, J.; Hunt, C.; Strein, J.; Berrebi, A.; Sisk, J.M.; et al. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA 2015, 112, 8738–8743. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Garron, T.; Agrawal, A.S.; Algaissi, A.; Peng, B.H.; Wakamiya, M.; Chan, T.S.; Lu, L.; Du, L.; Jiang, S.; et al. Characterization and Demonstration of the Value of a Lethal Mouse Model of Middle East Respiratory Syndrome Coronavirus Infection and Disease. J. Virol. 2016, 90, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wohlford-Lenane, C.L.; Channappanavar, R.; Park, J.E.; Earnest, J.T.; Bair, T.B.; Bates, A.M.; Brogden, K.A.; Flaherty, H.A.; Gallagher, T.; et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. USA 2017, 114, E3119–E3128. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Jiang, S.; Du, L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccines 2014, 13, 761–774. [Google Scholar] [CrossRef]
- Surjit, M.; Lal, S.K. The SARS-CoV nucleocapsid protein: A protein with multifarious activities. Infect. Genet. Evolut. 2008, 8, 397–405. [Google Scholar] [CrossRef]
- Chang, C.K.; Lo, S.C.; Wang, Y.S.; Hou, M.H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today 2016, 21, 562–572. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hulswit, R.J.G.; Widjaja, I.; Raj, V.S.; McBride, R.; Peng, W.; Widagdo, W.; Tortorici, M.A.; van Dieren, B.; Lang, Y.; et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, N.; Zuo, T.; Shi, X.; Poon, K.M.; Wu, Y.; Gao, F.; Li, D.; Wang, R.; Guo, J.; et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014, 6, 234ra59. [Google Scholar] [CrossRef] [PubMed]
- Irigoyen, N.; Firth, A.E.; Jones, J.D.; Chung, B.Y.; Siddell, S.G.; Brierley, I. High-Resolution Analysis of Coronavirus Gene Expression by RNA Sequencing and Ribosome Profiling. PLoS Pathog. 2016, 12, e1005473. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, U.J.; Bukreyev, A.; Yang, L.; Lamirande, E.W.; Murphy, B.R.; Subbarao, K.; Collins, P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA 2004, 101, 9804–9809. [Google Scholar] [CrossRef] [PubMed]
- Agnihothram, S.; Gopal, R.; Yount, B.L., Jr.; Donaldson, E.F.; Menachery, V.D.; Graham, R.L.; Scobey, T.D.; Gralinski, L.E.; Denison, M.R.; Zambon, M.; et al. Evaluation of serologic and antigenic relationships between middle eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J. Infect. Dis. 2014, 209, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.S.; Pan, Y.; Chen, H.Q.; Shen, Y.; Wang, X.C.; Sun, Y.J.; Tao, K.H. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol. Lett. 2004, 92, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Zhao, J.; Perlman, S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014, 59, 118–128. [Google Scholar] [CrossRef]
- Wang, Y.D.; Sin, W.Y.; Xu, G.B.; Yang, H.H.; Wong, T.Y.; Pang, X.W.; He, X.Y.; Zhang, H.G.; Ng, J.N.; Cheng, C.S.; et al. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J. Virol. 2004, 78, 5612–5618. [Google Scholar] [CrossRef]
- Zhao, J.; Alshukairi, A.N.; Baharoon, S.A.; Ahmed, W.A.; Bokhari, A.A.; Nehdi, A.M.; Layqah, L.A.; Alghamdi, M.G.; Al Gethamy, M.M.; Dada, A.M.; et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Deming, D.; Sheahan, T.; Heise, M.; Yount, B.; Davis, N.; Sims, A.; Suthar, M.; Harkema, J.; Whitmore, A.; Pickles, R.; et al. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006, 3, e525. [Google Scholar] [CrossRef]
- Tseng, C.T.; Sbrana, E.; Iwata-Yoshikawa, N.; Newman, P.C.; Garron, T.; Atmar, R.L.; Peters, C.J.; Couch, R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE 2012, 7, e35421. [Google Scholar] [CrossRef]
- Weihofen, W.A.; Liu, J.; Reutter, W.; Saenger, W.; Fan, H. Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface. J. Biol. Chem. 2004, 279, 43330–43335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wong, G.; Lu, G.; Yan, J.; Gao, G.F. MERS-CoV spike protein: Targets for vaccines and therapeutics. Antivir. Res. 2016, 133, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Hansson, M.; Nygren, P.A.; Stahl, S. Design and production of recombinant subunit vaccines. Biotechnol. Appl. Biochem. 2000, 32 (Pt 2), 95–107. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Liu, L.; Du, L.; Zhang, C.; Jiang, S.; Li, T.; He, Y. Potent and persistent antibody responses against the receptor-binding domain of SARS-CoV spike protein in recovered patients. Virol. J. 2010, 7, 299. [Google Scholar] [CrossRef]
- Du, L.; Zhao, G.; Kou, Z.; Ma, C.; Sun, S.; Poon, V.K.; Lu, L.; Wang, L.; Debnath, A.K.; Zheng, B.J.; et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013, 87, 9939–9942. [Google Scholar] [CrossRef]
- Mou, H.; Raj, V.S.; van Kuppeveld, F.J.; Rottier, P.J.; Haagmans, B.L.; Bosch, B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013, 87, 9379–9383. [Google Scholar] [CrossRef]
- Burton, D.R.; Williamson, R.A.; Parren, P.W. Antibody and virus: Binding and neutralization. Virology 2000, 270, 1–3. [Google Scholar] [CrossRef]
- Ma, C.; Li, Y.; Wang, L.; Zhao, G.; Tao, X.; Tseng, C.T.; Zhou, Y.; Du, L.; Jiang, S. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: Implication for designing novel mucosal MERS vaccines. Vaccine 2014, 32, 2100–2108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Wang, Y.; Mankowski, M.K.; Ptak, R.G.; Dimitrov, D.S. Cross-reactive HIV-1-neutralizing activity of serum IgG from a rabbit immunized with gp41 fused to IgG1 Fc: Possible role of the prolonged half-life of the immunogen. Vaccine 2009, 27, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, L.; Tao, X.; Zhang, N.; Yang, Y.; Tseng, C.K.; Li, F.; Zhou, Y.; Jiang, S.; Du, L. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments—The importance of immunofocusing in subunit vaccine design. Vaccine 2014, 32, 6170–6176. [Google Scholar] [CrossRef] [PubMed]
- Nyon, M.P.; Du, L.; Tseng, C.K.; Seid, C.A.; Pollet, J.; Naceanceno, K.S.; Agrawal, A.; Algaissi, A.; Peng, B.H.; Tai, W.; et al. Engineering a stable CHO cell line for the expression of a MERS-coronavirus vaccine antigen. Vaccine 2018, 36, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Letarov, A.V.; Londer, Y.Y.; Boudko, S.P.; Mesyanzhinov, V.V. The carboxy-terminal domain initiates trimerization of bacteriophage T4 fibritin. Biochem. Biokhimiia 1999, 64, 817–823. [Google Scholar]
- Tai, W.; Zhao, G.; Sun, S.; Guo, Y.; Wang, Y.; Tao, X.; Tseng, C.K.; Li, F.; Jiang, S.; Du, L.; et al. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology 2016, 499, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Yao, Y.; Deng, Y.; Chen, H.; Lu, G.; Wang, W.; Bao, L.; Deng, W.; Wei, Q.; Gao, G.F.; et al. Recombinant Receptor Binding Domain Protein Induces Partial Protective Immunity in Rhesus Macaques Against Middle East Respiratory Syndrome Coronavirus Challenge. EBioMedicine 2015, 2, 1438–1446. [Google Scholar] [CrossRef]
- Tai, W.; Wang, Y.; Fett, C.A.; Zhao, G.; Li, F.; Perlman, S.; Jiang, S.; Zhou, Y.; Du, L. Recombinant Receptor-Binding Domains of Multiple Middle East Respiratory Syndrome Coronaviruses (MERS-CoVs) Induce Cross-Neutralizing Antibodies against Divergent Human and Camel MERS-CoVs and Antibody Escape Mutants. J. Virol. 2017, 91, e01651-16. [Google Scholar] [CrossRef]
- Coleman, C.M.; Liu, Y.V.; Mu, H.; Taylor, J.K.; Massare, M.; Flyer, D.C.; Smith, G.E.; Frieman, M.B. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 2014, 32, 3169–3174. [Google Scholar] [CrossRef]
- Coleman, C.M.; Venkataraman, T.; Liu, Y.V.; Glenn, G.M.; Smith, G.E.; Flyer, D.C.; Frieman, M.B. MERS-CoV spike nanoparticles protect mice from MERS-CoV infection. Vaccine 2017, 35, 1586–1589. [Google Scholar] [CrossRef]
- Kam, Y.W.; Kien, F.; Roberts, A.; Cheung, Y.C.; Lamirande, E.W.; Vogel, L.; Chu, S.L.; Tse, J.; Guarner, J.; Zaki, S.R.; et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine 2007, 25, 729–740. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.S.; Chen, M.; Joyce, M.G.; Sastry, M.; Stewart-Jones, G.B.; Yang, Y.; Zhang, B.; Chen, L.; Srivatsan, S.; Zheng, A.; et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 2013, 342, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Penaranda, S.; Bankamp, B.; Maher, K.; Chen, M.H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed]
- Jiaming, L.; Yanfeng, Y.; Yao, D.; Yawei, H.; Linlin, B.; Baoying, H.; Jinghua, Y.; Gao, G.F.; Chuan, Q.; Wenjie, T. The recombinant N-terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Vaccine 2017, 35, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Aihara, H.; Miyazaki, J. Gene transfer into muscle by electroporation in vivo. Nat. Biotechnol. 1998, 16, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Sardesai, N.Y.; Weiner, D.B. Electroporation delivery of DNA vaccines: Prospects for success. Curr. Opin. Immunol. 2011, 23, 421–429. [Google Scholar] [CrossRef]
- Liu, M.A. DNA vaccines: An historical perspective and view to the future. Immunol. Rev. 2011, 239, 62–84. [Google Scholar] [CrossRef]
- Nichols, W.W.; Ledwith, B.J.; Manam, S.V.; Troilo, P.J. Potential DNA vaccine integration into host cell genome. Ann. N. Y. Acad. Sci. 1995, 772, 30–39. [Google Scholar] [CrossRef]
- Sheets, R.L.; Stein, J.; Manetz, T.S.; Duffy, C.; Nason, M.; Andrews, C.; Kong, W.P.; Nabel, G.J.; Gomez, P.L. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile virus is similar, without integration, despite differing plasmid backbones or gene inserts. Toxicol. Sci. 2006, 91, 610–619. [Google Scholar] [CrossRef]
- Muthumani, K.; Falzarano, D.; Reuschel, E.L.; Tingey, C.; Flingai, S.; Villarreal, D.O.; Wise, M.; Patel, A.; Izmirly, A.; Aljuaid, A.; et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 2015, 7, 301ra132. [Google Scholar] [CrossRef]
- Barouch, D.H.; Yang, Z.Y.; Kong, W.P.; Korioth-Schmitz, B.; Sumida, S.M.; Truitt, D.M.; Kishko, M.G.; Arthur, J.C.; Miura, A.; Mascola, J.R.; et al. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 2005, 79, 8828–8834. [Google Scholar] [CrossRef] [PubMed]
- Cayabyab, M.J.; Kashino, S.S.; Campos-Neto, A. Robust immune response elicited by a novel and unique Mycobacterium tuberculosis protein using an optimized DNA/protein heterologous prime/boost protocol. Immunology 2012, 135, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, W.; Joyce, M.G.; Modjarrad, K.; Zhang, Y.; Leung, K.; Lees, C.R.; Zhou, T.; Yassine, H.M.; Kanekiyo, M.; et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015, 6, 7712. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Zheng, X.; Wang, X.; Wang, C.; Wang, H.; Gai, W.; Perlman, S.; Yang, S.; Zhao, J.; Xia, X. DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine 2017, 35, 2069–2075. [Google Scholar] [CrossRef] [PubMed]
- Lanzavecchia, A. Mechanisms of antigen uptake for presentation. Curr. Opin. Immunol. 1996, 8, 348–354. [Google Scholar] [CrossRef]
- Al-Amri, S.S.; Abbas, A.T.; Siddiq, L.A.; Alghamdi, A.; Sanki, M.A.; Al-Muhanna, M.K.; Alhabbab, R.Y.; Azhar, E.I.; Li, X.; Hashem, A.M. Immunogenicity of Candidate MERS-CoV DNA Vaccines Based on the Spike Protein. Sci. Rep. 2017, 7, 44875. [Google Scholar] [CrossRef]
- Stevens, T.L.; Bossie, A.; Sanders, V.M.; Fernandez-Botran, R.; Coffman, R.L.; Mosmann, T.R.; Vitetta, E.S. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature 1988, 334, 255–258. [Google Scholar] [CrossRef]
- Rollier, C.S.; Reyes-Sandoval, A.; Cottingham, M.G.; Ewer, K.; Hill, A.V. Viral vectors as vaccine platforms: Deployment in sight. Curr. Opin. Immunol. 2011, 23, 377–382. [Google Scholar] [CrossRef]
- Pushko, P.; Parker, M.; Ludwig, G.V.; Davis, N.L.; Johnston, R.E.; Smith, J.F. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: Expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 1997, 239, 389–401. [Google Scholar] [CrossRef]
- Agnihothram, S.; Menachery, V.D.; Yount, B.L., Jr.; Lindesmith, L.C.; Scobey, T.; Whitmore, A.; Schafer, A.; Heise, M.T.; Baric, R.S. Development of a Broadly Accessible Venezuelan Equine Encephalitis Virus Replicon Particle Vaccine Platform. J. Virol. 2018, 92, e00027-18. [Google Scholar] [CrossRef]
- Zhao, J.; Mangalam, A.K.; Channappanavar, R.; Fett, C.; Meyerholz, D.K.; Agnihothram, S.; Baric, R.S.; David, C.S.; Perlman, S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 2016, 44, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Sutter, G.; Moss, B. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. USA 1992, 89, 10847–10851. [Google Scholar] [CrossRef] [PubMed]
- Stittelaar, K.J.; Kuiken, T.; de Swart, R.L.; van Amerongen, G.; Vos, H.W.; Niesters, H.G.; van Schalkwijk, P.; van der Kwast, T.; Wyatt, L.S.; Moss, B.; et al. Safety of modified vaccinia virus Ankara (MVA) in immune-suppressed macaques. Vaccine 2001, 19, 3700–3709. [Google Scholar] [CrossRef]
- Song, F.; Fux, R.; Provacia, L.B.; Volz, A.; Eickmann, M.; Becker, S.; Osterhaus, A.D.; Haagmans, B.L.; Sutter, G. Middle East respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J. Virol. 2013, 87, 11950–11954. [Google Scholar] [CrossRef] [PubMed]
- Volz, A.; Kupke, A.; Song, F.; Jany, S.; Fux, R.; Shams-Eldin, H.; Schmidt, J.; Becker, C.; Eickmann, M.; Becker, S.; et al. Protective Efficacy of Recombinant Modified Vaccinia Virus Ankara Delivering Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein. J. Virol. 2015, 89, 8651–8656. [Google Scholar] [CrossRef] [PubMed]
- Langenmayer, M.C.; Lulf-Averhoff, A.T.; Adam-Neumair, S.; Fux, R.; Sutter, G.; Volz, A. Distribution and absence of generalized lesions in mice following single dose intramuscular inoculation of the vaccine candidate MVA-MERS-S. Biologicals 2018, 54, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Haagmans, B.L.; van den Brand, J.M.; Raj, V.S.; Volz, A.; Wohlsein, P.; Smits, S.L.; Schipper, D.; Bestebroer, T.M.; Okba, N.; Fux, R.; et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science 2016, 351, 77–81. [Google Scholar] [CrossRef]
- Hammer, S.M.; Sobieszczyk, M.E.; Janes, H.; Karuna, S.T.; Mulligan, M.J.; Grove, D.; Koblin, B.A.; Buchbinder, S.P.; Keefer, M.C.; Tomaras, G.D.; et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N. Engl. J. Med. 2013, 369, 2083–2092. [Google Scholar] [CrossRef]
- Chirmule, N.; Propert, K.; Magosin, S.; Qian, Y.; Qian, R.; Wilson, J. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999, 6, 1574–1583. [Google Scholar] [CrossRef]
- Mast, T.C.; Kierstead, L.; Gupta, S.B.; Nikas, A.A.; Kallas, E.G.; Novitsky, V.; Mbewe, B.; Pitisuttithum, P.; Schechter, M.; Vardas, E.; et al. International epidemiology of human pre-existing adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies: Correlates of high Ad5 titers and implications for potential HIV vaccine trials. Vaccine 2010, 28, 950–957. [Google Scholar] [CrossRef]
- Mercier, S.; Rouard, H.; Delfau-Larue, M.H.; Eloit, M. Specific antibodies modulate the interactions of adenovirus type 5 with dendritic cells. Virology 2004, 322, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Okada, K.; Kenniston, T.; Raj, V.S.; AlHajri, M.M.; Farag, E.A.; AlHajri, F.; Osterhaus, A.D.; Haagmans, B.L.; Gambotto, A. Immunogenicity of an adenoviral-based Middle East Respiratory Syndrome coronavirus vaccine in BALB/c mice. Vaccine 2014, 32, 5975–5982. [Google Scholar] [CrossRef]
- Guo, X.; Deng, Y.; Chen, H.; Lan, J.; Wang, W.; Zou, X.; Hung, T.; Lu, Z.; Tan, W. Systemic and mucosal immunity in mice elicited by a single immunization with human adenovirus type 5 or 41 vector-based vaccines carrying the spike protein of Middle East respiratory syndrome coronavirus. Immunology 2015, 145, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Lemiale, F.; Haddada, H.; Nabel, G.J.; Brough, D.E.; King, C.R.; Gall, J.G. Novel adenovirus vaccine vectors based on the enteric-tropic serotype 41. Vaccine 2007, 25, 2074–2084. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.Y.; Kang, K.W.; Lee, E.Y.; Seo, D.W.; Kim, H.L.; Kim, H.; Kwon, T.; Park, H.L.; Lee, S.M.; Nam, J.H. Heterologous prime-boost vaccination with adenoviral vector and protein nanoparticles induces both Th1 and Th2 responses against Middle East respiratory syndrome coronavirus. Vaccine 2018, 36, 3468–3476. [Google Scholar] [CrossRef] [PubMed]
- Farina, S.F.; Gao, G.P.; Xiang, Z.Q.; Rux, J.J.; Burnett, R.M.; Alvira, M.R.; Marsh, J.; Ertl, H.C.; Wilson, J.M. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 2001, 75, 11603–11613. [Google Scholar] [CrossRef]
- Ledgerwood, J.E.; DeZure, A.D.; Stanley, D.A.; Coates, E.E.; Novik, L.; Enama, M.E.; Berkowitz, N.M.; Hu, Z.; Joshi, G.; Ploquin, A.; et al. Chimpanzee Adenovirus Vector Ebola Vaccine. N. Engl. J. Med. 2017, 376, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, N.K.; Padron-Regalado, E.; Thompson, C.P.; Kupke, A.; Wells, D.; Sloan, M.A.; Grehan, K.; Temperton, N.; Lambe, T.; Warimwe, G.; et al. ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine 2017, 35, 3780–3788. [Google Scholar] [CrossRef]
- Li, Z.; Howard, A.; Kelley, C.; Delogu, G.; Collins, F.; Morris, S. Immunogenicity of DNA vaccines expressing tuberculosis proteins fused to tissue plasminogen activator signal sequences. Infect. Immunity 1999, 67, 4780–4786. [Google Scholar]
- Zhang, Y.; Feng, L.; Li, L.; Wang, D.; Li, C.; Sun, C.; Li, P.; Zheng, X.; Liu, Y.; Yang, W.; et al. Effects of the fusion design and immunization route on the immunogenicity of Ag85A-Mtb32 in adenoviral vectored tuberculosis vaccine. Hum. Vaccines Immunother. 2015, 11, 1803–1813. [Google Scholar] [CrossRef]
- Munster, V.J.; Wells, D.; Lambe, T.; Wright, D.; Fischer, R.J.; Bushmaker, T.; Saturday, G.; van Doremalen, N.; Gilbert, S.C.; de Wit, E.; et al. Protective efficacy of a novel simian adenovirus vaccine against lethal MERS-CoV challenge in a transgenic human DPP4 mouse model. NPJ Vaccines 2017, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, A.; Wang, Z.; Liniger, M.; Hangartner, L.; Caballero, M.; Pavlovic, J.; Wild, P.; Viret, J.F.; Glueck, R.; Billeter, M.A.; et al. Attenuated measles virus as a vaccine vector. Vaccine 2007, 25, 2974–2983. [Google Scholar] [CrossRef] [PubMed]
- Malczyk, A.H.; Kupke, A.; Prufer, S.; Scheuplein, V.A.; Hutzler, S.; Kreuz, D.; Beissert, T.; Bauer, S.; Hubich-Rau, S.; Tondera, C.; et al. A Highly Immunogenic and Protective Middle East Respiratory Syndrome Coronavirus Vaccine Based on a Recombinant Measles Virus Vaccine Platform. J. Virol. 2015, 89, 11654–11667. [Google Scholar] [CrossRef]
- Bodmer, B.S.; Fiedler, A.H.; Hanauer, J.R.H.; Prufer, S.; Muhlebach, M.D. Live-attenuated bivalent measles virus-derived vaccines targeting Middle East respiratory syndrome coronavirus induce robust and multifunctional T cell responses against both viruses in an appropriate mouse model. Virology 2018, 521, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Samal, S.K. Newcastle Disease Virus as a Vaccine Vector for Development of Human and Veterinary Vaccines. Viruses 2016, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.-Q.; Ge, J.-Y.; Wang, J.-L.; Yu, S.; Zhang, H.-L.; Wang, J.-L.; Wen, Z.-Y.; Bu, Z.-G. Newcastle disease virus-based MERS-CoV candidate vaccine elicits high-level and lasting neutralizing antibodies in Bactrian camels. J. Integr. Agric. 2017, 16, 2264–2273. [Google Scholar] [CrossRef]
- Lichty, B.D.; Power, A.T.; Stojdl, D.F.; Bell, J.C. Vesicular stomatitis virus: Re-inventing the bullet. Trends Mol. Med. 2004, 10, 210–216. [Google Scholar] [CrossRef]
- Liu, R.; Wang, J.; Shao, Y.; Wang, X.; Zhang, H.; Shuai, L.; Ge, J.; Wen, Z.; Bu, Z. A recombinant VSV-vectored MERS-CoV vaccine induces neutralizing antibody and T cell responses in rhesus monkeys after single dose immunization. Antivir. Res. 2018, 150, 30–38. [Google Scholar] [CrossRef]
- Willet, M.; Kurup, D.; Papaneri, A.; Wirblich, C.; Hooper, J.W.; Kwilas, S.A.; Keshwara, R.; Hudacek, A.; Beilfuss, S.; Rudolph, G.; et al. Preclinical Development of Inactivated Rabies Virus-Based Polyvalent Vaccine Against Rabies and Filoviruses. J. Infect. Dis. 2015, 212 (Suppl. 2), S414–S424. [Google Scholar] [CrossRef][Green Version]
- Wirblich, C.; Coleman, C.M.; Kurup, D.; Abraham, T.S.; Bernbaum, J.G.; Jahrling, P.B.; Hensley, L.E.; Johnson, R.F.; Frieman, M.B.; Schnell, M.J. One-Health: A Safe, Efficient, Dual-Use Vaccine for Humans and Animals against Middle East Respiratory Syndrome Coronavirus and Rabies Virus. J. Virol. 2017, 91, e02040-16. [Google Scholar] [CrossRef]
- Zeltins, A. Construction and characterization of virus-like particles: A review. Mol. Biotechnol. 2013, 53, 92–107. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, X.; Gai, W.; Zhao, Y.; Wang, H.; Feng, N.; Chi, H.; Qiu, B.; Li, N.; Wang, T.; et al. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget 2017, 8, 12686–12694. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zheng, X.; Gai, W.; Wong, G.; Wang, H.; Jin, H.; Feng, N.; Zhao, Y.; Zhang, W.; Li, N.; et al. Novel chimeric virus-like particles vaccine displaying MERS-CoV receptor-binding domain induce specific humoral and cellular immune response in mice. Antivir. Res. 2017, 140, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Bolles, M.; Deming, D.; Long, K.; Agnihothram, S.; Whitmore, A.; Ferris, M.; Funkhouser, W.; Gralinski, L.; Totura, A.; Heise, M.; et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011, 85, 12201–12215. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.S.; Tao, X.; Algaissi, A.; Garron, T.; Narayanan, K.; Peng, B.H.; Couch, R.B.; Tseng, C.T. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vaccines Immunother. 2016, 12, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Lan, J.; Bao, L.; Huang, B.; Ye, F.; Chen, Y.; Yao, Y.; Wang, W.; Qin, C.; Tan, W. Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus. Emerg. Microbes Infect. 2018, 7, 60. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Uda, A.; Suzuki, T.; Tsunetsugu-Yokota, Y.; Sato, Y.; Morikawa, S.; Tashiro, M.; Sata, T.; Hasegawa, H.; Nagata, N. Effects of Toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-inactivated severe acute respiratory syndrome-related coronavirus vaccine. J. Virol. 2014, 88, 8597–8614. [Google Scholar] [CrossRef]
- Plotkin, S.A. Vaccines: Past, present and future. Nat. Med. 2005, 11 (Suppl. 4), S5–S11. [Google Scholar] [CrossRef]
- Ruch, T.R.; Machamer, C.E. The coronavirus E protein: Assembly and beyond. Viruses 2012, 4, 363–382. [Google Scholar] [CrossRef]
- Lamirande, E.W.; DeDiego, M.L.; Roberts, A.; Jackson, J.P.; Alvarez, E.; Sheahan, T.; Shieh, W.J.; Zaki, S.R.; Baric, R.; Enjuanes, L.; et al. A live attenuated severe acute respiratory syndrome coronavirus is immunogenic and efficacious in golden Syrian hamsters. J. Virol. 2008, 82, 7721–7724. [Google Scholar] [CrossRef]
- Almazan, F.; DeDiego, M.L.; Sola, I.; Zuniga, S.; Nieto-Torres, J.L.; Marquez-Jurado, S.; Andres, G.; Enjuanes, L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 2013, 4, e00650-13. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Becker, M.M.; Eckerle, L.D.; Bolles, M.; Denison, M.R.; Baric, R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012, 18, 1820–1826. [Google Scholar] [CrossRef] [PubMed]
- Zust, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Yount, B.L., Jr.; Josset, L.; Gralinski, L.E.; Scobey, T.; Agnihothram, S.; Katze, M.G.; Baric, R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J. Virol. 2014, 88, 4251–4264. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Gralinski, L.E.; Mitchell, H.D.; Dinnon, K.H., III; Leist, S.R.; Yount, B.L., Jr.; McAnarney, E.T.; Graham, R.L.; Waters, K.M.; Baric, R.S. Combination Attenuation Offers Strategy for Live Attenuated Coronavirus Vaccines. J. Virol. 2018, 92, e00710-18. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Gralinski, L.E.; Mitchell, H.D.; Dinnon, K.H., III; Leist, S.R.; Yount, B.L., Jr.; Graham, R.L.; McAnarney, E.T.; Stratton, K.G.; Cockrell, A.S.; et al. Middle East Respiratory Syndrome Coronavirus Nonstructural Protein 16 Is Necessary for Interferon Resistance and Viral Pathogenesis. mSphere 2017, 2, e00346-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Fung, T.S.; Chong, K.K.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014, 109, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Mitchell, H.D.; Cockrell, A.S.; Gralinski, L.E.; Yount, B.L., Jr.; Graham, R.L.; McAnarney, E.T.; Douglas, M.G.; Scobey, T.; Beall, A.; et al. MERS-CoV Accessory ORFs Play Key Role for Infection and Pathogenesis. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS); World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Hotez, P.J.; Bottazzi, M.E.; Tseng, C.T.; Zhan, B.; Lustigman, S.; Du, L.; Jiang, S. Calling for rapid development of a safe and effective MERS vaccine. Microbes Infect. 2014, 16, 529–531. [Google Scholar] [CrossRef]
- Prescott, J.; Falzarano, D.; de Wit, E.; Hardcastle, K.; Feldmann, F.; Haddock, E.; Scott, D.; Feldmann, H.; Munster, V.J. Pathogenicity and Viral Shedding of MERS-CoV in Immunocompromised Rhesus Macaques. Front. Immunol. 2018, 9, 205. [Google Scholar] [CrossRef]
| Vaccine Type | Humoral Response in: | Cell-Mediated Response in: | Protective in: | Clinical Trial | Source (s) |
|---|---|---|---|---|---|
| Subunit | |||||
| RBD | M, P | M, P | M, P | [50,55,58] | |
| S nanoparticles | M | M | [61] | ||
| Prefusion-locked S | M | [34] | |||
| NTD | M | M | M | [65] | |
| DNA | |||||
| pVax1-S | M, P, C | M, P | P | Phase I | [71] |
| pVRC8400-S 1 | M, P | P | [74] | ||
| pcDNA3.1(+)-S1 or S | M | M | M | [75,77] | |
| Viral Vector | |||||
| VEEV-S | M | [38] | |||
| VEEV-N | M | M | [82] | ||
| MVA-S | M, C | M | M, C | Phase I | [88] |
| Ad5-S or S1 | M | M | [93,94] | ||
| Ad5-S 2 | M | M | M | [96] | |
| Ad41-S | M | M | [94] | ||
| ChAdOx1-S | M | M | M | Phase I | [102] |
| MVvac2-S | M | M | M | [104] | |
| Newcastle-S | M, C | [107] | |||
| VSV-S | M, P | P | [109] | ||
| Rabies-S1 | M | M | [111] | ||
| Bac-S,E,M | P | P | [113] | ||
| Bac-RBD+VP2 | M | M | [114] | ||
| Whole | |||||
| Formalin inactivated | M | M | [117] | ||
| MERS-ΔE | [122] | ||||
| MERS-dNSP16 | M | M | [127] | ||
| MERS-dORF3-5 | M | M | [129] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schindewolf, C.; Menachery, V.D. Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism. Viruses 2019, 11, 74. https://doi.org/10.3390/v11010074
Schindewolf C, Menachery VD. Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism. Viruses. 2019; 11(1):74. https://doi.org/10.3390/v11010074
Chicago/Turabian StyleSchindewolf, Craig, and Vineet D. Menachery. 2019. "Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism" Viruses 11, no. 1: 74. https://doi.org/10.3390/v11010074
APA StyleSchindewolf, C., & Menachery, V. D. (2019). Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism. Viruses, 11(1), 74. https://doi.org/10.3390/v11010074





