Abstract
Influenza A Virus (IAV) is a respiratory virus that causes seasonal outbreaks annually and pandemics occasionally. The main targets of the virus are epithelial cells in the respiratory tract. Like many other viruses, IAV employs the host cell’s machinery to enter cells, synthesize new genomes and viral proteins, and assemble new virus particles. The cytoskeletal system is a major cellular machinery, which IAV exploits for its entry to and exit from the cell. However, in some cases, the cytoskeleton has a negative impact on efficient IAV growth. In this review, we highlight the role of cytoskeletal elements in cellular processes that are utilized by IAV in the host cell. We further provide an in-depth summary of the current literature on the roles the cytoskeleton plays in regulating specific steps during the assembly of progeny IAV particles.
1. IAV life cycle
IAV is an enveloped virus with an eight-segmented negative sense RNA genome. On the surface of the virus are three glycoproteins: hemagglutinin (HA), neuraminidase (NA), and the ion channel protein (M2). Immediately below the viral membrane, the matrix (M1) protein oligomerizes to coat the inside of the virus particle [1]. The viral RNA segments incorporated into the virus particle are present as a viral ribonucleoprotein (vRNP) complex between the viral RNA, nucleoprotein (NP), and three polymerase subunits: PA, PB1, and PB2 [2]. For entry, IAV first binds to the target cell via interactions between HA on the viral membrane and sialic acids on the host cell membrane [3,4]. Following this interaction, the virus is internalized via the host cell’s endocytic pathway. The acidic pH of the endosome causes a conformational change in HA, exposing a fusion peptide that allows the fusion of the endosomal membrane with the viral membrane [5], following which the vRNPs can be released in the cytosol and imported into the nucleus. Viral RNA replication and transcription of mRNAs take place in the nucleus, followed by viral protein translation in the cytoplasm. Assembly of new virus particles requires the trafficking of at least four viral structural proteins, HA, NA, M1, and M2, as well as vRNPs, to the plasma membrane [6,7]. After incorporation of the viral genome into budding virus particles, the M2 protein mediates scission and thus release of the nascent particles from the cell [8].
2. Overview of the Cytoskeleton
The dynamic nature of the cytoskeleton allows for the regulation of a wide range of cellular functions [9,10]. The cytoskeleton is comprised of three main cytoskeletal components: microtubules, actin filaments, and intermediate filaments. All three form filament-like structures that are organized to carry out three main functions: spatial organization within the cell; connecting the cell to the external environment; and regulation of cell motility and shape.
Microtubules self-assemble from tubulin dimers and form long filaments, which function in chromosome segregation during cell division, the transport of cargo such as intracellular vesicles and organelles, and the maintenance of cell polarity [11,12]. They are in most, but not all, cell types anchored to an organizing center, i.e., a centrosome in most cases, in the cell interior, and radiate outwards to the periphery. The minus end of the microtubule is anchored to the centrosome and the growth and shrinkage of the polymer takes place at the plus end. Two motors, dynein and kinesin, associate with microtubules and drive the transport of intracellular cargo towards and away from the center of the cell, respectively [13].
Actin monomers (G-actin) polymerize to form double-helical filaments (F-actin), which are thinner and less stiff than microtubules [10,14]. Actin filaments are asymmetric in nature, with a barbed end that grows faster and a pointed end that loses actin monomers faster. Unlike microtubules, actin filaments can form more complex structures, such as branched networks, bundled networks, and non-aligned networks. These different conformations of F-actin require different actin-nucleating proteins [15,16]. While the Arp2/3 complex nucleates branched F-actin structures, formins nucleate unbranched actin filaments and serve as elongation factors for the growing actin filament. In addition to nucleating factors, the organization of F-actin is dependent on cofilin, an actin-binding protein that severs filaments to generate free ends for the addition of G-actin [17]. Additionally, unlike microtubules, actin filaments do not grow outwards from an organizing center, but polymerize and depolymerize locally in response to different stimuli. The actin cytoskeleton functions in cell migration, providing the structure and strength of the plasma membrane, trafficking of cargo within the cell, organization of organelles, and regulation of cytokinesis during cell division. These functions are carried out with the help of myosin motors, which play key roles in regulating the dynamics of F-actin networks [18]. The Rho family of small GTPases also regulates actin assembly and disassembly. The most well-characterized Rho GTPases are RhoA, Rac, and Cdc42. RhoA stimulates the activity of formins and promotes the assembly of linear F-actin bundles. On the other hand, Rac and Cdc42 promote branching of the actin cytoskeleton by activating the Arp2/3 complex [19]. The activity of Rho GTPases is also regulated by microtubules and in turn, they exert their effect on the microtubule ends and fine-tune microtubule dynamics [20]. Therefore, their influence on F-actin and microtubule dynamics, as well as their regulation by microtubules, also make Rho GTPases important players in the crosstalk between these two cytoskeletal elements.
Intermediate filaments are the least stiff of the three major cytoskeletal elements. They are cross-linked to each other, as well as to actin filaments and microtubules, and help in resisting tensile and shear forces. Unlike microtubules and actin filaments, intermediate filaments do not support the movement of molecular motors [21,22]. More recently, a fourth group of cytoskeletal elements, septins, has been found in most eukaryotic cells [23,24,25]. Septins bind directly to membranes and polymerize into filaments, which allows them to help organize cell membranes [26]. In the context of IAV assembly, roles for microtubules and the actin cytoskeleton have been relatively well-described. Therefore, we focus on these cytoskeletal elements in this review.
3. Cytoskeletal Functions Relevant to IAV Infection
This section describes the roles of the actin cytoskeleton and microtubules in driving cellular processes relevant to IAV infection, in particular the processes implicated in the latter half of the IAV life cycle.
3.1. Endocytic Transport
Endocytic trafficking within the cell plays an important role in many cellular functions, such as receptor signaling, cell adhesion and migration, membrane protein turnover, and nutrient uptake. Trafficking of endosomes is highly dependent on the actin cytoskeleton and microtubules, as well as the motors that associate with them [27]. The first step in the endocytic pathway involves the uptake of material from the plasma membrane, which mainly occurs via clathrin- or non-clathrin-mediated endocytosis, macropinocytosis, and phagocytosis [28]. F-actin plays an important role in remodeling the membrane and providing support to stabilize and elongate the newly formed vesicle during these early steps [29,30,31]. The myosin VI motor is involved in the transport of vesicles away from the cell cortex [32,33], following which the cargo switches over to microtubule-based movement. After this switch, the dynein motor drives the trafficking of cargo towards the interior of the cell, where the cargo is sorted for delivery to different organelles [34,35].
IAV harnesses the endocytic pathway for two crucial steps in its lifecycle: genome entry into the target cell and genome packaging into assembling virions. Genome entry is mediated by either endocytosis (clathrin-dependent or clathrin- and caveolin- independent pathways) [36] or by macropinocytosis [37], depending on the size of the entering virus particle. Trafficking of the newly replicated genome to the plasma membrane and its packaging into assembling virus particles is thought to be mediated by recycling endosomes [38,39,40]. In the cell, recycling to the plasma membrane may be “fast” or “slow”. Fast recycling is regulated by the GTPases Rab4 and Rab35, while slow recycling is mediated by Rab11 [41]. Rab11 also localizes in the Golgi and post-Golgi vesicles and may serve as a link between the endocytic and exocytic pathways [42]. The ubiquitous localization of Rab11 in the cell also makes it difficult to understand the exact nature of the recycling compartment in the cell. Currently, it is known that the slow recycling pathway involves the formation of an endocytic recycling compartment (ERC), which is a collection of tubular structures associated with microtubules. Depending on the cell, the ERC may be condensed around the microtubule-organizing center (MTOC) or be more dispersed in the cytoplasm. The dynein motors support the sorting of cargo into the ERC, while the kinesin motors drive the movement of ERC-derived vesicles to the cell periphery [43,44,45]. The budding of ERC-derived vesicles is also assisted by the actin cytoskeleton [46]. In addition, once cargo travels via Rab11-positive vesicles and reaches the cell periphery, it is transported to the plasma membrane by F-actin and its associating motors, mainly myosin Vb, which associates with Rab11 and other proteins associated with these vesicles [47,48]. Overall, the cytoskeleton and the motors that associate with it play a crucial role in organizing and driving the transport of cargo via endocytic pathways in the cell.
3.2. ER-Golgi Transport
The IAV transmembrane proteins HA, NA, and M2 traffic from the ER to the plasma membrane via the anterograde transport pathway. The distribution of the ER within the cell is regulated by the actin cytoskeleton and microtubules [49]. After synthesis, translocation, and initial modifications at the ER, secretory and transmembrane proteins are packaged into vesicles, which are then transported to the cis Golgi. These ER-derived vesicles move along microtubules just like endosomes using the dynein motor [50]. A crosstalk between the actin cytoskeleton and microtubules is also known to be important for the ER-to-Golgi transport [51]. These cargo-carrying vesicles fuse with the cis Golgi, after which the proteins are further post-translationally modified and sorted for delivery. F-actin and microtubules associate with the Golgi and are thought to be important for its structural integrity [49]. The cargo processed in the Golgi exits the trans Golgi network in vesicles, which fuse with the plasma membrane or other organelle membranes. In the case of trafficking to the plasma membrane, these vesicles move along microtubules using kinesin motors [52]. In addition, F-actin associates with post-Golgi vesicles via the actin-binding protein cortactin and supports their transport [53]. Altogether, the current literature highlights the important role that the actin cytoskeleton and microtubules play in the transport of proteins via the ER-Golgi pathway.
3.3. Maintenance of Cell Polarity
Differentiated epithelial cells such as those lining the respiratory tract are tightly packed to form a monolayer, which is organized and strengthened by cell-to-cell adhesion and the maintenance of cell polarity. Cell polarity manifests in the form of the asymmetric distribution of cellular components, such as trafficking cargo, plasma membrane-associated proteins, organelles, and the cytoskeleton between the apical and basolateral sides of the cell [54]. Viruses that infect polarized cells (such as IAV) utilize this polarity and restrict their entry and subsequent spread to either, but not both, sides of the cell [55].
The cytoskeleton plays a key role in the creation and maintenance of cell polarity [54]. In epithelial cells, microtubules are not anchored to the centrosome, but are organized in parallel arrays along the apico-basolateral axis, with plus ends enriched close to the basolateral side and minus ends enriched close to the apical side [56]. Consequently, microtubules play a key role in the sorting and selective delivery of proteins to the apical and basolateral surfaces [57,58,59], a process that is important for maintaining epithelial cell polarity. Selective delivery of IAV structural components to the apical membrane and the role of microtubules in this process are discussed below. In the case of the actin cytoskeleton, F-actin and actin-binding proteins are non-uniformly distributed in polarized cells, with the apical side being more heavily enriched than the basolateral side [60,61]. Since IAV encounters this apical actin network during its entry and assembly in epithelial cells (discussed below and [62]), the actin cytoskeleton could regulate these stages of the IAV life cycle via its role in maintaining the polarity of epithelial cells.
3.4. Organization and Maintenance of Plasma Membrane Microdomains
The plasma membrane is heterogeneous in composition and fluidic in nature. It is composed of discrete, but often dynamic, domains, with unique physical and biological properties. These specialized regions are referred to as microdomains. Of these, the most extensively studied are lipid rafts or membrane rafts, which are enriched in cholesterol and sphingolipids [63,64]. The smallest microdomains span a 10-nm diameter, but merge together to form larger domains that are several micrometers in diameter. Larger micron-size microdomains are assembled in cells in response to activation stimuli, such as in the case of receptor engagement. While lipid compositions were initially thought to be the main determinant for the formation and stability of the microdomains, it has been suggested more recently that membrane-associated proteins also play important roles in the formation and maintenance of plasma membrane microdomains [64]. IAV structural proteins also localize to these microdomains during viral assembly and drive their coalescence into larger domains, which could lead to the stabilization of these domains [65].
The cortical actin cytoskeleton, which underlies the plasma membrane, plays an important role in stabilizing microdomains [66]. In addition, increases in local F-actin concentration in response to activation signals drive the large-scale clustering of microdomains [67,68,69,70]. These functions of the actin cytoskeleton, which are important for membrane rigidity, protein organization, and lateral movement of proteins [66], are dependent on plasma membrane-associated lipids and proteins. Specialized lipids phosphatidylinositol (4,5)-bisphosphate (PIP2) and phosphatidylinositol (3,4,5)-trisphosphate (PIP3) bind to and regulate proteins involved in actin polymerization, cross-linking of filaments, and filament capping [71,72,73]. The ERM (Ezrin, Radixin and Moesin) proteins [74,75,76] and talin [77,78], all of which tether the actin cytoskeleton to the transmembrane proteins, mediate the protein-based regulation of the plasma membrane structure and function by the actin cytoskeleton. Overall, the actin cytoskeleton and the plasma membrane microdomains are intricately linked with each other.
4. Relationships between Virus Growth and Cytoskeleton
Various lines of evidence support multifaceted relationships between the cytoskeleton F-actin and microtubules and the IAV life cycle. First, both actin and tubulin are incorporated into released IAV particles [79,80]. Second, viral components, specifically M1 and proteins comprising the vRNPs, associate with actin and/or tubulin in host cells [81,82,83,84]. Third, IAV infection alters the levels, structures, and functions of F-actin and microtubules in host cells. Several studies show an enhancement in total actin [85,86] and tubulin [87] levels upon infection. However, one study showed no difference in cellular actin levels [88], and another showed a reduction in tubulin levels [85] upon IAV infection. In addition to total actin and tubulin levels, IAV infection can also modulate the levels or activities of proteins involved in the regulation of F-actin and microtubule dynamics. While some key proteins involved in F-actin dynamics, such as cofilin-1 [86,89], ERM proteins [86], talin [86], and the regulatory light chain of myosin II motor [86], are upregulated in response to IAV infection, others, such as Arp2/3 [88], formins [88], cortactin [90], and myosin Vb [88], are downregulated. With respect to microtubule dynamics, dynactin, which serves as a co-factor for dynein motors, is upregulated in response to IAV, while specific kinesin motors are downregulated [88]. RhoA, which regulates the functions of both microtubules and the actin cytoskeleton, was downregulated in infected epithelial cells [91]. In addition, the activity of RhoA [86,91] and another Rho GTPase, Rac1 [92], is reduced upon IAV infection but the activity of a third Rho GTPase, Cdc42 [93], is enhanced upon infection. Therefore, there seems to be some specificity with which IAV infection modulates the expression or activity of proteins engaged in F-actin and microtubule dynamics.
Fourth, the disruption of cytoskeletal dynamics has either a negative or a positive effect on viral replication. Disruption of F-actin dynamics, by drugs inhibiting polymerization (cytochalasin D), enhancing depolymerization (latrunculin A), or stabilizing actin filaments (jasplakinolide), leads to an increase in released viral titers in non-polarized cells [94,95]. In epithelial cells, the effects of these drugs on viral replication differ between different studies. In the Madin-Darby Canine Kidney (MDCK) cell line, the disruption of F-actin dynamics has no [94], a positive [95], or a negative [96,97] effect on released viral titers. In polarized rhesus monkey kidney cells, which are inefficient in supporting the infection of a laboratory adapted IAV strain, the inhibition of F-actin polymerization [98] drastically increases released viral titers. Cofilin-1 [89] and cortactin [90], which can promote the disassembly and assembly of F-actin, respectively, are both thought to play positive roles during IAV infection. With respect to microtubules, the disruption of microtubules has no [99] to a moderately negative [39,100] effect on virus propagation, whereas a reduction in efficiency of microtubule assembly increases virus titers in a rhesus monkey kidney cell line [101]. In addition, an increase in tubulin acetylation status, which promotes microtubule function in trafficking [102], was observed to correlate with an increase in virus titers in one study [103], but not in the other [99].
Among the studies listed above, studies showing infection-induced changes in cytoskeletal proteins or their association with viral proteins or particles are consistent with a role for the cytoskeleton in IAV infection. Obviously, however, studies conducted using specific inhibitors or genetic ablation should provide more definitive evidence on the role of specific cytoskeletal components in the virus life cycle. Nonetheless, some studies using these strategies show contradictory data, even as to whether the cytoskeleton of interest plays a positive role or not [39,89,94,95,96,98,99,100,101]. This could partially be attributed to the use of viral titers as readout for IAV infection. Measurement of viral titers may not allow for identification of the role(s) of the cytoskeleton at different stages of the IAV life cycle, especially if the cytoskeleton plays opposing roles in the early and late stages of IAV infection. Even with studies focusing on the late stages of the virus life cycle, the disruption of F-actin can have contradictory effects on particle assembly [94,95,96,104]. This could be because F-actin can play negative and positive roles at different steps in IAV assembly. Depending on the conditions, such as host cell types, viral strains, and the time points at which analyses are performed, cumulative effects of cytoskeletal disruption may lead to different final outcomes on virus production. Therefore, to advance our understanding of the roles played by cytoskeletons in virus growth, it is necessary to determine the effects of cytoskeleton disruption on each of the single defined steps of the virus life cycle, in addition to the overall viral titers. In the following sections of this review, we describe the individual steps of the IAV assembly process, as well as the role of different cytoskeletal components during these steps.
5. IAV Assembly
IAV is thought to assemble in cholesterol-enriched microdomains, or membrane rafts, of the plasma membrane of host cells [6,7,65,105,106,107]. HA and NA accumulate and co-cluster at these microdomains and form sites of virus assembly known as budozones [105,106,108,109,110,111], while the third transmembrane protein, M2, is suggested to localize at the edge of the budozone [8,106,112,113]. M1, which is predominantly cytosolic, associates with membranes containing HA, NA, and M2, either during the ER-Golgi transport or at the plasma membrane [114,115,116,117]. Expression of HA, NA, M1, and M2 in cells is sufficient to drive the assembly and budding of virus-like particles at the plasma membrane [108,118]. A subset of the M1 population is also imported into the nucleus [119,120,121,122], where it mediates the export of vRNPs via the Crm1 pathway along with another viral non-structural protein, NS2 [123,124]. After nuclear export, vRNPs are thought to co-opt the cellular recycling compartment to traffic to assembly sites at the plasma membrane [38,39,40,125]. Arrival of vRNPs at virus assembly sites further promotes the assembly and budding of virus particles [126,127,128]. While HA, NA, and likely M1 induce and stabilize membrane curvature [108,118,129], membrane scission and release of virus buds requires M2. M2 is enriched at the neck of the virus bud [8,112,113] and is thought to induce positive membrane curvature, which may be sufficient for membrane scission [8,130]. NA prevents the retention of nascent particles that have undergone the scission by cleaving cell-surface sialic acid moieties, which could otherwise bind virus-associated HA [131,132].
IAV is pleomorphic, possessing two distinct morphologies: spherical virions that are ~100 nm in diameter and filamentous particles that are ~100 nm in diameter and up to 20 um in length [133,134,135,136]. Most commonly used laboratory-adapted strains, such as A/Puerto Rico/8/1934 (H1N1) (PR8) and A/WSN/1933 (H1N1) (WSN), are solely spherical [137]; however, in vivo human infection produces both spherical and filamentous virions [133,135]. In addition, virions with a filamentous morphology emerge after the passaging of spherical viral strains in guinea pigs, suggesting the selective advantages of this morphology [136]. The key genetic determinant of virion morphology is the M1 protein, since specific mutations in the M1 protein confer the ability to form filamentous virions [136,138,139,140,141,142], although M2 [141,143,144] and NP [128] also play some roles.
F-actin and microtubules could regulate at least the following steps in the IAV assembly process: trafficking of viral transmembrane proteins to the plasma membrane, trafficking of cytoplasmic viral components to the assembly sites, association between viral components at the plasma membrane, and morphogenesis of nascent particles. In the subsequent section, we describe the roles played by the actin cytoskeleton and microtubules at these individual steps of the IAV assembly process. The different steps of IAV assembly and the roles (positive or negative) of the cytoskeleton during these steps are depicted in Figure 1. In addition, the roles of specific cytoskeletal components at different steps of IAV assembly are summarized in Table 1.
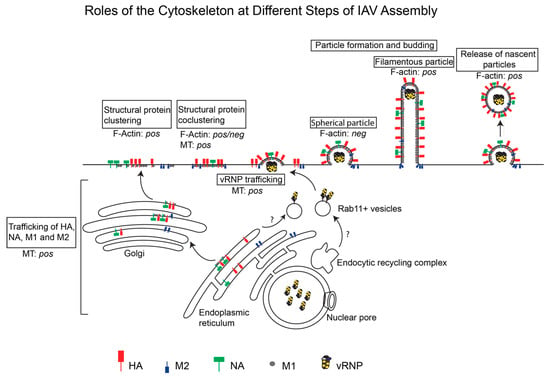
Figure 1.
Roles of the actin cytoskeleton and microtubules at different steps of IAV Assembly. IAV assembly is initiated at the plasma membrane after the arrival of three transmembrane proteins, HA, NA, and M2, and the cytoplasmic protein M1 through the cytoplasm. Clustering of these viral proteins drives assembly of the virus particle, which could have a spherical or filamentous morphology. vRNPs are transported across the cytoplasm on Rab11+ vesicles and are incorporated into the assembling virus particle. Whether the actin cytoskeleton and/or microtubules (MT) promote (pos) or suppress (neg) individual IAV assembly steps is depicted.

Table 1.
Roles of specific cytoskeletal components at different steps of IAV Assembly.
6. Roles Played by the Cytoskeleton at Specific Steps of IAV Assembly
6.1. Trafficking of Viral Transmembrane Proteins to the Apical Membrane
As mentioned above, IAV assembles at the apical surface of polarized epithelial cells, which requires the targeting of virus components to the apical plasma membrane [152,153,154,155,156,157,158,159,160,161,162,163,164]. F-actin disruption has no obvious effect on the apical targeting of transmembrane proteins HA and NA or virus assembly in polarized MDCK cells [97,145]. In one study, impairment in glycosylation of HA and NA was observed upon F-actin disruption; however, this did not have an effect on apical targeting of the proteins [97]. With respect to microtubules, earlier studies showed that their disruption reduces the apical targeting of HA in infected MDCK cells without a loss of polarity in the cell monolayer [145,146,147]. This apical transport of HA is dependent on the acetylation status of microtubules, since the deacetylation of microtubules reduces the efficiency of this transport [103]. Efficient apical targeting of HA in infected cells requires the association of a kinesin protein KIFC3 with trans Golgi-derived vesicles, further supporting the role of microtubules in this process [148]. Of note, increased mistargeting of HA to the basolateral membrane upon microtubule disruption does not inhibit virus particle assembly per se [145,146]; however, in one study, virus assembly was still restricted to the apical surface [145], whereas in the other, it took place at both apical and basolateral membranes [146]. More recent studies have shown that even when HA is intentionally mistargeted to the basolateral membrane, virus assembly, likely mediated by some residual apically targeted HA and/or other viral proteins, still takes place at the apical surface [165,166]. Similarly, mistargeting of M2 to the basolateral membrane still allows for particle assembly and release specifically at the apical surface, although it likely disrupts apical targeting and the incorporation of vRNPs into assembled particles [167]. Therefore, if microtubule disruption does alter the site of IAV assembly [146], it is likely due to either the mistargeting of one of the other viral components (such as NA and vRNPs) or mistargeting of multiple viral proteins, but not just HA or M2, to the basolateral membrane.
6.2. Trafficking of Viral Cytoplasmic Components to the Assembly Sites
In addition to viral transmembrane protein trafficking, M1 protein trafficking to the plasma membrane is also thought to at least partially rely on the ER-Golgi transport pathway, since M1 associates with HA- and NA-enriched membranes at both the ER-Golgi and the plasma membrane [114,115]. In the cytoplasm, M1 may also associate with actin, since it remains insoluble after a detergent treatment that disrupts M1-lipid interactions, but still allows M1-actin interactions [81]. However, it is not clear whether this association of M1 with F-actin mediates the trafficking of M1 to HA- and/or NA-enriched membranes or is involved in the cytoplasmic transport of the M1-vRNP complex.
Trafficking of the viral genome or vRNPs to the apical membrane is also a key step in the assembly of infectious IAV particles. Upon export from the nucleus, vRNPs associate with both the actin cytoskeleton [82] and microtubules [103,168]. While the association with F-actin is proposed to govern the intracellular localization of vRNPs [82] and partially drive their movement in the cytoplasm [169], their association with microtubules appears to be a key driver for vRNP trafficking [100,103,168,169,170]. vRNPs rely on Rab11-positive vesicles for apical targeting to assembly sites [39,40,99,170,171,172,173,174,175]. While these vesicles were thought to be derived from the ERC in earlier studies [39,40,100,127,169,171,175], a more recent study suggested that they are derived from the ER [125]. While their origin remains to be determined, the Rab11-positive vesicles, which are enriched in cholesterol [126], are proposed to promote IAV particle assembly [126,176]. Rab11-dependent transport is also important for the bundling of vRNPs so that eight unique segments can be incorporated into assembling virus particles [171,172]. The kinesin motor KIF13A is also involved in driving the Rab11-dependent apical trafficking of vRNPs [149]. While microtubule disruption reduces the association of vRNPs with Rab11 [99] and slows down their movement in the cytoplasm [170], the role of microtubules in the incorporation of vRNPs into assembling particles is not clear. Earlier studies showed a moderate reduction in infectious particle assembly upon microtubule disruption [39,100], while a more recent study showed no effect of microtubule disruption on the production of infectious virus particles [99]. The most recent understanding in the field is that intact microtubules are required for the association of vRNPs with Rab11-positive vesicles [99], but not for the bundling of vRNPs [171] or for their apical transport and incorporation into virus particles [99]. Thus, it remains to be understood how vRNPs traffic to assembly sites across the cytoplasm. A potential clue to this transport mechanism was provided by a recent study, which suggested that Rab11-dependent transport of vRNPs is mediated by the ER [125]. The sliding dynamics of ER membranes, which is dependent on microtubules resistant to nocodazole (which was used to determine the role of microtubules in vRNP trafficking [39,99,100]) [177], may conceivably drive the trafficking of vRNPs.
Overall, the findings from previous studies support roles for the actin cytoskeleton and microtubules in the apical targeting of IAV components. However, in most cases, the disruption of cytoskeletal elements has a minimal to modest effect on the overall infectious particle assembly process, suggesting that the virus may employ multiple pathways for apical targeting of its proteins.
6.3. Association between Viral Components at the Plasma Membrane
Viral proteins have been shown to co-cluster in microdomains of the plasma membrane [104,106,178]. However, it is not known whether these associations between viral proteins are initiated during the ER-Golgi transport or occur post-arrival at the plasma membrane. Since HA and NA accelerate each other’s apical trafficking [110], they are likely to co-traffic and associate with each other prior to their arrival at the plasma membrane. In contrast, M2 is suggested not to co-traffic with HA [110], and its association with HA (and likely NA) is a discrete step that takes place at the plasma membrane in a cell-type-dependent manner [104]. Despite this knowledge, we currently do not know the exact sequence of association between the transmembrane proteins, as well as their association with M1 and vRNPs. In addition, very little is known about the host cell mechanisms that regulate these associations.
The cortical actin cytoskeleton is likely to be involved in the regulation of co-clustering of viral transmembrane proteins through its ability to regulate the structure of plasma membrane microdomains. In fact, the concentration of cortical actin is apparently increased close to the plasma membrane in response to IAV infection, and this reorganization of F-actin is proposed to be important for efficient virus assembly and budding [89]. The cortical actin lowers the mobility of HA at the plasma membrane and enhances the clustering of HA molecules [150]. While HA movement is restricted to regions of the plasma membrane with an underlying F-actin network, the HA mobility negatively correlates with the density of the underlying F-actin [150]. A role for the actin cytoskeleton in modulating HA clustering is further supported by the presence of HA aggregates at the plasma membrane when the activity of myosin II motors is inhibited [96]. Association or co-clustering between HA with M2 is also regulated by the actin cytoskeleton. Thaa et al. used fluorescence resonance energy transfer (FRET) approaches, where fluorescent probes were fused to the cytoplasmic domains of HA and M2. They observed that cytochalasin D treatment reduces FRET between fluorescent protein fusions of HA and M2, indicating that the actin cytoskeleton plays a positive role in the association between the two proteins in the absence of any other viral proteins [151]. In an apparent contrast, we recently showed using a proximity-ligation approach that in infected primary human macrophages, which do not support efficient IAV assembly, the F-actin network suppresses the association between HA and M2. Disruption of the actin cytoskeleton with cytochalsin D restores HA-M2 association and particle assembly in macrophages, indicating that F-actin plays inhibitory roles in viral protein association/IAV assembly in these cells [104] (possible mechanisms discussed later). The discrepancies in the role of F-actin in HA-M2 association between the two above-mentioned studies might be due to differences in the cell types used in these studies (Chinese Hamster Ovary cells in [151] versus primary human blood derived macrophages in [104]). Cell-type dependent differences for the role of the F-actin and microtubules at different stages of the IAV life cycle have also been reported before [94,98,101,179,180,181]. In addition, technical differences could account for the discrepancies; for example, the presence of other viral components in infection-based experiments [104] or attachment of fluorescent proteins to the cytoplasmic domains of HA and M2 in the FRET study [151] may affect the interaction with subcortical actin. With respect to microtubules, a positive role for the microtubule-mediated transport of vRNPs on co-clustering between HA and M2 in infected HeLa cells has been described [126]. However, in that study, it is not clear whether microtubules support co-clustering between HA and M2 by changing the organization of plasma membrane microdomains directly or indirectly via cholesterol transport or by promoting the trafficking of vRNPs to the assembly sites.
6.4. Morphogenesis of Nascent Particles at the Plasma Membrane
In addition to regulating the association between viral proteins, the actin cytoskeleton also plays direct roles in particle morphogenesis. As described above, the formation of a spherical IAV particle is negatively regulated by F-actin in primary human macrophages. Intriguingly, spherical particle formation occurs regardless of the state of the actin cytoskeleton in a monocytic cell line that is differentiated into a macrophage-like morphology [104]. In epithelial cell lines, the effect of F-actin disruption on particle morphogenesis varies, depending on the virus morphology, with no obvious effects of F-actin disruption observed for IAV strains that assemble solely spherical particles (WSN or PR8) [95,97]. However, one caveat of previous studies with epithelial cells is that they were all performed using epithelial cell lines and not primary epithelial cells. It is becoming increasingly clear that some steps in the IAV particle assembly process are more stringently regulated in primary human cells than in cells lines [104,167], highlighting the need for more studies looking at the role of the actin cytoskeleton in primary cells.
In the case of an IAV strain that forms filamentous particles (A/Udorn/72 [H3N2]), particle morphogenesis is highly dependent on F-actin. Both the stabilization and disruption of F-actin impede filamentous particle formation at the apical surface [94,95]. The mechanisms by which the actin cytoskeleton supports filamentous IAV assembly are not understood. The actin cytoskeleton may provide mechanical support at the base of or inside the viral filament. Of note, while proteomics studies showed the incorporation of actin molecules into released virions [79,80], F-actin has been observed to localize only at the base of the viral filament [95]. In such locations, it is possible that the actin cytoskeleton interacts with one or more viral structural proteins and drives their incorporation into the growing filamentous particle. This role of the actin cytoskeleton might not be so important for spherical particle assembly due to a requirement for the incorporation of a fewer number of copies of the structural proteins, such as HA, NA, M1, and M2, into spherical particles relative to filamentous particles [1]. Of note, the role of F-actin in particle morphogenesis has thus far been examined for only a limited number of the laboratory strains; whether the observations above hold true for the morphogenesis of a broad range of IAV strains with a varied tendency to form filamentous versus spherical particles remains to be tested.
7. Potential Mechanisms for F-Actin-Dependent Restriction of IAV Assembly
As described earlier, in non-permissive primary human macrophages, the actin cytoskeleton plays a negative role in the assembly of spherical (and likely filamentous) IAV particles, likely by restricting HA-M2 association at assembly sites (Figure 2A). However, the mechanism of action of the actin cytoskeleton during this step is not understood. The restrictive role of F-actin in primary human macrophages could either be due to the distinct structure and/or function of the actin network or due to the differential function or expression of additional actin-dependent host factors in this cell type. We propose three different possible mechanisms by which the actin cytoskeleton suppresses IAV assembly in macrophages:
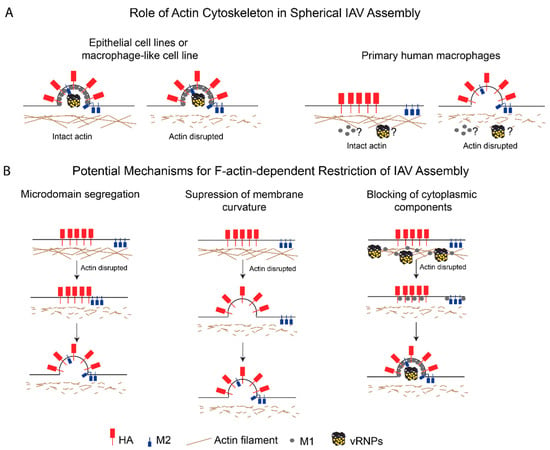
Figure 2.
The role of the actin cytoskeleton in spherical IAV assembly in host cells. (A) Contrasting roles for the actin cytoskeleton in spherical IAV assembly in different cell types. In IAV-permissive cells, such as epithelial cell lines and a macrophage-like cell line, the actin cytoskeleton either promotes or has no effects on IAV particle production. In contrast, the actin cytoskeleton restricts HA-M2 association and spherical IAV assembly in primary human macrophages. (B) Proposed mechanisms by which actin restricts spherical IAV assembly in primary human macrophages. Microdomain segregation: F-actin partitions HA- and M2-enriched plasma membrane microdomains. Suppression of membrane curvature: F-actin restricts HA-mediated curvature induction at the plasma membrane, which is required for M2 recruitment to the assembling particle. Blocking of cytoplasmic components: F-actin restricts trafficking and incorporation of other curvature-inducing structural components, that is, M1 and vRNP, to virus assembly sites.
- Microdomain segregation: In this model, the actin cytoskeleton restricts the movement of HA- and M2-enriched microdomains and keeps the microdomains (and hence, HA and M2) segregated from each other. In fact, M2 is present in microdomains distinct from HA-enriched microdomains early on in the assembly process [106,109], but is later recruited to these assembly sites [112,178,182]. The cortical actin network in primary macrophages may keep these plasma membrane microdomains apart via interactions with either lipids [183,184] or cytoplasmic tails of transmembrane proteins [185,186,187]. As discussed above, the ERM proteins and talin are likely to be involved in linking the cortical actin cytoskeleton to the plasma membrane microdomains [74,76,77]. ERM-mediated tethering of transmembrane proteins can allow for these tethered proteins to form pickets, which restrict the mobility of other proteins associated with these microdomains [186,187];
- Suppression of membrane curvature: According to this model, the actin cytoskeleton suppresses HA- and/or NA-induced membrane curvature by modulating the plasma membrane stiffness [188,189]. This possibility is consistent with several studies that have shown that M2 is not required for the induction of membrane curvature during particle assembly [8,112,143,190] and that M2 may be recruited after the induction of membrane curvature [191,192], which is likely mediated by HA, NA, or M1. Therefore, F-actin may be modulating membrane curvature in a manner that is independent of the recruitment of M2 to assembly sites;
- Blocking of cytoplasmic components: In this model, the actin network inhibits IAV assembly by restricting the trafficking, incorporation, or function of additional components essential for IAV assembly, that is, M1 and vRNPs. Both M1 [81] and NP [82] are reported to associate with F-actin, and this association might suppress their mobility and trafficking to assembly sites. In addition, the dense F-actin cortex could also serve as a physical barrier to the diffusion of proteins or vesicles carrying these proteins [193,194].
These three potential mechanisms of action for the actin cytoskeleton in IAV assembly in primary human macrophages are depicted in Figure 2B. In some cases, the disruption of F-actin in epithelial cells modestly increases spherical virus production [94,95]. Therefore, it is possible that the mechanisms described above may also operate in IAV-permissive cells, depending on the condition.
8. Concluding Remarks and Outstanding Questions
In sum, in this review, we summarize evidence in support of multiple roles for the cytoskeleton in IAV assembly. The current evidence suggests that F-actin and microtubules can play either positive or negative roles at particular steps during IAV assembly, e.g., trafficking of viral components or protein-protein interactions. However, for most of these steps, we have yet to gain a clear understanding of the mechanisms involved. Some of the most pressing questions that remain unanswered in the field are: What is the role of the cytoskeleton in restricting IAV assembly to the apical surface of epithelial cells? What is the exact nature of the vesicular network involved in Rab11-dependent trafficking of vRNPs? Which cytoskeletal component(s) is involved in the trafficking of vRNPs to the apical membrane? What are the mechanisms by which F-actin regulates the association between HA and M2? What are the mechanisms by which F-actin promotes filamentous IAV assembly? What properties of F-actin allow it to suppress spherical particle assembly in primary human macrophages? Additional studies that address these questions using novel approaches to examine the behaviors of viral structural proteins and RNAs are likely to elucidate the exact roles of the cytoskeleton at specific steps in IAV assembly.
Funding
A.O. was supported by funding from the National Institutes of Health (R01 AI071727 and R21 AI143276). S.B. was supported by the Clayton Willison- and Emma Elizabeth Willison-Endowed Graduate Fellowship.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Calder, L.J.; Wasilewski, S.; Berriman, J.A.; Rosenthal, P.B. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. USA 2010, 107, 10685–10690. [Google Scholar] [CrossRef] [PubMed]
- Compans, R.W.; Content, J.; Duesberg, P.H. Structure of the ribonucleoprotein of influenza virus. J. Virol. 1972, 10, 795–800. [Google Scholar] [PubMed]
- Greber, U.F. Virus and Host Mechanics Support Membrane Penetration and Cell Entry. J. Virol. 2016, 90, 3802–3805. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Smrt, S.T.; Lorieau, J.L. Membrane Fusion and Infection of the Influenza Hemagglutinin. Adv. Exp. Med. Biol. 2017, 966, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Lamb, R.A. Influenza virus assembly and budding. Virology 2011, 411, 229–236. [Google Scholar] [CrossRef]
- Nayak, D.P.; Balogun, R.A.; Yamada, H.; Zhou, Z.H.; Barman, S. Influenza virus morphogenesis and budding. Virus Res. 2009, 143, 147–161. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef]
- Bezanilla, M.; Gladfelter, A.S.; Kovar, D.R.; Lee, W.L. Cytoskeletal dynamics: A view from the membrane. J. Cell Biol. 2015, 209, 329–337. [Google Scholar] [CrossRef]
- Akhmanova, A.; Steinmetz, M.O. Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015, 16, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Borisy, G.; Heald, R.; Howard, J.; Janke, C.; Musacchio, A.; Nogales, E. Microtubules: 50 years on from the discovery of tubulin. Nat. Rev. Mol. Cell Biol. 2016, 17, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Gelfand, V.I. Moonlighting Motors: Kinesin, Dynein, and Cell Polarity. Trends Cell Biol. 2017, 27, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.J.; Wood, W. Unravelling the Actin Cytoskeleton: A New Competitive Edge? Trends Cell Biol. 2016, 26, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Chesarone, M.A.; Goode, B.L. Actin nucleation and elongation factors: Mechanisms and interplay. Curr. Opin. Cell Biol. 2009, 21, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Firat-Karalar, E.N.; Welch, M.D. New mechanisms and functions of actin nucleation. Curr. Opin. Cell Biol. 2011, 23, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Magalhaes, M.A.; Eddy, R.J.; Hodgson, L.; Condeelis, J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013, 14, 405–415. [Google Scholar] [CrossRef]
- Hartman, M.A.; Spudich, J.A. The myosin superfamily at a glance. J. Cell Sci. 2012, 125, 1627–1632. [Google Scholar] [CrossRef]
- Sit, S.T.; Manser, E. Rho GTPases and their role in organizing the actin cytoskeleton. J. Cell Sci. 2011, 124, 679–683. [Google Scholar] [CrossRef]
- Wojnacki, J.; Quassollo, G.; Marzolo, M.P.; Cáceres, A. Rho GTPases at the crossroad of signaling networks in mammals: Impact of Rho-GTPases on microtubule organization and dynamics. Small GTPases 2014, 5, e28430. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Cytoplasmic Intermediate Filaments in Cell Biology. Annu. Rev. Cell Dev. Biol. 2018, 34, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Leduc, C.; Etienne-Manneville, S. Intermediate filaments join the action. Cell Cycle 2017, 16, 1389–1390. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Ding, X.; Yu, W.; Yang, X.; Shen, S.; Yu, L. Phylogenetic and evolutionary analysis of the septin protein family in metazoan. FEBS Lett. 2007, 581, 5526–5532. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Malmberg, R.L.; Momany, M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 2007, 7, 103. [Google Scholar] [CrossRef]
- Nishihama, R.; Onishi, M.; Pringle, J.R. New insights into the phylogenetic distribution and evolutionary origins of the septins. Biol. Chem. 2011, 392, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, S.; Cossart, P. Septins: The fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012, 13, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Granger, E.; McNee, G.; Allan, V.; Woodman, P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 2014, 31, 20–29. [Google Scholar] [CrossRef]
- Doherty, G.J.; McMahon, H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef]
- Mooren, O.L.; Galletta, B.J.; Cooper, J.A. Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 2012, 81, 661–686. [Google Scholar] [CrossRef]
- Collins, A.; Warrington, A.; Taylor, K.A.; Svitkina, T. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr. Biol. 2011, 21, 1167–1175. [Google Scholar] [CrossRef]
- Humphries, A.C.; Way, M. The non-canonical roles of clathrin and actin in pathogen internalization, egress and spread. Nat. Rev. Microbiol. 2013, 11, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Buss, F.; Arden, S.D.; Lindsay, M.; Luzio, J.P.; Kendrick-Jones, J. Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. EMBO J. 2001, 20, 3676–3684. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.M.; Arden, S.D.; Roberts, R.C.; Kendrick-Jones, J.; Cooper, J.A.; Luzio, J.P.; Buss, F. Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic 2002, 3, 331–341. [Google Scholar] [CrossRef]
- Driskell, O.J.; Mironov, A.; Allan, V.J.; Woodman, P.G. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat. Cell Biol. 2007, 9, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Flores-Rodriguez, N.; Rogers, S.S.; Kenwright, D.A.; Waigh, T.A.; Woodman, P.G.; Allan, V.J. Roles of dynein and dynactin in early endosome dynamics revealed using automated tracking and global analysis. PLoS ONE 2011, 6, e24479. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.J.; Lakadamyali, M.; Zhang, F.; Zhuang, X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 2004, 11, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Leser, G.P.; Lamb, R.A. Filamentous influenza virus enters cells via macropinocytosis. J. Virol. 2012, 86, 10950–10960. [Google Scholar] [CrossRef]
- Jo, S.; Kawaguchi, A.; Takizawa, N.; Morikawa, Y.; Momose, F.; Nagata, K. Involvement of vesicular trafficking system in membrane targeting of the progeny influenza virus genome. Microbes Infect. 2010, 12, 1079–1084. [Google Scholar] [CrossRef]
- Amorim, M.J.; Bruce, E.A.; Read, E.K.; Foeglein, A.; Mahen, R.; Stuart, A.D.; Digard, P. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 2011, 85, 4143–4156. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Kawakami, E.; Watanabe, T.; Neumann, G.; Kawaoka, Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 2011, 85, 6117–6126. [Google Scholar] [CrossRef]
- Grant, B.D.; Donaldson, J.G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Welz, T.; Wellbourne-Wood, J.; Kerkhoff, E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 2014, 24, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.X.; Gundersen, G.G.; Maxfield, F.R. Export from pericentriolar endocytic recycling compartment to cell surface depends on stable, detyrosinated (glu) microtubules and kinesin. Mol. Biol. Cell 2002, 13, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.R.; Maritzen, T.; Kukhtina, V.; Higman, V.A.; Doglio, L.; Barak, N.N.; Strauss, H.; Oschkinat, H.; Dotti, C.G.; Haucke, V. Regulation of endosomal membrane traffic by a Gadkin/AP-1/kinesin KIF5 complex. Proc. Natl. Acad. Sci. USA 2009, 106, 15344–15349. [Google Scholar] [CrossRef] [PubMed]
- Schonteich, E.; Wilson, G.M.; Burden, J.; Hopkins, C.R.; Anderson, K.; Goldenring, J.R.; Prekeris, R. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J. Cell Sci. 2008, 121, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Puthenveedu, M.A.; Lauffer, B.; Temkin, P.; Vistein, R.; Carlton, P.; Thorn, K.; Taunton, J.; Weiner, O.D.; Parton, R.G.; von Zastrow, M. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell 2010, 143, 761–773. [Google Scholar] [CrossRef]
- Lapierre, L.A.; Kumar, R.; Hales, C.M.; Navarre, J.; Bhartur, S.G.; Burnette, J.O.; Provance, D.W.; Mercer, J.A.; Bähler, M.; Goldenring, J.R. Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 2001, 12, 1843–1857. [Google Scholar] [CrossRef]
- Hales, C.M.; Vaerman, J.P.; Goldenring, J.R. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 2002, 277, 50415–50421. [Google Scholar] [CrossRef]
- Gurel, P.S.; Hatch, A.L.; Higgs, H.N. Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr. Biol. 2014, 24, R660–R672. [Google Scholar] [CrossRef]
- Watson, P.; Forster, R.; Palmer, K.J.; Pepperkok, R.; Stephens, D.J. Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat. Cell Biol. 2005, 7, 48–55. [Google Scholar] [CrossRef]
- Campellone, K.G.; Webb, N.J.; Znameroski, E.A.; Welch, M.D. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 2008, 134, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Luini, A.; Mironov, A.A.; Polishchuk, E.V.; Polishchuk, R.S. Morphogenesis of post-Golgi transport carriers. Histochem. Cell Biol. 2008, 129, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Weller, S.; Orth, J.D.; Chen, J.; Huang, B.; Chen, J.L.; Stamnes, M.; McNiven, M.A. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat. Cell Biol. 2005, 7, 483–492. [Google Scholar] [CrossRef]
- Rodriguez-Boulan, E.; Macara, I.G. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 2014, 15, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Bergelson, J.M. Intercellular junctional proteins as receptors and barriers to virus infection and spread. Cell Host Microbe 2009, 5, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Müsch, A. Microtubule organization and function in epithelial cells. Traffic 2004, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Saunders, C.; Limbird, L.E. Disruption of microtubules reveals two independent apical targeting mechanisms for G-protein-coupled receptors in polarized renal epithelial cells. J. Biol. Chem. 1997, 272, 19035–19045. [Google Scholar] [CrossRef] [PubMed]
- Kreitzer, G.; Schmoranzer, J.; Low, S.H.; Li, X.; Gan, Y.; Weimbs, T.; Simon, S.M.; Rodriguez-Boulan, E. Three-dimensional analysis of post-Golgi carrier exocytosis in epithelial cells. Nat. Cell Biol. 2003, 5, 126–136. [Google Scholar] [CrossRef]
- Jaulin, F.; Xue, X.; Rodriguez-Boulan, E.; Kreitzer, G. Polarization-dependent selective transport to the apical membrane by KIF5B in MDCK cells. Dev. Cell 2007, 13, 511–522. [Google Scholar] [CrossRef]
- Pan, J.; You, Y.; Huang, T.; Brody, S.L. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J. Cell Sci. 2007, 120, 1868–1876. [Google Scholar] [CrossRef]
- Shaw, R.J.; Henry, M.; Solomon, F.; Jacks, T. RhoA-dependent phosphorylation and relocalization of ERM proteins into apical membrane/actin protrusions in fibroblasts. Mol. Biol. Cell 1998, 9, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Delorme-Axford, E.; Coyne, C.B. The actin cytoskeleton as a barrier to virus infection of polarized epithelial cells. Viruses 2011, 3, 2462–2477. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Kerviel, A.; Thomas, A.; Chaloin, L.; Favard, C.; Muriaux, D. Virus assembly and plasma membrane domains: Which came first? Virus Res. 2013, 171, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Chichili, G.R.; Rodgers, W. Cytoskeleton-membrane interactions in membrane raft structure. Cell Mol. Life Sci. 2009, 66, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- Chichili, G.R.; Rodgers, W. Clustering of membrane raft proteins by the actin cytoskeleton. J. Biol. Chem. 2007, 282, 36682–36691. [Google Scholar] [CrossRef]
- Winter, P.W.; van Orden, A.K.; Roess, D.A.; Barisas, B.G. Actin-dependent clustering of insulin receptors in membrane microdomains. Biochim. Biophys. Acta 2012, 1818, 467–473. [Google Scholar] [CrossRef]
- Goswami, D.; Gowrishankar, K.; Bilgrami, S.; Ghosh, S.; Raghupathy, R.; Chadda, R.; Vishwakarma, R.; Rao, M.; Mayor, S. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 2008, 135, 1085–1097. [Google Scholar] [CrossRef]
- Lillemeier, B.F.; Pfeiffer, J.R.; Surviladze, Z.; Wilson, B.S.; Davis, M.M. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. USA 2006, 103, 18992–18997. [Google Scholar] [CrossRef]
- Yin, H.L.; Janmey, P.A. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 2003, 65, 761–789. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.; Bi, K.; Rodriguez, F.; Tanaka, Y.; Schoenberger, S.; Altman, A. Vav1/Rac-dependent actin cytoskeleton reorganization is required for lipid raft clustering in T cells. J. Cell Biol. 2001, 155, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Inabe, K.; Ishiai, M.; Scharenberg, A.M.; Freshney, N.; Downward, J.; Kurosaki, T. Vav3 modulates B cell receptor responses by regulating phosphoinositide 3-kinase activation. J. Exp. Med. 2002, 195, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Hirao, M.; Sato, N.; Kondo, T.; Yonemura, S.; Monden, M.; Sasaki, T.; Takai, Y.; Tsukita, S. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: Possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol. 1996, 135, 37–51. [Google Scholar] [CrossRef]
- Heiska, L.; Alfthan, K.; Grönholm, M.; Vilja, P.; Vaheri, A.; Carpén, O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 1998, 273, 21893–21900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Bohlson, S.S.; Dy, M.; Tenner, A.J. Modulated interaction of the ERM protein, moesin, with CD93. Immunology 2005, 115, 63–73. [Google Scholar] [CrossRef]
- Kaufmann, S.; Käs, J.; Goldmann, W.H.; Sackmann, E.; Isenberg, G. Talin anchors and nucleates actin filaments at lipid membranes. A direct demonstration. FEBS Lett. 1992, 314, 203–205. [Google Scholar] [CrossRef]
- Martel, V.; Racaud-Sultan, C.; Dupe, S.; Marie, C.; Paulhe, F.; Galmiche, A.; Block, M.R.; Albiges-Rizo, C. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 2001, 276, 21217–21227. [Google Scholar] [CrossRef]
- Shaw, M.L.; Stone, K.L.; Colangelo, C.M.; Gulcicek, E.E.; Palese, P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008, 4, e1000085. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; Charles, P.D.; Hester, S.S.; Thomas, B.; Trudgian, D.; Martínez-Alonso, M.; Fodor, E. Conserved and host-specific features of influenza virion architecture. Nat. Commun. 2014, 5, 4816. [Google Scholar] [CrossRef]
- Avalos, R.T.; Yu, Z.; Nayak, D.P. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J. Virol. 1997, 71, 2947–2958. [Google Scholar] [PubMed]
- Digard, P.; Elton, D.; Bishop, K.; Medcalf, E.; Weeds, A.; Pope, B. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J. Virol. 1999, 73, 2222–2231. [Google Scholar] [PubMed]
- Mayer, D.; Molawi, K.; Martínez-Sobrido, L.; Ghanem, A.; Thomas, S.; Baginsky, S.; Grossmann, J.; García-Sastre, A.; Schwemmle, M. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 2007, 6, 672–682. [Google Scholar] [CrossRef]
- Bradel-Tretheway, B.G.; Mattiacio, J.L.; Krasnoselsky, A.; Stevenson, C.; Purdy, D.; Dewhurst, S.; Katze, M.G. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J. Virol. 2011, 85, 8569–8581. [Google Scholar] [CrossRef]
- Yu, G.; Liang, W.; Liu, J.; Meng, D.; Wei, L.; Chai, T.; Cai, Y. Proteomic Analysis of Differential Expression of Cellular Proteins in Response to Avian H9N2 Virus Infection of A549 Cells. Front. Microbiol. 2016, 7, 1962. [Google Scholar] [CrossRef]
- Mindaye, S.T.; Ilyushina, N.A.; Fantoni, G.; Alterman, M.A.; Donnelly, R.P.; Eichelberger, M.C. Impact of Influenza A Virus Infection on the Proteomes of Human Bronchoepithelial Cells from Different Donors. J. Proteome Res. 2017, 16, 3287–3297. [Google Scholar] [CrossRef]
- Ohman, T.; Rintahaka, J.; Kalkkinen, N.; Matikainen, S.; Nyman, T.A. Actin and RIG-I/MAVS signaling components translocate to mitochondria upon influenza A virus infection of human primary macrophages. J. Immunol. 2009, 182, 5682–5692. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M.; Berard, A.; Xu, W.; Krokhin, O.; Meng, X.; Cortens, J.P.; Kobasa, D.; Wilkins, J.; Brown, E.G. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 2010, 84, 10888–10906. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Xiang, Y.; Guo, C.; Pei, Y.; Wang, Y.; Kitazato, K. Cofilin-1 is involved in regulation of actin reorganization during influenza A virus assembly and budding. Biochem. Biophys. Res. Commun. 2014, 453, 821–825. [Google Scholar] [CrossRef]
- Chen, D.Y.; Husain, M. Caspase-mediated degradation of host cortactin that promotes influenza A virus infection in epithelial cells. Virology 2016, 497, 146–156. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, Q.; Chen, S.; Gao, S.; Song, L.; Liu, P.; Huang, W. Influenza A virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein. J. Virol. 2013, 87, 3039–3052. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Sheng, C.; Gu, X.; Liu, D.; Yao, C.; Gao, S.; Chen, S.; Huang, Y.; Huang, W.; Fang, M. Suppression of Rac1 Signaling by Influenza A Virus NS1 Facilitates Viral Replication. Sci. Rep. 2016, 6, 35041. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Chen, Y.; Wei, H.; Gao, G.F.; Liu, H.; Huang, S.; Chen, J.L. Transport of influenza virus neuraminidase (NA) to host cell surface is regulated by ARHGAP21 and Cdc42 proteins. J. Biol. Chem. 2012, 287, 9804–9816. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.C.; Compans, R.W. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. USA 1998, 95, 5746–5751. [Google Scholar] [CrossRef] [PubMed]
- Simpson-Holley, M.; Ellis, D.; Fisher, D.; Elton, D.; McCauley, J.; Digard, P. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 2002, 301, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Kumakura, M.; Kawaguchi, A.; Nagata, K. Actin-myosin network is required for proper assembly of influenza virus particles. Virology 2015, 476, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.A.; Compans, R.W. Effect of cytochalasin B on the maturation of enveloped viruses. J. Exp. Med. 1979, 150, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Arcangeletti, M.C.; de Conto, F.; Ferraglia, F.; Pinardi, F.; Gatti, R.; Orlandini, G.; Covan, S.; Motta, F.; Rodighiero, I.; Dettori, G.; et al. Host-cell-dependent role of actin cytoskeleton during the replication of a human strain of influenza A virus. Arch. Virol. 2008, 153, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Nturibi, E.; Bhagwat, A.R.; Coburn, S.; Myerburg, M.M.; Lakdawala, S.S. Intracellular Colocalization of Influenza Viral RNA and Rab11A Is Dependent upon Microtubule Filaments. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Momose, F.; Kikuchi, Y.; Komase, K.; Morikawa, Y. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect. 2007, 9, 1422–1433. [Google Scholar] [CrossRef]
- De Conto, F.; Di Lonardo, E.; Arcangeletti, M.C.; Chezzi, C.; Medici, M.C.; Calderaro, A. Highly dynamic microtubules improve the effectiveness of early stages of human influenza A/NWS/33 virus infection in LLC-MK2 cells. PLoS ONE 2012, 7, e41207. [Google Scholar] [CrossRef] [PubMed]
- Reed, N.A.; Cai, D.; Blasius, T.L.; Jih, G.T.; Meyhofer, E.; Gaertig, J.; Verhey, K.J. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 2006, 16, 2166–2172. [Google Scholar] [CrossRef] [PubMed]
- Husain, M.; Cheung, C.Y. Histone deacetylase 6 inhibits influenza A virus release by downregulating the trafficking of viral components to the plasma membrane via its substrate, acetylated microtubules. J. Virol. 2014, 88, 11229–11239. [Google Scholar] [CrossRef] [PubMed]
- Bedi, S.; Noda, T.; Kawaoka, Y.; Ono, A. A Defect in Influenza A Virus Particle Assembly Specific to Primary Human Macrophages. mBio 2018, 9. [Google Scholar] [CrossRef]
- Takeda, M.; Leser, G.P.; Russell, C.J.; Lamb, R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 2003, 100, 14610–14617. [Google Scholar] [CrossRef] [PubMed]
- Leser, G.P.; Lamb, R.A. Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 2005, 342, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Hess, S.T.; Kumar, M.; Verma, A.; Farrington, J.; Kenworthy, A.; Zimmerberg, J. Quantitative electron microscopy and fluorescence spectroscopy of the membrane distribution of influenza hemagglutinin. J. Cell Biol. 2005, 169, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Leser, G.P.; Morita, E.; Lamb, R.A. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 2007, 81, 7111–7123. [Google Scholar] [CrossRef]
- Zhang, J.; Pekosz, A.; Lamb, R.A. Influenza virus assembly and lipid raft microdomains: A role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 2000, 74, 4634–4644. [Google Scholar] [CrossRef]
- Ohkura, T.; Momose, F.; Ichikawa, R.; Takeuchi, K.; Morikawa, Y. Influenza A virus hemagglutinin and neuraminidase mutually accelerate their apical targeting through clustering of lipid rafts. J. Virol. 2014, 88, 10039–10055. [Google Scholar] [CrossRef]
- Scolari, S.; Engel, S.; Krebs, N.; Plazzo, A.P.; De Almeida, R.F.; Prieto, M.; Veit, M.; Herrmann, A. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 2009, 284, 15708–15716. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Balannik, V.; Pinto, L.H.; Lamb, R.A. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 2010, 84, 5078–5088. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.L.; Leser, G.P.; Ma, C.; Lamb, R.A. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J. Virol. 2013, 87, 9973–9982. [Google Scholar] [CrossRef] [PubMed]
- Barman, S.; Ali, A.; Hui, E.K.; Adhikary, L.; Nayak, D.P. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 2001, 77, 61–69. [Google Scholar] [CrossRef]
- Ali, A.; Avalos, R.T.; Ponimaskin, E.; Nayak, D.P. Influenza virus assembly: Effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 2000, 74, 8709–8719. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Leser, G.P.; Zhang, J.; Lamb, R.A. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997, 16, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lamb, R.A. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology 1996, 225, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Chlanda, P.; Schraidt, O.; Kummer, S.; Riches, J.; Oberwinkler, H.; Prinz, S.; Kräusslich, H.G.; Briggs, J.A. Structural Analysis of the Roles of Influenza A Virus Membrane-Associated Proteins in Assembly and Morphology. J. Virol. 2015, 89, 8957–8966. [Google Scholar] [CrossRef]
- Elster, C.; Larsen, K.; Gagnon, J.; Ruigrok, R.W.; Baudin, F. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J. Gen. Virol. 1997, 78 Pt 7, 1589–1596. [Google Scholar] [CrossRef]
- Bui, M.; Wills, E.G.; Helenius, A.; Whittaker, G.R. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 2000, 74, 1781–1786. [Google Scholar] [CrossRef]
- Baudin, F.; Petit, I.; Weissenhorn, W.; Ruigrok, R.W. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 2001, 281, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, A.; Hirayama, E.; Hiraki, A.; Ishida, Y.; Kim, J. Nuclear export of influenza viral ribonucleoprotein is temperature-dependently inhibited by dissociation of viral matrix protein. Virology 2003, 306, 244–253. [Google Scholar] [CrossRef]
- Elton, D.; Simpson-Holley, M.; Archer, K.; Medcalf, L.; Hallam, R.; McCauley, J.; Digard, P. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 2001, 75, 408–419. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.E.; Talon, J.; Palese, P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998, 17, 288–296. [Google Scholar] [CrossRef] [PubMed]
- De Castro Martin, I.F.; Fournier, G.; Sachse, M.; Pizarro-Cerda, J.; Risco, C.; Naffakh, N. Influenza virus genome reaches the plasma membrane via a modified endoplasmic reticulum and Rab11-dependent vesicles. Nat. Commun. 2017, 8, 1396. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, A.; Hirohama, M.; Harada, Y.; Osari, S.; Nagata, K. Influenza Virus Induces Cholesterol-Enriched Endocytic Recycling Compartments for Budozone Formation via Cell Cycle-Independent Centrosome Maturation. PLoS Pathog. 2015, 11, e1005284. [Google Scholar] [CrossRef] [PubMed]
- Amorim, M.J.; Kao, R.Y.; Digard, P. Nucleozin targets cytoplasmic trafficking of viral ribonucleoprotein-Rab11 complexes in influenza A virus infection. J. Virol. 2013, 87, 4694–4703. [Google Scholar] [CrossRef] [PubMed]
- Bialas, K.M.; Bussey, K.A.; Stone, R.L.; Takimoto, T. Specific nucleoprotein residues affect influenza virus morphology. J. Virol. 2014, 88, 2227–2234. [Google Scholar] [CrossRef]
- Chlanda, P.; Mekhedov, E.; Waters, H.; Sodt, A.; Schwartz, C.; Nair, V.; Blank, P.S.; Zimmerberg, J. Palmitoylation Contributes to Membrane Curvature in Influenza A Virus Assembly and Hemagglutinin-Mediated Membrane Fusion. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Martyna, A.; Bahsoun, B.; Badham, M.D.; Srinivasan, S.; Howard, M.J.; Rossman, J.S. Membrane remodeling by the M2 amphipathic helix drives influenza virus membrane scission. Sci. Rep. 2017, 7, 44695. [Google Scholar] [CrossRef]
- Palese, P.; Tobita, K.; Ueda, M.; Compans, R.W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 1974, 61, 397–410. [Google Scholar] [CrossRef]
- Palese, P.; Compans, R.W. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): Mechanism of action. J. Gen. Virol. 1976, 33, 159–163. [Google Scholar] [CrossRef]
- Kilbourne, E.D.; Murphy, J.S. Genetic studies of influenza viruses. I. Viral morphology and growth capacity as exchangeable genetic traits. Rapid in ovo adaptation of early passage Asian strain isolates by combination with PR8. J. Exp. Med. 1960, 111, 387–406. [Google Scholar] [CrossRef] [PubMed]
- Choppin, P.W.; Murphy, J.S.; Tamm, I. Studies of two kinds of virus particles which comprise influenza A2 virus strains. III. Morphological characteristics: Independence to morphological and functional traits. J. Exp. Med. 1960, 112, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.M.; Dawson, I.M.; Elford, W.J. Filamentous forms associated with newly isolated influenza virus. Lancet 1949, 1, 602. [Google Scholar] [CrossRef]
- Seladi-Schulman, J.; Steel, J.; Lowen, A.C. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. J. Virol. 2013, 87, 13343–13353. [Google Scholar] [CrossRef]
- Mosley, V.M.; Wyckoff, R.W. Electron micrography of the virus of influenza. Nature 1946, 157, 263. [Google Scholar] [CrossRef]
- Burleigh, L.M.; Calder, L.J.; Skehel, J.J.; Steinhauer, D.A. Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 2005, 79, 1262–1270. [Google Scholar] [CrossRef]
- Bourmakina, S.V.; García-Sastre, A. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 2003, 84, 517–527. [Google Scholar] [CrossRef]
- Elleman, C.J.; Barclay, W.S. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 2004, 321, 144–153. [Google Scholar] [CrossRef]
- Roberts, P.C.; Lamb, R.A.; Compans, R.W. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 1998, 240, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.J.; Danzy, S.; Kyriakis, C.S.; Deymier, M.J.; Lowen, A.C.; Steel, J. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J. Virol. 2014, 88, 3802–3814. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki-Horimoto, K.; Horimoto, T.; Noda, T.; Kiso, M.; Maeda, J.; Watanabe, S.; Muramoto, Y.; Fujii, K.; Kawaoka, Y. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 2006, 80, 5233–5240. [Google Scholar] [CrossRef] [PubMed]
- Grantham, M.L.; Stewart, S.M.; Lalime, E.N.; Pekosz, A. Tyrosines in the influenza A virus M2 protein cytoplasmic tail are critical for production of infectious virus particles. J. Virol. 2010, 84, 8765–8776. [Google Scholar] [CrossRef] [PubMed]
- Salas, P.J.; Misek, D.E.; Vega-Salas, D.E.; Gundersen, D.; Cereijido, M.; Rodriguez-Boulan, E. Microtubules and actin filaments are not critically involved in the biogenesis of epithelial cell surface polarity. J. Cell Biol. 1986, 102, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Rindler, M.J.; Ivanov, I.E.; Sabatini, D.D. Microtubule-acting drugs lead to the nonpolarized delivery of the influenza hemagglutinin to the cell surface of polarized Madin-Darby canine kidney cells. J. Cell Biol. 1987, 104, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Van Zeijl, M.J.; Matlin, K.S. Microtubule perturbation inhibits intracellular transport of an apical membrane glycoprotein in a substrate-dependent manner in polarized Madin-Darby canine kidney epithelial cells. Cell Regul. 1990, 1, 921–936. [Google Scholar] [CrossRef]
- Noda, Y.; Okada, Y.; Saito, N.; Setou, M.; Xu, Y.; Zhang, Z.; Hirokawa, N. KIFC3, a microtubule minus end-directed motor for the apical transport of annexin XIIIb-associated Triton-insoluble membranes. J. Cell Biol. 2001, 155, 77–88. [Google Scholar] [CrossRef]
- Ramos-Nascimento, A.; Kellen, B.; Ferreira, F.; Alenquer, M.; Vale-Costa, S.; Raposo, G.; Delevoye, C.; Amorim, M.J. KIF13A mediates trafficking of influenza A virus ribonucleoproteins. J. Cell Sci. 2017, 130, 4038–4050. [Google Scholar] [CrossRef]
- Gudheti, M.V.; Curthoys, N.M.; Gould, T.J.; Kim, D.; Gunewardene, M.S.; Gabor, K.A.; Gosse, J.A.; Kim, C.H.; Zimmerberg, J.; Hess, S.T. Actin mediates the nanoscale membrane organization of the clustered membrane protein influenza hemagglutinin. Biophys. J. 2013, 104, 2182–2192. [Google Scholar] [CrossRef]
- Thaa, B.; Herrmann, A.; Veit, M. Intrinsic cytoskeleton-dependent clustering of influenza virus M2 protein with hemagglutinin assessed by FLIM-FRET. J. Virol. 2010, 84, 12445–12449. [Google Scholar] [CrossRef] [PubMed]
- Rindler, M.J.; Ivanov, I.E.; Plesken, H.; Sabatini, D.D. Polarized delivery of viral glycoproteins to the apical and basolateral plasma membranes of Madin-Darby canine kidney cells infected with temperature-sensitive viruses. J. Cell Biol. 1985, 100, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Boulan, E.; Sabatini, D.D. Asymmetric budding of viruses in epithelial monlayers: A model system for study of epithelial polarity. Proc. Natl. Acad. Sci. USA 1978, 75, 5071–5075. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Boulan, E.; Pendergast, M. Polarized distribution of viral envelope proteins in the plasma membrane of infected epithelial cells. Cell 1980, 20, 45–54. [Google Scholar] [CrossRef]
- Rodriguez-Boulan, E.; Paskiet, K.T.; Sabatini, D.D. Assembly of enveloped viruses in Madin-Darby canine kidney cells: Polarized budding from single attached cells and from clusters of cells in suspension. J. Cell Biol. 1983, 96, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Boulan, E.; Paskiet, K.T.; Salas, P.J.; Bard, E. Intracellular transport of influenza virus hemagglutinin to the apical surface of Madin-Darby canine kidney cells. J. Cell Biol. 1984, 98, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Barman, S.; Nayak, D.P. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 2000, 74, 6538–6545. [Google Scholar] [CrossRef] [PubMed]
- Barman, S.; Adhikary, L.; Chakrabarti, A.K.; Bernas, C.; Kawaoka, Y.; Nayak, D.P. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J. Virol. 2004, 78, 5258–5269. [Google Scholar] [CrossRef] [PubMed]
- Hughey, P.G.; Compans, R.W.; Zebedee, S.L.; Lamb, R.A. Expression of the influenza A virus M2 protein is restricted to apical surfaces of polarized epithelial cells. J. Virol. 1992, 66, 5542–5552. [Google Scholar]
- Lin, S.; Naim, H.Y.; Rodriguez, A.C.; Roth, M.G. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 1998, 142, 51–57. [Google Scholar] [CrossRef]
- Bonilha, V.L.; Marmorstein, A.D.; Cohen-Gould, L.; Rodriguez-Boulan, E. Apical sorting of influenza hemagglutinin by transcytosis in retinal pigment epithelium. J. Cell Sci. 1997, 110 Pt 15, 1717–1727. [Google Scholar]
- Guerriero, C.J.; Lai, Y.; Weisz, O.A. Differential sorting and Golgi export requirements for raft-associated and raft-independent apical proteins along the biosynthetic pathway. J. Biol. Chem. 2008, 283, 18040–18047. [Google Scholar] [CrossRef] [PubMed]
- Cresawn, K.O.; Potter, B.A.; Oztan, A.; Guerriero, C.J.; Ihrke, G.; Goldenring, J.R.; Apodaca, G.; Weisz, O.A. Differential involvement of endocytic compartments in the biosynthetic traffic of apical proteins. EMBO J. 2007, 26, 3737–3748. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, M.; Amorim, M.J.; Digard, P. Lipid raft-dependent targeting of the influenza A virus nucleoprotein to the apical plasma membrane. Traffic 2004, 5, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Mora, R.; Rodriguez-Boulan, E.; Palese, P.; García-Sastre, A. Apical budding of a recombinant influenza A virus expressing a hemagglutinin protein with a basolateral localization signal. J. Virol. 2002, 76, 3544–3553. [Google Scholar] [CrossRef] [PubMed]
- Barman, S.; Adhikary, L.; Kawaoka, Y.; Nayak, D.P. Influenza A virus hemagglutinin containing basolateral localization signal does not alter the apical budding of a recombinant influenza A virus in polarized MDCK cells. Virology 2003, 305, 138–152. [Google Scholar] [CrossRef]
- Wohlgemuth, N.; Lane, A.P.; Pekosz, A. Influenza A Virus M2 Protein Apical Targeting Is Required for Efficient Virus Replication. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Matsumoto, K.; Nagata, K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J. Virol. 2012, 86, 11086–11095. [Google Scholar] [CrossRef]
- Avilov, S.V.; Moisy, D.; Munier, S.; Schraidt, O.; Naffakh, N.; Cusack, S. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J. Virol. 2012, 86, 1433–1448. [Google Scholar] [CrossRef]
- Avilov, S.V.; Moisy, D.; Naffakh, N.; Cusack, S. Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein. Vaccine 2012, 30, 7411–7417. [Google Scholar] [CrossRef]
- Chou, Y.Y.; Heaton, N.S.; Gao, Q.; Palese, P.; Singer, R.H.; Singer, R.; Lionnet, T. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 2013, 9, e1003358. [Google Scholar] [CrossRef]
- Lakdawala, S.S.; Wu, Y.; Wawrzusin, P.; Kabat, J.; Broadbent, A.J.; Lamirande, E.W.; Fodor, E.; Altan-Bonnet, N.; Shroff, H.; Subbarao, K. Influenza a virus assembly intermediates fuse in the cytoplasm. PLoS Pathog. 2014, 10, e1003971. [Google Scholar] [CrossRef]
- Momose, F.; Sekimoto, T.; Ohkura, T.; Jo, S.; Kawaguchi, A.; Nagata, K.; Morikawa, Y. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS ONE 2011, 6, e21123. [Google Scholar] [CrossRef] [PubMed]
- Avilov, S.; Magnus, J.; Cusack, S.; Naffakh, N. Time-Resolved Visualisation of Nearly-Native Influenza A Virus Progeny Ribonucleoproteins and Their Individual Components in Live Infected Cells. PLoS ONE 2016, 11, e0149986. [Google Scholar] [CrossRef] [PubMed]
- Vale-Costa, S.; Alenquer, M.; Sousa, A.L.; Kellen, B.; Ramalho, J.; Tranfield, E.M.; Amorim, M.J. Influenza A virus ribonucleoproteins modulate host recycling by competing with Rab11 effectors. J. Cell Sci. 2016, 129, 1697–1710. [Google Scholar] [CrossRef]
- Bruce, E.A.; Digard, P.; Stuart, A.D. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 2010, 84, 5848–5859. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Webster, B.M.; Mastronarde, D.N.; Verhey, K.J.; Voeltz, G.K. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 2010, 190, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Leser, G.P.; Lamb, R.A. Lateral Organization of Influenza Virus Proteins in the Budozone Region of the Plasma Membrane. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- De Conto, F.; Fazzi, A.; Razin, S.V.; Arcangeletti, M.C.; Medici, M.C.; Belletti, S.; Chezzi, C.; Calderaro, A. Mammalian Diaphanous-related formin-1 restricts early phases of influenza A/NWS/33 virus (H1N1) infection in LLC-MK2 cells by affecting cytoskeleton dynamics. Mol. Cell. Biochem. 2018, 437, 185–201. [Google Scholar] [CrossRef]
- Sun, X.; Whittaker, G.R. Role of the actin cytoskeleton during influenza virus internalization into polarized epithelial cells. Cell. Microbiol. 2007, 9, 1672–1682. [Google Scholar] [CrossRef]
- Gujuluva, C.N.; Kundu, A.; Murti, K.G.; Nayak, D.P. Abortive replication of influenza virus A/WSN/33 in HeLa229 cells: Defective viral entry and budding processes. Virology 1994, 204, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Leser, G.P.; Jackson, D.; Lamb, R.A. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 2008, 82, 10059–10070. [Google Scholar] [CrossRef] [PubMed]
- Honigmann, A.; Sadeghi, S.; Keller, J.; Hell, S.W.; Eggeling, C.; Vink, R. A lipid bound actin meshwork organizes liquid phase separation in model membranes. eLife 2014, 3, e01671. [Google Scholar] [CrossRef] [PubMed]
- Vogel, S.K.; Greiss, F.; Khmelinskaia, A.; Schwille, P. Control of lipid domain organization by a biomimetic contractile actomyosin cortex. eLife 2017, 6. [Google Scholar] [CrossRef]
- Haggie, P.M.; Kim, J.K.; Lukacs, G.L.; Verkman, A.S. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol. Biol. Cell 2006, 17, 4937–4945. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Veracini, L.; Benistant, C.; Jacobson, K. The transmembrane protein CBP plays a role in transiently anchoring small clusters of Thy-1, a GPI-anchored protein, to the cytoskeleton. J. Cell Sci. 2009, 122, 3966–3972. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.A.; Vega, A.; Riedl, M.; Collins, R.F.; Ostrowski, P.P.; Woods, E.C.; Bertozzi, C.R.; Tammi, M.I.; Lidke, D.S.; Johnson, P.; et al. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 2018, 172, 305–317.e310. [Google Scholar] [CrossRef] [PubMed]
- Tinevez, J.Y.; Schulze, U.; Salbreux, G.; Roensch, J.; Joanny, J.F.; Paluch, E. Role of cortical tension in bleb growth. Proc. Natl. Acad. Sci. USA 2009, 106, 18581–18586. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.S.; Nowak, R.B.; Zhou, S.; Giannetto, M.; Gokhin, D.S.; Papoin, J.; Ghiran, I.C.; Blanc, L.; Wan, J.; Fowler, V.M. Myosin IIA interacts with the spectrin-actin membrane skeleton to control red blood cell membrane curvature and deformability. Proc. Natl. Acad. Sci. USA 2018, 115, E4377–E4385. [Google Scholar] [CrossRef] [PubMed]
- McCown, M.F.; Pekosz, A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 2005, 79, 3595–3605. [Google Scholar] [CrossRef]
- Madsen, J.J.; Grime, J.M.A.; Rossman, J.S.; Voth, G.A. Entropic forces drive clustering and spatial localization of influenza A M2 during viral budding. Proc. Natl. Acad. Sci. USA 2018, 115, E8595–E8603. [Google Scholar] [CrossRef] [PubMed]
- Martyna, A.; Gómez-Llobregat, J.; Lindén, M.; Rossman, J.S. Curvature Sensing by a Viral Scission Protein. Biochemistry 2016, 55, 3493–3496. [Google Scholar] [CrossRef] [PubMed]
- Giner, D.; Neco, P.; Francés, M.e.M.; López, I.; Viniegra, S.; Gutiérrez, L.M. Real-time dynamics of the F-actin cytoskeleton during secretion from chromaffin cells. J. Cell Sci. 2005, 118, 2871–2880. [Google Scholar] [CrossRef] [PubMed]
- Giner, D.; López, I.; Villanueva, J.; Torres, V.; Viniegra, S.; Gutiérrez, L.M. Vesicle movements are governed by the size and dynamics of F-actin cytoskeletal structures in bovine chromaffin cells. Neuroscience 2007, 146, 659–669. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).