S1 Subunit of Spike Protein from a Current Highly Virulent Porcine Epidemic Diarrhea Virus Is an Important Determinant of Virulence in Piglets
Abstract
1. Introduction
2. Material and Methods
2.1. Cells and Viruses
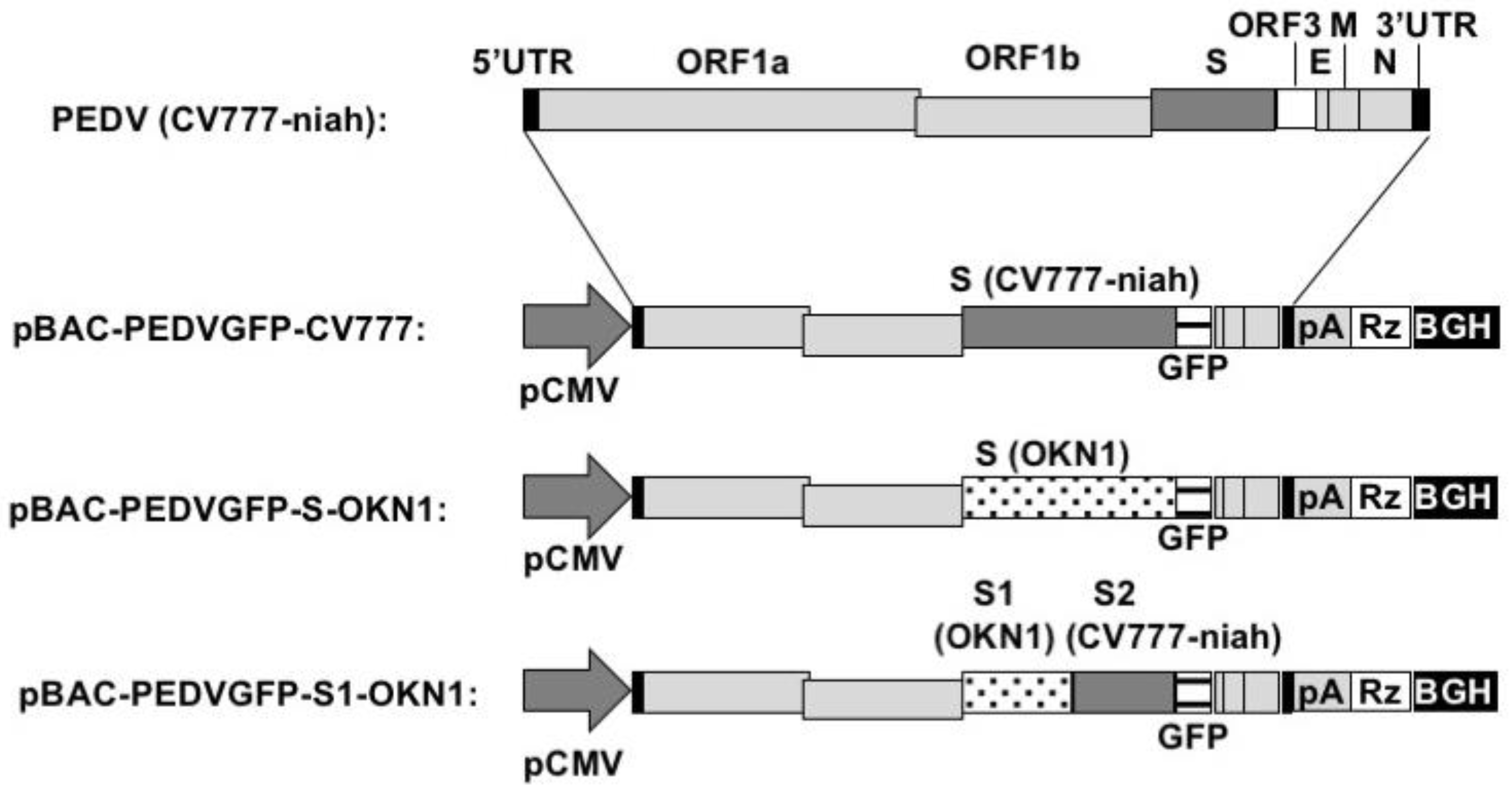
2.2. Construction of Bacterial Artificial Chromosomes (BAC)
2.3. Recovery of Recombinant PEDVs
2.4. Animal Studies
2.4.1. Experimental Infection in 7-Day-Old Gnotobiotic Piglets
2.4.2. Experimental Infection in Five-Day-Old Gnotobiotic Piglets
2.5. Virus Isolation and Sequencing
2.6. Antibody Detection by Immunofluorescence Assay
3. Results
3.1. Recovery of Recombinant PEDVs Derived from CV777
3.2. Experimental Infection in Seven-Day-Old Gnotobiotic Piglets
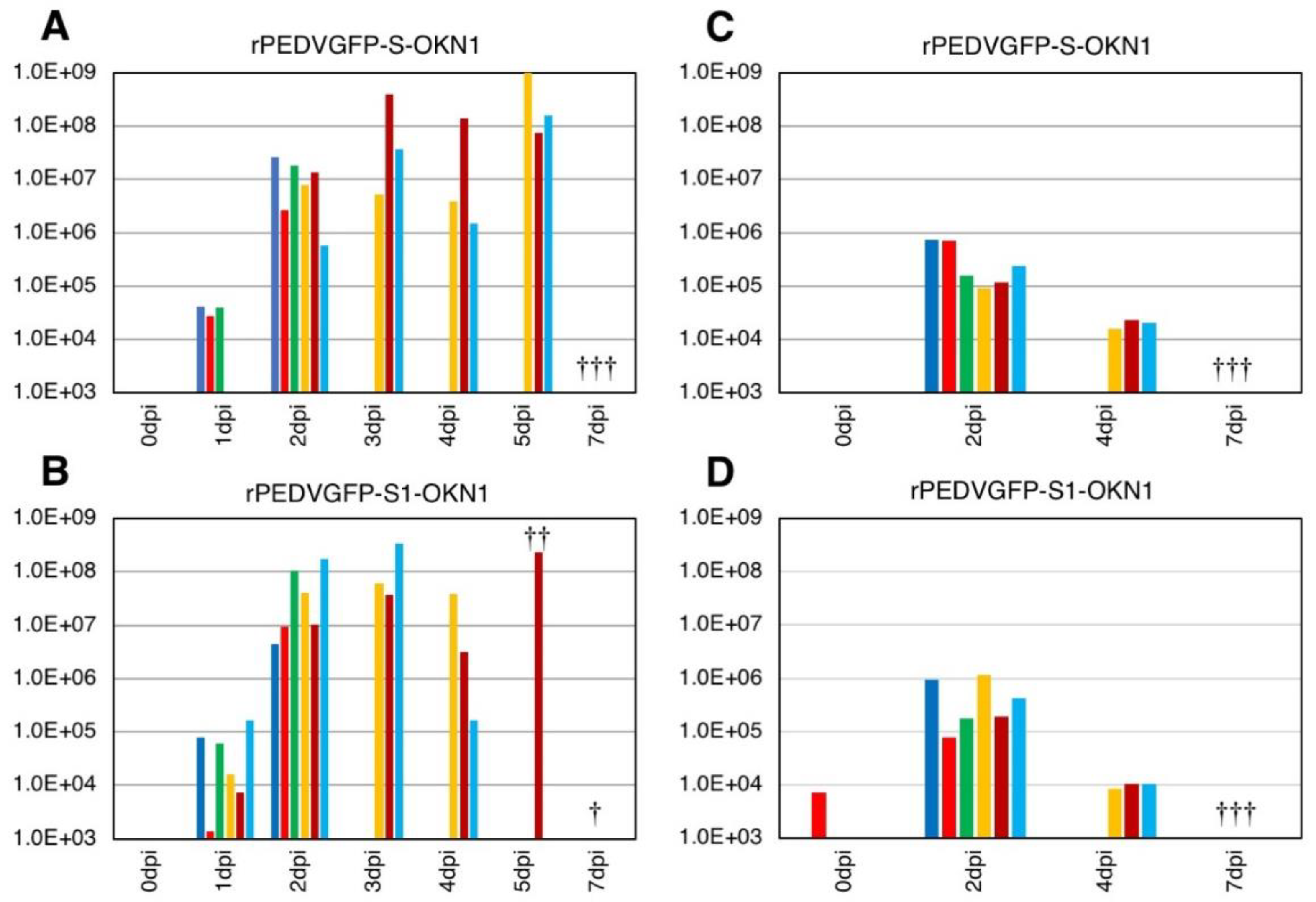
3.3. Experimental Infection in Five-Day-Old Gnotobiotic Piglets
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.N. An apparently new syndrome of porcine epidemic diarrhea. Vet. Rec. 1977, 100, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Chasey, D.; Cartwright, S.F. Virus-like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978, 25, 255–256. [Google Scholar] [PubMed]
- Takahashi, K.; Okada, K.; Ohshima, K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Jpn. J. Vet. Sci. 1983, 45, 829–832. [Google Scholar] [CrossRef]
- Kweon, C.H.; Kwon, B.J.; Jung, T.S.; Kee, Y.J.; Hur, D.H.; Hwang, E.K.; Rhee, J.C.; An, S.H. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J. Vet. Res. 1993, 33, 249–254. [Google Scholar]
- Van Reeth, K.; Pensaert, M.B. Prevalence of infectious with enzootic respiratory and enteric viruses in feeder pigs entering fattening herds. Vet. Rec. 1994, 135, 594–597. [Google Scholar] [PubMed]
- Nagy, B.; Nagy, G.; Meder, M.; Mocsari, E. Enterotoxigenic Escherichia coli, rotavirus, porcine epidemic diarrhoea virus, adenovirus and calici-like virus in porcine postweaning diarrhoea in Hungary. Acta Vet. Hung. 1996, 44, 9–19. [Google Scholar] [PubMed]
- Song, D.; Park, B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis and vaccine. Virus Genes 2012, 44, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, C.; Shi, H.; Qiu, H.; Liu, S.; Chen, X.; Zhang, Z.; Feng, L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010, 155, 1471–1476. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, J.; Yu, F.; Ge, J.; Lin, T.; Song, T. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) samples from field cases in Fujian, China. Virus Genes 2012, 45, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.L.; Zhu, L.; Ma, J.Y.; Zhou, Q.F.; Song, Y.H.; Sun, B.L.; Chen, R.A.; Xie, Q.M.; Bee, Y.Z. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field cases in south China. Virus Genes 2012, 45, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Dickerman, A.W.; Pineyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio 2013, 4, e00737-13. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, G.W.; Hoang, H.; Schwartz, K.J.; Burrough, E.R.; Sun, D.; Madson, D.; Cooper, V.L.; Pillatzki, A.; Gauger, P.; Schmitt, B.J.; et al. Emergence of porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 2013, 25, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Byrum, B.; Zhang, Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014, 20, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Marthaler, D.; Wang, Q.; Culhane, M.R.; Rossow, K.D.; Rovira, A.R.; Collins, J.; Saif, L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014, 20, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Pasick, J.; Berhane, Y.; Ojkic, D.; Maxie, G.; Embury-Hyatt, C.; Swekla, K.; Handel, K.; Fairles, J.; Alexandersen, S. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound. Emerg. Dis. 2014, 61, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014, 20, 1223–1226. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Lee, K.; Pi, J.H.; Park, S.; Song, C.; Choi, I.; Lee, J.; Lee, D.; Lee, S. Comparative genome analysis and molecular epidemiclogy of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infect. Genet. Evol. 2014, 26, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Chiou, M.; Yu, C.; Chang, C.; Chung, W.; Wu, H.; Lin, C.; Lin, C. Molecular characterization of the porcine epidemic diarrhea virus TW4/2014 in Taiwan. Austin Virol. Retrovirol. 2014, 1, 2–5. [Google Scholar]
- Hanke, D.; Jenckel, M.; Petrov, A.; Pitzmann, M.; Stadler, J.; Akimkin, V.; Blome, S.; Pohlmann, A.; Schirrmeier, H.; Beer, M.; et al. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg. Infect. Dis. 2015, 21, 4893–4896. [Google Scholar] [CrossRef] [PubMed]
- Theuns, S.; Conceição-Neto, N.; Christiaens, I.; Zeller, M.; Desmarets, L.M.; Roukaerts, I.D.; Acar, D.D.; Heylen, E.; Matthijnssens, J.; Nauwynck, H.J. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in Belgium, January 2015. Genome Announc. 2015, 3, e00506-15. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Miyazaki, A.; Takahashi, O.; Hashizume, O.; Hase, Y.; Ohashi, S.; Suzuki, T. Complete genome sequence of the porcine epidemic diarrhoea virus variant Tottori2/JPN/2014. Genome Announc. 2015, 3, e00877-15. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Murakami, S.; Takahashi, O.; Kodera, A.; Masuda, T.; Itou, S.; Miyazaki, A.; Ohashi, S.; Tsutsui, T. Molecular characterization of pig epidemic diarrhoea viruses isolated in Japan from 2013 to 2014. Infect. Genet. Evol. 2015, 36, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Murakami, S.; Takahashi, O.; Miyazaki, A.; Ohashi, S.; Yamasato, H.; Suzuki, T. Discovery of a new PEDV variant with a large deletion in the spike gene identified in domestic pigs. Arch. Virol. 2015, 160, 2565–2568. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Shibahara, T.; Yamaguchi, R.; Nakade, K.; Yamamoto, T.; Miyazaki, A.; Ohashi, S. Pig epidemic diarrhoea virus S gene variant with a large deletion non-lethal to colostrum-deprived newborn piglet. J. Gen. Virol. 2016, 97, 1823–1826. [Google Scholar] [CrossRef] [PubMed]
- De Haan, C.A.M.; Haijema, B.J.; Schellen, P.; Wichgers Schreur, P.; te Lintelo, E.; Vennema, H.; Rottier, P.J.M. Cleavage of group 1 coronavirus spike proteins: How furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 2008, 82, 6078–6083. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tang, J.; Ma, Y.; Liang, X.; Yang, Y.; Peng, G.; Jiang, S.; Li, J.; Du, L.; Li, F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015, 89, 6121–6125. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, P.; Correa, I.; Pensaert, M.; Jiménez, G.; Enjuanes, L. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J. Gen. Virol. 1988, 69, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, T.M.; Buchmeier, M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology 2001, 279, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Hulswit, R.J.G.; de Haan, C.A.M.; Bosch, B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar] [PubMed]
- Hou, Y.; Lin, C.; Yokoyama, M.; Yount, B.L.; Marthaler, D.; Douglas, A.L.; Ghimire, S.; Qin, Y.; Baric, R.S.; Saif, L.J.; et al. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017, 91, e00227-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ge, X.; Chen, D.; Li, J.; Cal, Y.; Deng, J.; Zhou, L.; Guo, X.; Han, J.; Yang, H. The S is necessary but not sufficient for the virulence of porcine epidemic diarrhea virus novel variant strain BC2011C. J. Virol. 2018, 92, e00603-18. [Google Scholar] [CrossRef] [PubMed]
- Almazán, F.; Sola, I.; Zuñiga, S.; Marquez-Jurado, S.; Morales, L.; Becares, M.; Enjuanes, L. Coronavirus reverse genetic systems; infectious clones and replicons. Virus Res. 2014, 189, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Scobey, T.; Yount, B.L.; Sims, A.C.; Donaldson, E.F.; Agnihotharm, S.S.; Menachey, V.D.; Graham, R.L.; Swanstrom, J.; Bove, P.F.; Kim, J.D.; et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2013, 110, 16157–16162. [Google Scholar] [CrossRef] [PubMed]
- Almazán, F.; Dediego, M.L.; Galán, C.; Escors, D.; Alvarez, E.; Ortego, J.; Sola, I.; Zuñiga, S.; Alonso, S.; Moreno, J.L.; et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006, 80, 10900–10906. [Google Scholar]
- Rasmussen, T.B.; Boniotti, M.B.; Papetti, A.; Papetti, A.; Grasland, B.; Frossard, J.; Dastjerdi, A.; Hulst, M.; Hanke, D.; Pohlmann, A.; et al. Full-length genome sequences of porcine epidemic diarrhoea virus strain CV777; Use of NGS to analyse genomic and sub-genomic RNAs. PLoS ONE 2018, 13, e0193682. [Google Scholar] [CrossRef] [PubMed]
- Pensaert, M.B.; Martelli, P. Porcine epidemic diarrhea: A retrospect from Europe and matters of debate. Virus Res. 2016, 226, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D. Efficacy of a transmissible gastroenteritis coronavirus with an altered ORF-3 gene. Can. J. Vet. Res. 2001, 65, 28–32. [Google Scholar]
- Song, D.S.; Yang, J.S.; Oh, J.S.; Han, J.H.; Park, B.K. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF3. Vaccine 2003, 21, 1833–1842. [Google Scholar] [CrossRef]
- Park, S.J.; Moon, H.J.; Luo, Y.; Kim, H.K.; Kim, E.M.; Yang, J.S.; Song, D.S.; Kang, B.K.; Lee, C.S.; Park, B.K. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes 2008, 36, 95–104. [Google Scholar] [CrossRef] [PubMed]


| Region | Position of Nucleotides in Reference CV777 | Nucleotide in Reference CV777 | Amino Acid in Reference CV777 | Nucleotide in CV777-Niah | Amino Acid in CV777-Niah |
|---|---|---|---|---|---|
| 5’-UTR | 72 | a | Deletion | ||
| 5’-UTR | 82–85 | tcct | Deletion | ||
| ORF1a/1b | 1667 | t | A | c | A |
| ORF1a/1b | 6052 | t | V | c | A |
| ORF1a/1b | 6593 | c | H | t | H |
| ORF1a/1b | 6630 | t | F | c | L |
| ORF1a/1b | 10,542 | g | V | a | I |
| ORF1a/1b | 11,887 | g | G | a | D |
| ORF1a/1b | 12,257 | t | D | c | D |
| Spike | 22,145 | c | S | t | L |
| Spike | 23,323 | g | D | a | N |
| Spike | 24,461 | a | N | c | T |
| Spike/ORF3 | 24,765–24,816 | * | Deletion | ||
| Nucleocapsid | 26,940 | c | N | t | N |
| Inoculum | n | % Mortality | Clinical Signs | Antibodies (GMT) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Diarrhea | Anorexia | Lethargy | Dehydration | Weight Loss | Astasia | ||||
| CV777-niah | 3 | 0 (0/3) | Mild | + | - | - | - | - | 23.8 |
| rPEDVGFP-CV777 | 5 | 0 (0/5) | Mild | + | - | - | - | - | 44.9 |
| rPEDVGFP-S-OKN1 | 4 | 0 (0/4) | Severe | + | + | + | + | + | 160 |
| Inoculum | n a | % Mortality | Death at Days Post-Inoculation | Clinical Signs | |||||
|---|---|---|---|---|---|---|---|---|---|
| Diarrhea | Anorexia | Lethargy | Dehydration | Weight Loss | Astasia | ||||
| rPEDVGFP-S-OKN1 | 6 | 100 (3/3) | 3/3 (7 DPI) | Severe | + | + | + | + | + |
| rPEDVGFP-S1-OKN1 | 6 | 100 (3/3) | 2/3 (5DPI) 1/3 (7DPI) | Severe | + | + | + | + | + |
| DMEM (Mock) | 6 | 0 (0/3) | 0/3 (28 DPI) | - | - | - | - | - | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, T.; Terada, Y.; Enjuanes, L.; Ohashi, S.; Kamitani, W. S1 Subunit of Spike Protein from a Current Highly Virulent Porcine Epidemic Diarrhea Virus Is an Important Determinant of Virulence in Piglets. Viruses 2018, 10, 467. https://doi.org/10.3390/v10090467
Suzuki T, Terada Y, Enjuanes L, Ohashi S, Kamitani W. S1 Subunit of Spike Protein from a Current Highly Virulent Porcine Epidemic Diarrhea Virus Is an Important Determinant of Virulence in Piglets. Viruses. 2018; 10(9):467. https://doi.org/10.3390/v10090467
Chicago/Turabian StyleSuzuki, Tohru, Yutaka Terada, Luis Enjuanes, Seiichi Ohashi, and Wataru Kamitani. 2018. "S1 Subunit of Spike Protein from a Current Highly Virulent Porcine Epidemic Diarrhea Virus Is an Important Determinant of Virulence in Piglets" Viruses 10, no. 9: 467. https://doi.org/10.3390/v10090467
APA StyleSuzuki, T., Terada, Y., Enjuanes, L., Ohashi, S., & Kamitani, W. (2018). S1 Subunit of Spike Protein from a Current Highly Virulent Porcine Epidemic Diarrhea Virus Is an Important Determinant of Virulence in Piglets. Viruses, 10(9), 467. https://doi.org/10.3390/v10090467





