The Virology of Taterapox Virus In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cells and Viruses
2.3. Cytoskeleton Dynamics
2.4. Cytokine Analysis
2.5. Microscopy
2.6. Bioinformatics
2.7. Statistics
3. Results
3.1. Comparison of TATV Genome with Genomes of CPXV, VARV, and ECTV
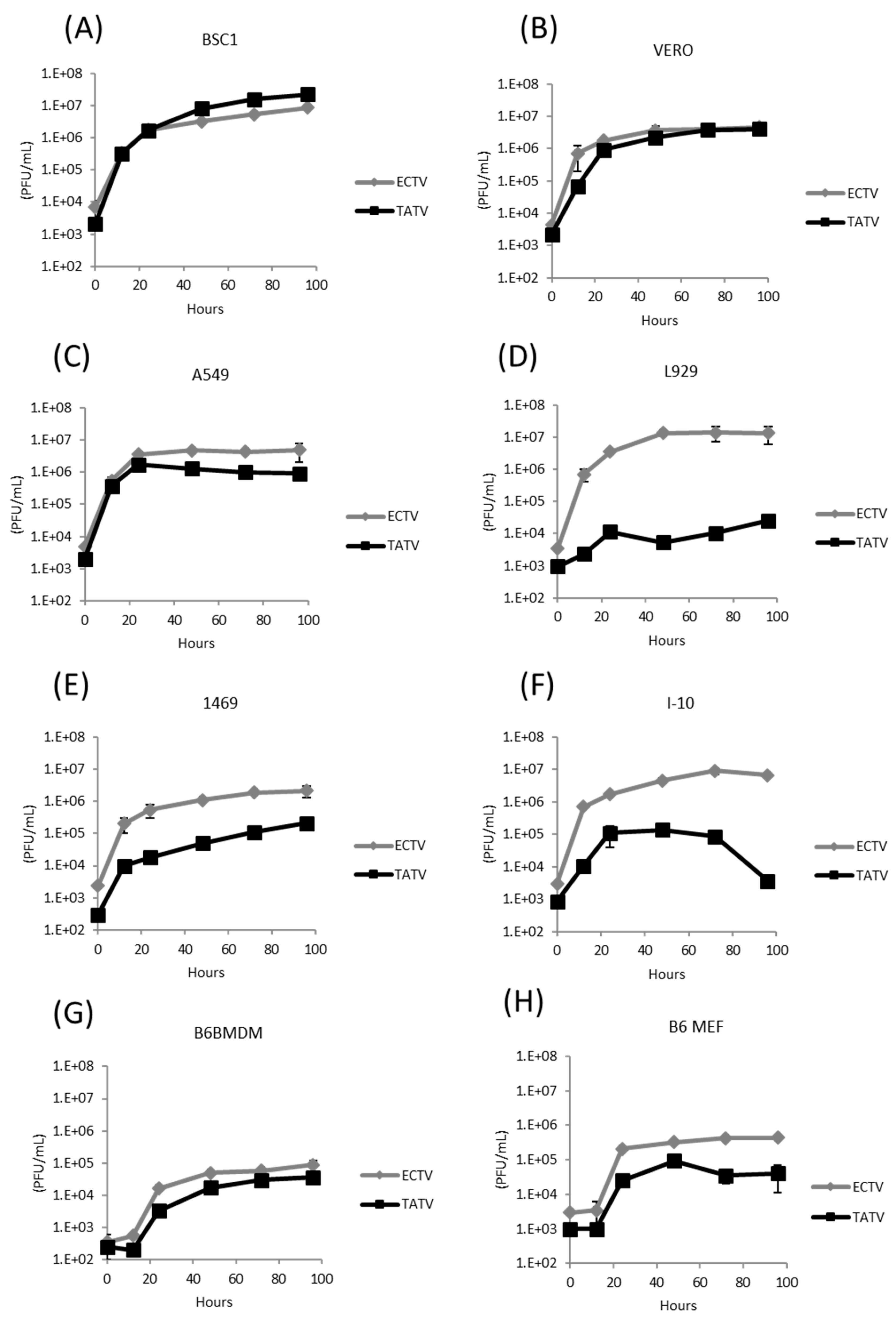
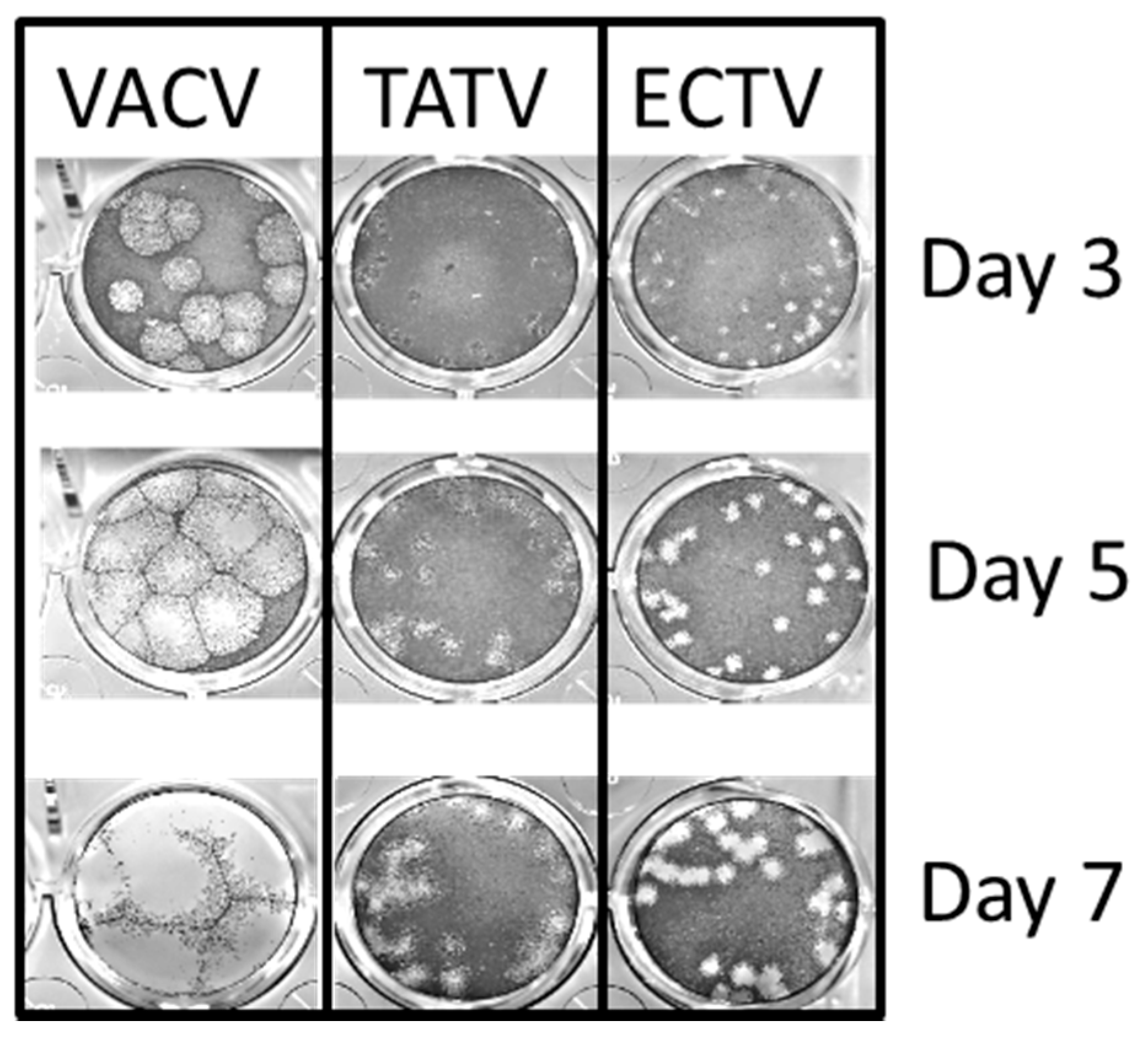
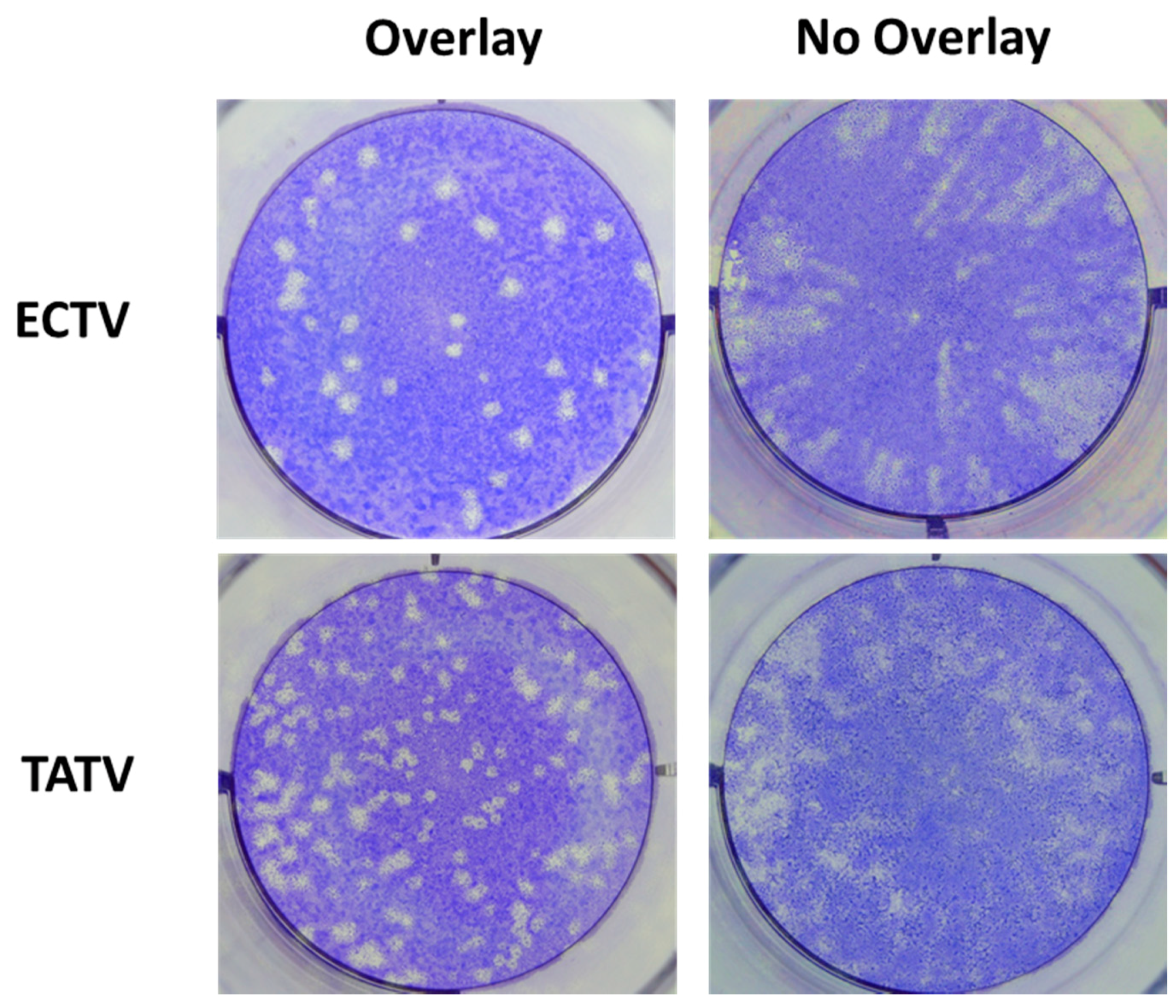
3.2. In Vitro Replication Studies of TATV
3.3. In Vitro Cytoskeletal Network Changes Following Infection
3.4. In Vitro Cytokine Changes in Splenocytes Infected with ECTV or TATV
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, N.; Bellone, C.J.; Schriewer, J.; Owens, G.; Fredrickson, T.; Parker, S.; Buller, R.M. Poxvirus interleukin-4 expression overcomes inherent resistance and vaccine-induced immunity: Pathogenesis, prophylaxis, and antiviral therapy. Virology 2011, 409, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Esposito, J.J.; Knight, J.C. Orthopoxvirus DNA: A comparison of restriction profiles and maps. Virology 1985, 143, 230–251. [Google Scholar] [CrossRef]
- Esposito, J.J.; Sammons, S.A.; Frace, A.M.; Osborne, J.D.; Olsen-Rasmussen, M.; Zhang, M.; Govil, D.; Damon, I.K.; Kline, R.; Laker, M.; et al. Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science 2006, 313, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Lourie, B.; Nakano, J.H.; Kemp, G.E.; Setzer, H.W. Isolation of poxvirus from an African Rodent. J. Infect. Dis. 1975, 132, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.; Crump, R.; Hartzler, H.; Buller, R.M. Evaluation of Taterapox Virus in Small Animals. Viruses 2017, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Moss, B.; Earl, P.L. Expression of proteins in mammalian cells using vaccinia virus vectors. In Overview of the Vaccinia Virus Expression System; Current Protocols in Molecular Biology; Wiley: Hoboken, NJ, USA, 1998; pp. 16.15.1–16.15.5. [Google Scholar]
- Esteban, D.; Parker, S.; Schriewer, J.; Hartzler, H.; Buller, R.M. Mousepox, a small animal model of smallpox. Methods Mol. Biol. 2012, 890, 177–198. [Google Scholar] [PubMed]
- Wallace, G.D.; Buller, R.M. Kinetics of ectromelia virus (mousepox) transmission and clinical response in C57BL/6j, BALB/cByj and AKR/J inbred mice. Lab. Anim. Sci. 1985, 35, 41–46. [Google Scholar] [PubMed]
- Hendrickson, R.C.; Wang, C.W.; Hatcher, E.L.; Lefkowitz, E.J. Orthopoxvirus genome evolution: The role of gene loss. Viruses 2010, 2, 1933–1967. [Google Scholar] [CrossRef] [PubMed]
- Odom, M.R.; Hendrickson, R.C.; Lefkowitz, E.J. Poxvirus protein evolution: Family wide assessment of possible horizontal gene transfer events. Virus Res. 2009, 144, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Li, G.; Liszewski, M.K.; Atkinson, J.P.; Jahrling, P.B.; Feng, Z.; Schriewer, J.; Buck, C.; Wang, C.; Lefkowitz, E.J.; et al. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 2005, 340, 46–63. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.G.; Shchelkunov, S.N.; Kotwal, G.J. The cowpox virus-encoded homolog of the vaccinia virus complement control protein is an inflammation modulatory protein. Virology 1997, 229, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Liszewski, M.K.; Leung, M.K.; Hauhart, R.; Buller, R.M.; Bertram, P.; Wang, X.; Rosengard, A.M.; Kotwal, G.J.; Atkinson, J.P. Structure and regulatory profile of the monkeypox inhibitor of complement: Comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 2006, 176, 3725–3734. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Liszewski, M.K.; Farries, T.C.; Lublin, D.M.; Rooney, I.A.; Atkinson, J.P. Control of the complement system. Adv. Immunol. 1996, 61, 201–283. [Google Scholar] [PubMed]
- Mercer, A.A.; Fleming, S.B.; Ueda, N. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 2005, 31, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Sperling, K.M.; Schwantes, A.; Staib, C.; Schnierle, B.S.; Sutter, G. The orthopoxvirus 68-kilodalton ankyrin-like protein is essential for DNA replication and complete gene expression of modified vaccinia virus Ankara in nonpermissive human and murine cells. J. Virol. 2009, 83, 6029–6038. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.L.; Smith, G.L. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008, 16, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.C.; Leite, F.G.; Brasil, B.S.; Soares-Martins, J.A.; Torres, A.A.; Pimenta, P.F.; Souto-Padron, T.; Traktman, P.; Ferreira, P.C.; Kroon, E.G.; et al. A vaccinia virus-driven interplay between the MKK4/7-JNK1/2 pathway and cytoskeleton reorganization. J. Virol. 2012, 86, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Schepis, A.; Schramm, B.; de Haan, C.A.; Locker, J.K. Vaccinia virus-induced microtubule-dependent cellular rearrangements. Traffic 2006, 7, 308–323. [Google Scholar] [CrossRef] [PubMed]
- Ploubidou, A.; Moreau, V.; Ashman, K.; Reckmann, I.; Gonzalez, C.; Way, M. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 2000, 19, 3932–3944. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, C.M.; Way, M.; Smith, G.L. Virus-induced cell motility. J. Virol. 1998, 72, 1235–1243. [Google Scholar] [PubMed]
- Valderrama, F.; Cordeiro, J.V.; Schleich, S.; Frischknecht, F.; Way, M. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science 2006, 311, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Stack, J.; Haga, I.R.; Schroder, M.; Bartlett, N.W.; Maloney, G.; Reading, P.C.; Fitzgerald, K.A.; Smith, G.L.; Bowie, A.G. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 2005, 201, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Ruiz-Arguello, M.B.; Ho, Y.; Smith, V.P.; Saraiva, M.; Alcami, A. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc. Natl. Acad. Sci. USA 2006, 103, 5995–6000. [Google Scholar] [CrossRef] [PubMed]
- Loparev, V.N.; Parsons, J.M.; Knight, J.C.; Panus, J.F.; Ray, C.A.; Buller, R.M.; Pickup, D.J.; Esposito, J.J. A third distinct tumor necrosis factor receptor of orthopoxviruses. Proc. Natl. Acad. Sci. USA 1998, 95, 3786–3791. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The complete DNA sequence of vaccinia virus. Virology 1990, 179, 247–266. [Google Scholar] [CrossRef]
- Panus, J.F.; Smith, C.A.; Ray, C.A.; Smith, T.D.; Patel, D.D.; Pickup, D.J. Cowpox virus encodes a fifth member of the tumor necrosis factor receptor family: A soluble, secreted CD30 homologue. Proc. Natl. Acad. Sci. USA 2002, 99, 8348–8353. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.C.; Bahar, M.W.; Abrescia, N.G.; Smith, G.L.; Stuart, D.I.; Grimes, J.M. Structure of CrmE, a virus-encoded tumour necrosis factor receptor. J. Mol. Biol. 2007, 372, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Reading, P.C.; Khanna, A.; Smith, G.L. Vaccinia virus CrmE encodes a soluble and cell surface tumor necrosis factor receptor that contributes to virus virulence. Virology 2002, 292, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Hu, F.Q.; Smith, T.D.; Richards, C.L.; Smolak, P.; Goodwin, R.G.; Pickup, D.J. Cowpox virus genome encodes a second soluble homologue of cellular TNF receptors, distinct from CrmB, that binds TNF but not LT alpha. Virology 1996, 223, 132–147. [Google Scholar] [CrossRef] [PubMed]
- Alcami, A.; Khanna, A.; Paul, N.L.; Smith, G.L. Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumour necrosis factor receptors. J. Gen. Virol. 1999, 80, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.M.; Dairaghi, D.J.; Deitz, M.; Tsang, M.; Schall, T.J. Comprehensive mapping of poxvirus vCCI chemokine-binding protein. Expanded range of ligand interactions and unusual dissociation kinetics. J. Biol. Chem. 2002, 277, 2785–2789. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Messauodi, I.; Estep, R.D.; Orzechowska, B.; Wong, S.W. Monkeypox virus viral chemokine inhibitor (MPV vCCI), a potent inhibitor of rhesus macrophage inflammatory protein-1. Cytokine 2008, 43, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Smith, T.D.; Smolak, P.J.; Friend, D.; Hagen, H.; Gerhart, M.; Park, L.; Pickup, D.J.; Torrance, D.; Mohler, K.; et al. Poxvirus genomes encode a secreted, soluble protein that preferentially inhibits beta chemokine activity yet lacks sequence homology to known chemokine receptors. Virology 1997, 236, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.L.; Fremont, D.H. Structural determinants of chemokine binding by an Ectromelia virus-encoded decoy receptor. J. Virol. 2006, 15, 7439–7449. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Derider, M.; McCornack, M.A.; Jao, S.C.; Isern, N.; Ness, T.; Moyer, R.; LiWang, P.J. Solution structure of the complex between poxvirus-encoded CC chemokine inhibitor vCCI and human MIP-1beta. Proc. Natl. Acad. Sci. USA 2006, 103, 13985–13990. [Google Scholar] [CrossRef] [PubMed]

| TATV | ECTV | |||||
|---|---|---|---|---|---|---|
| CPXV-GRI/GER Stop 2,3 | VACV-COP Ortholog (CPXV Protein Size) | Function (Motif) | Status | Protein Size (% Compared to CPXV) | Status | Protein Size (% Compared to CPXV) |
| 35,634 | C2L (512) | Unknown (Kelch-like) | Frag 4 | ORF018 | 512 (100) | |
| 38,413 | N2L (175) | Alpha amanitin sensitivity protein | Frag | ORF020 | 177 (101) | |
| 170,420 | A46R (240) | Interleukin 1 (IL-1) signalling inhibitor | Frag | ORF145 | 240 (100) | |
| 191,696 | B12R (283) | Serine/Threonine kinase | Frag | ORF152 | 286 (101) | |
| 9217 | (273) | Unknown | Miss | ORF004 | 273 (100) | |
| 10,160 | (655) | N-term ankyrin, C-term F-box | Miss | ORF005 | 650 (99) | |
| 12,346 | (96) | C-type lectin | Miss | ORF006 | 103 (107) | |
| 14,342 | (202) | Unknown (partial homology to TNFR CrmB) | Miss | ORF008 | 202 (100) | |
| 14,947 | (111) | Tumour necrosis factor receptor (vCD30) | Miss | ORF009 | 111 (100) | |
| 15,374 | (764) | Unknown (Ankyrin) | Miss | ORF010 | 763 (100) | |
| 32,306 | (29) | Unknown | Miss | ORF 7 | 29 (100) | |
| 213,858 | (322) | Tumour necrosis factor receptor (CrmD) | Miss | ORF003/170 | 320 (99) | |
| 23,924 | C10L (331) | Interleukin 1 (IL-1) receptor antagonist | Trnc 5 | 27 (8) | ORF011 | 331 (100) |
| 34,788 | C3L (259) | Complement binding protein (secreted) | Trnc | 194 (75) | ORF017 | 262 (101) |
| 41,155 | K1L (284) | NFkB inhibitor (Ankyrin) | Trnc | 88 (31) | ORF022 | 285 (100) |
| 73,579 | O1L (666) | Unknown | Trnc | 64 (10) | ORF052 | 666 (100) |
| 150,930 | A25L (1279) | A-type inclusion protein | Trnc | 736 (58) | ORF128 | 1113 (87) |
| 165,419 | A39R (402) | Semaphorin | Trnc | 134 (33) | ORF139 | 399 (99) |
| 168,244 | A44L (346) | Hydroxysteroid dehydrogenase | Trnc | 88 (25) | ORF143 | 346 (100) |
| 169,708 | A45R (125) | Superoxide dismutase (Cu-Zn) like protein | Trnc | 46 (37) | ORF144 | 125 (100) |
| 171,963 | A48R (204) | Thymidylate kinase | Trnc | 58 (28) | ORF147 | 204 (100) |
| 178,726 | A55R (564) | Kelch-like | Trnc | 219 (39) | ORF150 | 563 (100) |
| 197,359 | B18R (574) | N-term ankyrin, C-term F-box | Trnc | 147 (26) | ORF165/010 | 574 (100) |
| 222,274 | B29R/C23 6 (255) | Chemokine binding protein | Trnc | 61 (24) | ORF001/172 | 247 (97) |
| 172,501 | A49R (162) | Phosphotransferase anion transport protein (putative) | Trnc | 77 (48) | Trnc | 116 (72) |
| Target | ECTV | TATV | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Chemokines | Mock | 12 H | 24 H | 48 H | Mock | 12 H | 24 H | 48 H | |
| MIP-2 | Chemotactic for polymorphonuclear leukocytes and hematopoietic stem cells | 12.2 ± 4.3 | 2.6 ± 1.8 | 12.4 ± 2.6 | 23.5 ± 2.8 | 8.0 ± 3.8 | 14 ± 8.1 | 182.7 ± 4.4 2 | 260.0 ± 11.4 |
| MCP-1 | Recruits monocytes, memory T cells, and dendritic cells | 0.6 ± 0.2 | 3.5 ± 0.5 | 3.3 ± 0.1 | 1.7 ± 0.7 | 0.5 ± 0.1 | 0.3 ± 0.2 | 11.6 ± 0.1 | 22.9 ± 1.1 |
| MIP-1β | Activate neutrophils, eosinophils and basophils | 27.2 ± 2.3 | 0.0 ± 0.0 3 | 0.6 ± 0.3 | 0.7 ± 0.5 | 20.7 ± 0.1 | 23.1 ± 6.6 | 64.1 ± 5.4 | 93.5 ± 6.9 |
| MIP-1α | Activate neutrophils, eosinophils and basophils | 22.9 ± 0.1 | 1.2 ± 0.3 | 1.4 ± 0.3 | 1.3 ± 0.2 | 22.7 ± 2.1 | 24.2 ± 6.6 | 63.6 ± 2.8 | 91.0 ± 5.8 |
| MIG | Chemoattractant for stimulated T-cell | 9.5 ± 0.1 | 6.1 ± 0.7 | 15.0 ± 1.5 | 16.1 ± 1.7 | 8.6 ± 1.7 | 7.2 ± 1.6 | 14.7 ± 2.8 | 15.1 ± 2.1 |
| RANTES | Chemotatic for T-cells, monocyte lineages, eosinophils, and basophils | 8.4 ± 0.4 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 6.7 ± 0.8 | 6.8 ± 1.7 | 14.7 ± 1.2 | 16.4 ± 0.7 |
| Cytokine | |||||||||
| TNF-α | Systemic inflammation and stimulation of the acute phase reaction | 0.2 ± 0.0 | 0.0 ± 0.0 | 1.5 ± 0.2 | 1.8 ± 0.1 | 0.0 ± 0.1 | 0.4 ± 0.3 | 1.9 ± 0.2 | 3.4 ± 0.5 |
| Growth Factors | |||||||||
| VEGF | Highly specific mitogen for vascular endothelial cells | 0.6 ± 0.2 | 27.0 ± 1.1 | 39.8 ± 9.1 | 44.9 ± 9.1 | 0.1 ± 0.1 | 0.6 ± 0.3 | 6.3 ± 1.2 | 9.5 ± 1.5 |
| KC | Chemotaxis and cell activation of neutrophils | 2.3 ± 0 | 2.7 ± 0.1 | 9.2 ± 1.2 | 13.5 ± 1.3 | 2.4 ± 0.4 | 2.7 ± 1.5 | 30.8 ± 4.1 | 50.2 ± 3.9 |
| M-CSF | Proliferation and differentiation of macrophages and monocytes | 0.4 ± 0.2 | 3.7 ± 0.1 | 8.2 ± 2.7 | 7.5 ± 1.0 | 0.3 ± 0.2 | 0.0 ± 0.0 | 1.0 ± 0.2 | 2.3 ± 0.1 |
| CPXV-GRI/GER Stop | VACV-COP | TNFR | TATV | CPXV-GRI | CPXV-GER91 | CMLV-M96 | ECTV-MOS | MPXV-ZAR | VACV-WR | VARV-BRA66 |
|---|---|---|---|---|---|---|---|---|---|---|
| 14,947 | vCD30 | Miss 1 | Gene | Gene | Miss | Gene | Miss | Miss | Miss | |
| 213,858 | crmD | Miss | Gene | Trnc | Miss | Gene | Miss | Miss | Miss | |
| 214,439 | crmE | Miss | Gene | Gene | Trnc | Trnc | Miss | Miss | Miss | |
| 176,775 | A53R | crmC | Trnc 2 | Gene | Gene | Miss | Miss | Miss | Trnc | Miss |
| 221,381 1 | B28R/C22L | crmB | Trnc | Gene | Gene | Gene | Miss | Gene | Trnc | Gene |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parker, S.; Camilo de Oliveira, L.; Lefkowitz, E.J.; Hendrickson, R.C.; Bonjardim, C.A.; Wold, W.S.M.; Hartzler, H.; Crump, R.; Buller, R.M. The Virology of Taterapox Virus In Vitro. Viruses 2018, 10, 463. https://doi.org/10.3390/v10090463
Parker S, Camilo de Oliveira L, Lefkowitz EJ, Hendrickson RC, Bonjardim CA, Wold WSM, Hartzler H, Crump R, Buller RM. The Virology of Taterapox Virus In Vitro. Viruses. 2018; 10(9):463. https://doi.org/10.3390/v10090463
Chicago/Turabian StyleParker, Scott, Leonardo Camilo de Oliveira, Elliot J. Lefkowitz, Robert Curtis Hendrickson, Cláudio A. Bonjardim, William S. M. Wold, Hollyce Hartzler, Ryan Crump, and Robert Mark Buller. 2018. "The Virology of Taterapox Virus In Vitro" Viruses 10, no. 9: 463. https://doi.org/10.3390/v10090463
APA StyleParker, S., Camilo de Oliveira, L., Lefkowitz, E. J., Hendrickson, R. C., Bonjardim, C. A., Wold, W. S. M., Hartzler, H., Crump, R., & Buller, R. M. (2018). The Virology of Taterapox Virus In Vitro. Viruses, 10(9), 463. https://doi.org/10.3390/v10090463





