Neutralizing Epitopes and Residues Mediating the Potential Antigenic Drift of the Hemagglutinin-Esterase Protein of Influenza C Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Antibodies
2.3. Enzyme-Linked Immunosorbent Assay
2.4. Hemagglutination Inhibition Test
2.5. Neutralization Test
2.6. Selection of Escape Mutants
2.7. Nucleotide Sequencing and Phylogenetic Analysis
2.8. Structural Analysis
3. Results
3.1. Selection of Escape Mutants
3.2. Antigenic Sites Revealed by the Reactivities of MAbs with Escape Mutants in HI Tests
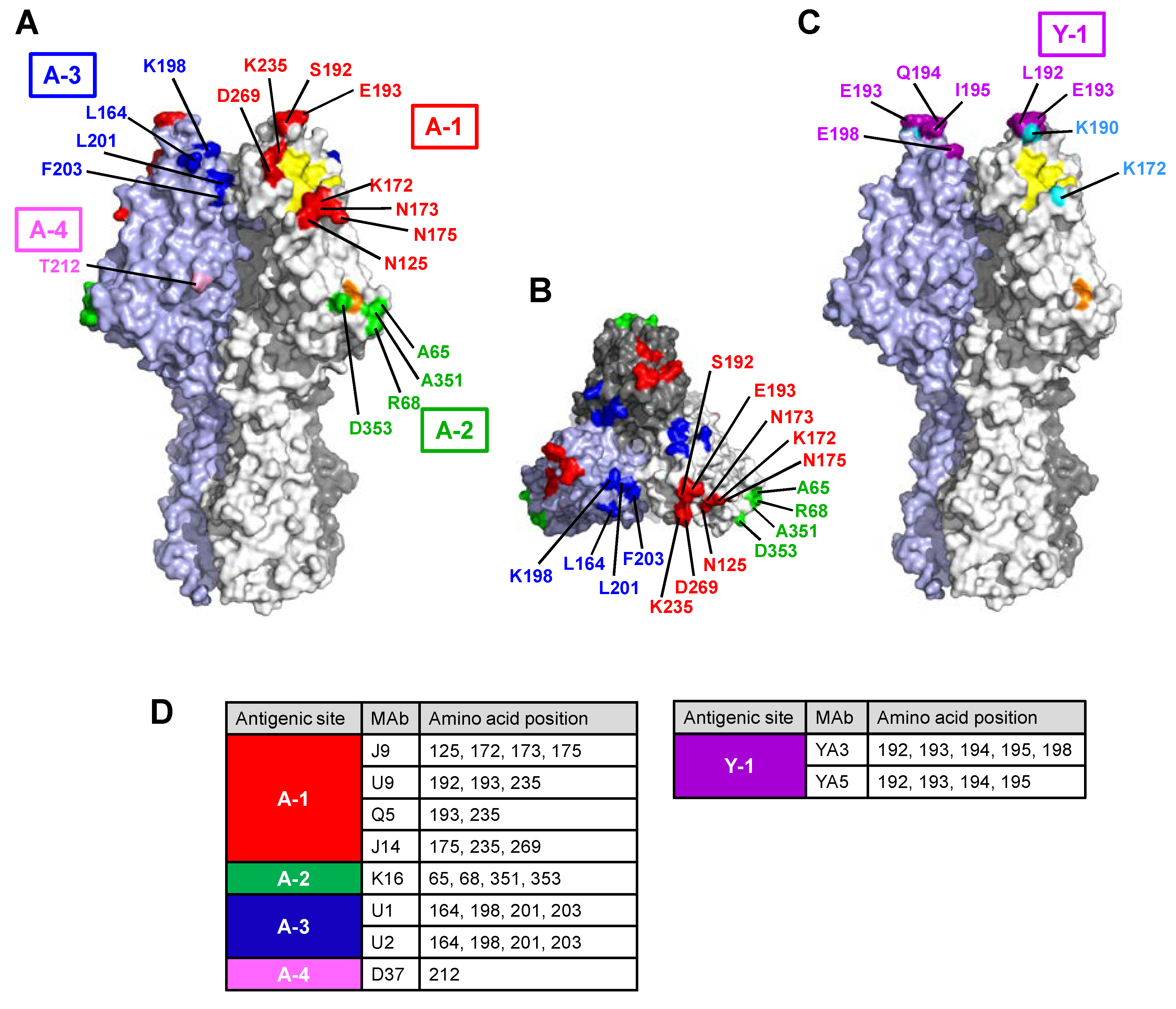
3.2.1. Site A-1
3.2.2. Site A-2
3.2.3. Site A-3
3.2.4. Site Y-1
3.3. Antigenic Structure of Influenza C Virus HE
3.4. Antigenic Changes in Influenza C Virus Strains Isolated from 1947 to 2016
3.4.1. C/Taylor Lineage
3.4.2. C/Mississippi Lineage
3.4.3. C/Aichi Lineage
3.4.4. C/Yamagata Lineage
3.4.5. C/Kanagawa Lineage
3.4.6. C/Sao Paulo Lineage
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pfeifer, J.B.; Compans, R.W. Structure of the influenza C glycoprotein gene as determined from cloned DNA. Virus Res. 1984, 1, 281–296. [Google Scholar] [CrossRef]
- Vlasak, R.; Krystal, M.; Nacht, M.; Palese, P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology 1987, 160, 419–425. [Google Scholar] [CrossRef]
- Herrler, G.; Dürkop, I.; Becht, H.; Klenk, H.D. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J. Gen. Virol. 1988, 69, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Formanowski, F.; Meier-Ewert, H. Isolation of the influenza C virus glycoprotein in a soluble form by bromelain digestion. Virus Res. 1988, 10, 177–191. [Google Scholar] [CrossRef]
- Kimura, H.; Abiko, C.; Peng, G.; Muraki, Y.; Sugawara, K.; Hongo, S.; Kitame, F.; Mizuta, K.; Numazaki, Y.; Suzuki, H.; et al. Interspecies transmission of influenza C virus between humans and pigs. Virus Res. 1997, 48, 71–79. [Google Scholar] [CrossRef]
- Katagiri, S.; Ohizumi, A.; Homma, M. An outbreak of type C influenza in a children’s home. J. Infect. Dis. 1983, 148, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Katsushima, N.; Nagai, Y.; Shoji, M.; Itagaki, T.; Sakamoto, M.; Kitaoka, S.; Mizuta, K.; Nishimura, H. Clinical features of influenza C virus infection in children. J. Infect. Dis. 2006, 193, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Thielen, B.K.; Friedlander, H.; Bistodeau, S.; Shu, B.; Lynch, B.; Martin, K.; Bye, E.; Como-Sabetti, K.; Boxrud, D.; Strain, A.K.; et al. Detection of influenza C viruses among outpatients and patients hospitalized for severe acute respiratory infection, Minnesota, 2013–2016. Clin. Infect. Dis. 2018, 66, 1092–1098. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Sugawara, K.; Furuse, Y.; Shimotai, Y.; Hongo, S.; Oshitani, H.; Mizuta, K.; Nishimura, H. Genetic lineage and reassortment of influenza C viruses circulating between 1947 and 2014. J. Virol. 2016, 90, 8251–8265. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Muraki, Y.; Sugawara, K.; Hongo, S.; Nishimura, H.; Kitame, F.; Katsushima, N.; Numazaki, Y.; Nakamura, K. Cocirculation of two distinct groups of influenza C virus in Yamagata City, Japan. Virology 1994, 202, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Mizuta, K.; Sugawara, K.; Tsuchiya, E.; Muraki, Y.; Hongo, S.; Suzuki, H.; Nishimura, H. Frequent reassortment among influenza C viruses. J. Virol. 2003, 77, 871–881. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Sugawara, K.; Abiko, C.; Ikeda, T.; Aoki, Y.; Mizuta, K.; Katsushima, N.; Katsushima, F.; Katsushima, Y.; Itagaki, T.; et al. Epidemiological information regarding the periodic epidemics of influenza C virus in Japan (1996–2013) and the seroprevalence of antibodies to different antigenic groups. J. Clin. Virol. 2014, 61, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, M.; Sugawara, K.; Adachi, K.; Hongo, S.; Nishimura, H.; Kitame, F.; Nakamura, K. Location of neutralizing epitopes on the hemagglutinin-esterase protein of influenza C virus. Virology 1992, 189, 79–87. [Google Scholar] [CrossRef]
- Rosenthal, P.B.; Zhang, X.; Formanowski, F.; Fitz, W.; Wong, C.H.; Meier-Ewert, H.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 1998, 396, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, K.; Nishimura, H.; Kitame, F.; Nakamura, K. Antigenic variation among human strains of influenza C virus detected with monoclonal antibodies to gp88 glycoprotein. Virus Res. 1986, 6, 27–32. [Google Scholar] [PubMed]
- Sugawara, K.; Kitame, F.; Nishimura, H.; Nakamura, K. Operational and topological analyses of antigenic sites on influenza C virus glycoprotein and their dependence on glycosylation. J. Gen. Virol. 1988, 69, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, K.; Nishimura, H.; Hongo, S.; Muraki, Y.; Kitame, F.; Nakamura, K. Construction of an antigenic map of the haemagglutinin-esterase protein of influenza C virus. J. Gen. Virol. 1993, 74, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Abiko, C.; Mizuta, K.; Sugawara, K.; Takashita, E.; Muraki, Y.; Suzuki, H.; Mikawa, M.; Shimada, S.; Sato, K.; et al. A nationwide epidemic of influenza C virus infection in Japan in 2004. J. Clin. Microbiol. 2007, 45, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Mizuta, K.; Kimura, H.; Sugawara, K.; Tsuchiya, E.; Suzuki, H.; Hongo, S.; Nakamura, K. Characterization of antigenically unique influenza C virus strains isolated in Yamagata and Sendai cities, Japan, during 1992–1993. J. Gen. Virol. 2000, 81, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Aoki, Y.; Matoba, Y.; Yahagi, K.; Mizuta, K.; Itagaki, T.; Katsushima, F.; Katsushima, Y.; Matsuzaki, Y. The dominant antigenic group of influenza C infections changed from C/Sao Paulo/378/82-lineage to C/Kanagawa/1/76-lineage in Yamagata, Japan, in 2014. Jpn. J. Infect. Dis. 2015, 68, 166–168. [Google Scholar] [CrossRef] [PubMed]
- Hongo, S.; Sugawara, K.; Homma, M.; Nakamura, K. The functions of oligosaccharide chains associated with influenza C viral glycoproteins. II. The role of carbohydrates in the antigenic properties of influenza C viral glycoproteins. Arch. Virol. 1986, 89, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, H.; Tashiro, M.; Kitame, F.; Homma, M.; Nakamura, K. Genetic variation among human strains of influenza C virus isolated in Japan. Virus Res. 1986, 4, 275–288. [Google Scholar] [CrossRef]
- Furuse, Y.; Matsuzaki, Y.; Nishimura, H.; Oshitani, H. Analyses of evolutionary characteristics of the hemagglutinin-esterase gene of influenza C virus during a period of 68 years reveals evolutionary patterns different from influenza A and B viruses. Viruses 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Sugawara, K.; Nakauchi, M.; Takahashi, Y.; Onodera, T.; Tsunetsugu-Yokota, Y.; Matsumura, T.; Ato, M.; Kobayashi, K.; Shimotai, Y.; et al. Epitope mapping of the hemagglutinin molecule of A/(H1N1)pdm09 influenza virus by using monoclonal antibody escape mutants. J. Virol. 2014, 88, 12364–12373. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, E.; Sugawara, K.; Hongo, S.; Matsuzaki, Y.; Muraki, Y.; Li, Z.N.; Nakamura, K. Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. J. Gen. Virol. 2001, 82, 2475–2484. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.; Hongo, S.; Sugawara, K.; Li, Z.N.; Tsuchiya, E.; Muraki, Y.; Matsuzaki, Y.; Nakamura, K. Role of individual oligosaccharide chains in antigenic properties, intracellular transport, and biological activities of influenza C virus hemagglutinin-esterase protein. Virology 2001, 285, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Kitame, F.; Sugawara, K.; Nishimura, H.; Nakamura, K. Antigenic and genetic characterization of three influenza C strains isolated in the Kinki district of Japan in 1982–1983. Virology 1989, 172, 125–133. [Google Scholar] [CrossRef]
- Ohyama, S.; Adachi, K.; Sugawara, K.; Hongo, S.; Nishimura, H.; Kitame, F.; Nakamura, K. Antigenic and genetic analyses of eight influenza C strains isolated in various areas of Japan during 1985–1989. Epidemiol. Infect. 1992, 108, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Takao, S.; Shimada, S.; Mizuta, K.; Sugawara, K.; Takashita, E.; Muraki, Y.; Hongo, S.; Nishimura, H. Characterization of antigenically and genetically similar influenza C viruses isolated in Japan during the 1999–2000 season. Epidemiol. Infect. 2004, 132, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Sugawara, K.; Mizuta, K.; Tsuchiya, E.; Muraki, Y.; Hongo, S.; Suzuki, H.; Nakamura, K. Antigenic and genetic characterization of influenza C viruses which caused two outbreaks in Yamagata City, Japan, in 1996 and 1998. J. Clin. Microbiol. 2002, 40, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.F.; Neumann, G.; Kawaoka, Y. Orthomyxoviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 1186–1243, ISBN-13 978-145-110-5636. [Google Scholar]
- Air, G.M. Influenza neuraminidase. Influenza Other Respir. Viruses 2012, 6, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Hachinohe, S.; Sugawara, K.; Nishimura, H.; Kitame, F.; Nakamura, K. Effect of anti-haemagglutinin-esterase glycoprotein monoclonal antibodies on the receptor-destroying activity of influenza C virus. J. Gen. Virol. 1989, 70, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Umetsu, Y.; Sugawara, K.; Nishimura, H.; Hongo, S.; Matsuzaki, M.; Kitame, F.; Nakamura, K. Selection of antigenically distinct variants of influenza C viruses by the host cell. Virology 1992, 189, 740–744. [Google Scholar] [CrossRef]
| Antibody Titer | Selection Frequency (−log10) | ||||||
|---|---|---|---|---|---|---|---|
| MAb | Isotype 1 | ELISA (×104) 1 | HI | NT50 1 | CJP1 | CJP3 | CJP5 |
| J9 | IgG1 | 1600 | 128,000 | 160,000 | 6.18 | 6.53 | 6.69 |
| U9 | IgG1 | 200 | 128,000 | 200 | 5.87 | 5.30 | 6.10 |
| Q5 | IgG2a | 1600 | 64,000 | 16,000 | 5.82 | 6.37 | 6.13 |
| J14 | IgG1 | 1000 | 256,000 | 8,000,000 | 7.16 | 6.28 | 6.97 |
| K16 | IgG1 | 640 | 80 | 320,000 | 5.58 | 4.62 | 5.05 |
| U1 | IgG1 | 200 | 16,000 | 20,000 | 6.96 | 7.58 | 6.36 |
| U2 | IgG1 | 100 | 6400 | 4000 | 5.92 | 5.80 | 5.94 |
| D37 | IgG2a | 200 | 160 | 8000 | ― 2 | ― | ― |
| Antibody Titer | Selection Frequency (−log10) | ||||||
|---|---|---|---|---|---|---|---|
| MAb | Isotype | ELISA (×104) | HI | NT50 | CYP1 | CYP2 | CYP4 |
| YA3 | IgG1 | 800 | 25,600 | 80,000 | 8.42 | 8.22 | 9.25 |
| YA5 | IgG1 | 800 | 12,800 | 40,000 | 8.17 | 7.11 | 8.19 |
| Amino Acid Change of Escape Mutants | MAb(s) Used for Selection of Escape Mutants | No. of Escape Mutants Derived from Parent Virus | Hemagglutination Inhibition Titer of MAb 1 or of Chicken Antiserum | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAb against C/Ann Arbor/1/50 Virus at Site: | Chicken Antiserum against C/Ann Arbor/1/50 | ||||||||||||
| A-1 | A-2 | A-3 | |||||||||||
| CJP1 | CJP3 | CJP5 | Total | J9 | U9 | Q5 | J14 | K16 | U1 | U2 | |||
| None (Parent Virus) | 128,000 | 128,000 | 64,000 | 256,000 | 80 | 16,000 | 6400 | 1280 | |||||
| N125D | J9 | 1 | 1 | 2 | <20 | ― | ― | 3200 | ― | ― | ― | 2560 | |
| K172R | J9 | 1 | 1 | 2 | 4 | 160 | ― | 3200 | ― | ― | ― | ― | 2560 |
| K172Q | J9 | 7 | 1 | 1 | 9 | <20 | ― | 640 | ― | ― | ― | ― | 2560 |
| K172N | J9 | 2 | 2 | <20 | ― | 160 | ― | ― | ― | ― | 5120 | ||
| N173I | J9 | 2 | 6 | 4 | 12 | 320 | ― | ― | ― | ― | ― | ― | 2560 |
| N175S | J9, J14 | 9 | 2 | 3 | 14 | <20 | ― | ― | <20 | ― | ― | ― | 1280 |
| S192L | U9 | 9 | 10 | 4 | 23 | ― | 160 | ― | ― | ― | ― | ― | 2560 |
| E193K | U9, Q5 | 6 | 5 | 9 | 20 | ― | <20 | <20 | ― | <20 | ― | ― | 320 |
| K235R | U9, Q5 | 4 | 5 | 4 | 13 | ― | <20 | <20 | ― | ― | ― | ― | 2560 |
| K235I | U9, Q5, J14 | 3 | 3 | ― | <20 | <20 | <20 | ― | ― | ― | 1280 | ||
| K235E | J14 | 3 | 3 | ― | <20 | <20 | <20 | ― | ― | ― | 1280 | ||
| D269N | J14 | 5 | 6 | 11 | ― | ― | ― | <20 | <20 | ― | ― | 640 | |
| A65D | K16 | 1 | 1 | ― | ― | ― | ― | <20 | ― | ― | 2560 | ||
| R68W | K16 | 3 | 3 | ― | ― | ― | ― | <20 | ― | ― | 2560 | ||
| A351V | K16 | 7 | 7 | ― | ― | ― | ― | <20 | ― | ― | 1280 | ||
| D353G | K16 | 2 | 2 | 4 | ― | ― | ― | ― | <20 | ― | ― | 1280 | |
| D353N | K16 | 3 | 1 | 1 | 5 | ― | ― | ― | ― | <20 | ― | ― | 1280 |
| D353Y | K16 | 1 | 2 | 2 | ― | ― | ― | ― | <20 | ― | ― | 1280 | |
| L164P | U1, U2 | 4 | 10 | 2 | 16 | ― | ― | ― | ― | ― | 160 | <20 | 2560 |
| L164S | U2 | 1 | 1 | ― | ― | ― | ― | ― | ― | ― | 2560 | ||
| K198E | U1, U2 | 8 | 10 | 11 | 29 | ― | ― | ― | ― | ― | 320 | 20 | 1280 |
| L201H | U1, U2 | 1 | 1 | 2 | ― | ― | ― | ― | ― | 640 | 80 | 2560 | |
| L201I | U2 | 1 | 1 | ― | ― | ― | ― | ― | ― | ― | 2560 | ||
| L201R | U2 | 1 | 1 | ― | ― | ― | ― | ― | 80 | <20 | 1280 | ||
| F203S | U1, U2 | 4 | 5 | 9 | ― | ― | ― | ― | ― | 1280 | 160 | 1280 | |
| Total no. of mutants | 63 | 66 | 69 | 198 | |||||||||
 and
and  denote a greater than 10-fold lower HI titer of MAbs at sites A-1 and A-3 than that of the parent virus, respectively.
denote a greater than 10-fold lower HI titer of MAbs at sites A-1 and A-3 than that of the parent virus, respectively.  ,
,  , and
, and  denote undetermined (less than 20) HI titer of MAbs at sites A-1, A-2, and A-3, respectively.
denote undetermined (less than 20) HI titer of MAbs at sites A-1, A-2, and A-3, respectively.| Amino Acid Change of Escape Mutants | MAb(s) Used for Selection of Escape Mutants | No. of Escape Mutants Derived from Parent Virus | Hemagglutination Inhibition Titer of MAb 1 or of Chicken Antiserum | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mab against C/Yamagata/15/2004 Virus at Site: | MAb against C/Ann Arbor/1/50 Virus at Site: | Chicken Antiserum against C/Yamagata/10/89 2 | |||||||||||
| Y-1 | A-1 | A-3 | |||||||||||
| CYP1 | CYP2 | CYP4 | Total | YA3 | YA5 | U9 | Q5 | J14 | U1 | U2 | |||
| None (parent virus) | 12,800 | 12,800 | 1280 | 12,800 | 32,000 | 6400 | 640 | 5120 | |||||
| K172R | YA5 | 1 | 1 | ― | ― | ― | <20 | ― | ― | ― | 5120 | ||
| K190N | YA3, YA5 | 2 | 8 | 5 | 15 | ― | ― | 80 | ― | ― | ― | ― | 5120 |
| E193K | YA3, YA5 | 5 | 3 | 3 | 11 | 1280 | 640 | <20 | <20 | 320 | ― | ― | 640 |
| 192-195 deletion | YA3, YA5 | 9 | 7 | 16 | <20 | <20 | <20 | ― | ― | ― | ― | 2560 | |
| 198 deletion | YA3 | 1 | 4 | 5 | <20 | <20 | <20 | 80 | ― | ― | ― | 640 | |
| Total no. of mutants | 16 | 19 | 13 | 48 | |||||||||
 and
and  denote a greater than 10-fold lower HI titer of MAbs at sites Y-1 and A-1 than that of the parent virus, respectively.
denote a greater than 10-fold lower HI titer of MAbs at sites Y-1 and A-1 than that of the parent virus, respectively.  and
and  denote undetermined (less than 20) HI titer of MAbs at sites Y-1 and A-1, respectively. 2 The HE antigenicity of C/Yamagata/10/89 is identical to that of C/Yamagata/26/81 [10], which is a reference strain of the C/Yamagata lineage.
denote undetermined (less than 20) HI titer of MAbs at sites Y-1 and A-1, respectively. 2 The HE antigenicity of C/Yamagata/10/89 is identical to that of C/Yamagata/26/81 [10], which is a reference strain of the C/Yamagata lineage.| HE Lineage | Strain | HI Titer | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAbs of Respective Antigenic Site | Chicken Antiserum against: | ||||||||||
| A-1 | A-3 | Y-1 | |||||||||
| J9 | U9 | Q5 | J14 | U1 | U2 | YA3 | YA5 | C/Ann Arbor/1/50 | C/Yamagata/10/89 1 | ||
| C/Taylor | C/Taylor/1233/47 | 64,000 | 64,000 | 32,000 | 256,000 | 16,000 | 3200 | <2 | < | 1280 | 640 |
| C/Ann Arbor/1/50 | 64,000 | 64,000 | 16,000 | 25,6000 | 16,000 | 6400 | < | < | 1280 | 640 | |
| C/Paris/1/67 | 512,000 | < | < | 512,000 | 16,000 | 3200 | < | < | 1280 | 320 | |
| C/Mississippi | C/Greece/1/79 | < | < | < | 256,000 | 32,000 | 6400 | < | < | 320 | 160 |
| C/Mississippi/80 | < | < | < | 256,000 | 32,000 | 12,800 | < | < | 160 | 160 | |
| C/Yamagata/26/2004 | < | < | < | 256,000 | 32,000 | 12,800 | < | < | 160 | 160 | |
| C/Aichi | C/Johannesburg/1/66 | < | 16,000 | < | 512,000 | 16,000 | 3200 | < | < | 160 | 320 |
| C/Georgia/1/69 | < | 16,000 | < | 512,000 | < | 20 | < | < | 160 | 320 | |
| C/Aichi/1/81 | < | 16,000 | < | 256,000 | 640 | 80 | < | < | 80 | 320 | |
| C/Yamagata/4/92 | < | 16,000 | < | 256,000 | 640 | 80 | < | < | 160 | 320 | |
| C/Yamagata | C/Great Lakes/1167/54 | 6400 | 3200 | 320 | 16,000 | < | 20 | < | < | 1280 | 2560 |
| C/Sapporo/71 | 640 | 640 | 1600 | 128,000 | 3200 | 320 | 6400 | 6400 | 160 | 1280 | |
| C/Yamagata/26/81 | < | 640 | 1600 | 64,000 | 40 | 160 | 12,800 | 12,800 | 320 | 2560 | |
| C/Miyagi/2/92 | < | 640 | 320 | 16,000 | 32,000 | 12,800 | 25,600 | 25,600 | 320 | 2560 | |
| C/Yamagata/15/2004 | < | 640 | 1600 | 32,000 | 20 | 80 | 12,800 | 12,800 | 320 | 2560 | |
| C/Kanagawa | C/Yamagata/64 | 128,000 | 1280 | 32,000 | 256,000 | < | 20 | 3200 | 3200 | 320 | 320 |
| C/Aomori/74 | < | 160 | 320 | 128,000 | 3200 | 640 | 1600 | 800 | 320 | 640 | |
| C/Kanagawa/1/76 | < | 40 | 320 | 128,000 | < | < | < | < | 160 | 160 | |
| C/Miyagi/77 | < | 16,000 | 320 | 256,000 | < | < | < | < | 160 | 320 | |
| C/Miyagi/9/96 | < | 40 | 80 | 16,000 | < | < | < | < | 160 | 640 | |
| C/Yamagata/13/2014 | < | 320 | 160 | 256,000 | < | < | 160 | 80 | 160 | 320 | |
| C/Sao Paulo | C/Sao Paulo/378/82 | < | 16,000 | 800 | 64,000 | 32,000 | 12,800 | 12,800 | 6400 | 640 | 320 |
| C/Fukuoka/1/2005 | < | 32,000 | 1600 | 128,000 | 16,000 | 3200 | 6400 | 6400 | 320 | 320 | |
| C/Yamagata/33/2014 | < | 32,000 | 1600 | 32,000 | 16,000 | 3200 | 6400 | 3200 | 640 | 640 | |
| C/Yamagata/30/2014 | 1280 | 32,000 | 800 | 64,000 | 8000 | 1600 | < | < | 320 | 320 | |
| C/Yamagata/1/2016 | 128,000 | 32,000 | 1600 | 128,000 | 32,000 | 12,800 | < | < | 320 | 320 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuzaki, Y.; Sugawara, K.; Furuse, Y.; Shimotai, Y.; Hongo, S.; Mizuta, K.; Nishimura, H. Neutralizing Epitopes and Residues Mediating the Potential Antigenic Drift of the Hemagglutinin-Esterase Protein of Influenza C Virus. Viruses 2018, 10, 417. https://doi.org/10.3390/v10080417
Matsuzaki Y, Sugawara K, Furuse Y, Shimotai Y, Hongo S, Mizuta K, Nishimura H. Neutralizing Epitopes and Residues Mediating the Potential Antigenic Drift of the Hemagglutinin-Esterase Protein of Influenza C Virus. Viruses. 2018; 10(8):417. https://doi.org/10.3390/v10080417
Chicago/Turabian StyleMatsuzaki, Yoko, Kanetsu Sugawara, Yuki Furuse, Yoshitaka Shimotai, Seiji Hongo, Katsumi Mizuta, and Hidekazu Nishimura. 2018. "Neutralizing Epitopes and Residues Mediating the Potential Antigenic Drift of the Hemagglutinin-Esterase Protein of Influenza C Virus" Viruses 10, no. 8: 417. https://doi.org/10.3390/v10080417
APA StyleMatsuzaki, Y., Sugawara, K., Furuse, Y., Shimotai, Y., Hongo, S., Mizuta, K., & Nishimura, H. (2018). Neutralizing Epitopes and Residues Mediating the Potential Antigenic Drift of the Hemagglutinin-Esterase Protein of Influenza C Virus. Viruses, 10(8), 417. https://doi.org/10.3390/v10080417




