Small RNA NGS Revealed the Presence of Cherry Virus A and Little Cherry Virus 1 on Apricots in Hungary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Sample Preparation
2.2. Pipeline for Data Evaluation of NGS Results (Bioinformatics)
2.3. Validation of Predicted Virus Diagnostics by RT-PCR
2.4. Phylogenetic Analysis
2.5. Validation by Northern Blot
3. Results and Discussion
3.1. Small RNA NGS Revealed the Presence of CVA and LChV-1
3.1.1. Initial Statistics
3.1.2. Small RNA NGS-Based Virus Diagnostics
3.2. Validation of the Small RNA NGS Virus Diagnostics
3.2.1. Validation of the Presence of Cherry Virus A
3.2.2. Validation of the Presence of Little Cherry Virus 1
3.2.3. Phylogenetic Relationship of Hungarian CVA and LChV-1 Isolates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Varveri, C.; Maliogka, V.I.; Kapari-Isaia, T. Chapter One—Principles for Supplying Virus-Tested Material. In Advances in Virus Research; Loebenstein, G., Katis, N.I., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 91, pp. 1–32. [Google Scholar]
- Barba, M.; Ilardi, V.; Pasquini, G. Chapter Three—Control of Pome and Stone Fruit Virus Diseases. In Advances in Virus Research; Loebenstein, G., Katis, N.I., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 91, pp. 47–83. [Google Scholar]
- Barba, M.; Czosnek, H.; Hadidi, A. Historical Perspective, Development and Applications of Next-Generation Sequencing in Plant Virology. Viruses 2014, 6, 106–136. [Google Scholar] [CrossRef] [PubMed]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A.; Flores, R.; Candresse, T.; Barba, M. Next-Generation Sequencing and Genome Editing in Plant Virology. Front. Microbiol. 2016, 7, 1325. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral Diagnostics in Plants Using Next Generation Sequencing: Computational Analysis in Practice. Front. Plant Sci. 2017, 8, 1770. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus Res. 2017, 239, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant Virus Metagenomics: Advances in Virus Discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Giampetruzzi, A.; De Stradis, A.; Digiaro, M. Deep-sequencing analysis of an apricot tree with vein clearing symptoms reveals the presence of a novel betaflexivirus. Virus Res. 2014, 181, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Marais, A.; Faure, C.; Candresse, T. New Insights into Asian Prunus Viruses in the Light of NGS-Based Full Genome Sequencing. PLoS ONE 2016, 11, e0146420. [Google Scholar] [CrossRef] [PubMed]
- Marais, A.; Faure, C.; Mustafayev, E.; Barone, M.; Alioto, D.; Candresse, T. Characterization by Deep Sequencing of Prunus virus T, a Novel Tepovirus Infecting Prunus Species. Phytopathology 2014, 105, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Villamor, D.E.V.; Pillai, S.S.; Eastwell, K.C. High throughput sequencing reveals a novel fabavirus infecting sweet cherry. Arch. Virol. 2017, 162, 811–816. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cai, L.; Zhou, L.; Yang, Z.; Hong, N.; Wang, G.; Li, S.; Xu, W. Deep sequencing reveals the first fabavirus infecting peach. Sci. Rep. 2017, 7, 11329. [Google Scholar] [CrossRef] [PubMed]
- Marais, A.; Faure, C.; Mustafayev, E.; Candresse, T. Characterization of New Isolates of Apricot vein clearing-associated virus and of a New Prunus-Infecting Virus: Evidence for Recombination as a Driving Force in Betaflexiviridae Evolution. PLoS ONE 2015, 10, e0129469. [Google Scholar] [CrossRef] [PubMed]
- Koloniuk, I.; Sarkisova, T.; Petrzik, K.; Lenz, O.; Přibylová, J.; Fránová, J.; Špak, J.; Lotos, L.; Beta, C.; Katsiani, A.; et al. Variability Studies of Two Prunus-Infecting Fabaviruses with the Aid of High-Throughput Sequencing. Viruses 2018, 10, 204. [Google Scholar] [CrossRef] [PubMed]
- Šafářová, D.; Faure, C.; Marais, A.; Suchá, J.; Paprštein, F.; Navrátil, M.; Candresse, T. First Report of Prunus virus F Infecting Sour Cherry in the Czech Republic. Plant Dis. 2017, 101, 1828. [Google Scholar] [CrossRef]
- Šafářová, D.; Faure, C.; Candresse, T.; Navrátil, M.; Nečas, T.; Marais, A. First Report of Little cherry virus 1 Infecting Apricot in the Czech Republic. Plant Dis. 2016, 101, 845. [Google Scholar] [CrossRef]
- James, D.; Phelan, J.; Jesperson, G. First Report of Prunus virus F infecting sweet cherry (Prunus avium cv. ‘StaccatoTM’) in Canada. Plant Dis. 2018. [Google Scholar] [CrossRef]
- Donaire, L.; Wang, Y.; Gonzalez-Ibeas, D.; Mayer, K.F.; Aranda, M.A.; Llave, C. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 2009, 392, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Parent, J.-S.; Martinez de Alba, A.E.; Vaucheret, H. The origin and effect of small RNA signaling in plants. Front. Plant Sci. 2012, 3, 179. [Google Scholar] [CrossRef] [PubMed]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pecman, A.; Kutnjak, D.; Gutierrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next Generation Sequencing for Detection and Discovery of Plant Viruses and Viroids: Comparison of Two Approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef] [PubMed]
- Santala, J.; Valkonen, J.P.T. Sensitivity of Small RNA-Based Detection of Plant Viruses. Front. Microbiol. 2018, 9, 939. [Google Scholar] [CrossRef] [PubMed]
- Navarro, B.; Pantaleo, V.; Gisel, A.; Moxon, S.; Dalmay, T.; Bisztray, G.; Di Serio, F.; Burgyan, J. Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PLoS ONE 2009, 4, e7686. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, V.; Saldarelli, P.; Miozzi, L.; Giampetruzzi, A.; Gisel, A.; Moxon, S.; Dalmay, T.; Bisztray, G.; Burgyan, J. Deep sequencing analysis of viral short RNAs from an infected Pinot Noir grapevine. Virology 2010, 408, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Giampetruzzi, A.; Roumi, V.; Roberto, R.; Malossini, U.; Yoshikawa, N.; La Notte, P.; Terlizzi, F.; Credi, R.; Saldarelli, P. A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small RNAs in Cv Pinot gris. Virus Res. 2012, 163, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, Y.; Cao, M.; Pantaleo, V.; Burgyan, J.; Li, W.X.; Ding, S.W. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. USA 2012, 109, 3938–3943. [Google Scholar] [CrossRef] [PubMed]
- Czotter, N.; Molnar, J.; Szabó, E.; Demian, E.; Kontra, L.; Baksa, I.; Szittya, G.; Kocsis, L.; Deak, T.; Bisztray, G.; et al. NGS of Virus-Derived Small RNAs as a Diagnostic Method Used to Determine Viromes of Hungarian Vineyards. Front. Microbiol. 2018, 9, 122. [Google Scholar] [CrossRef]
- Jelkmann, W. Cherry virus A: CDNA cloning of dsRNA, nucleotide sequence analysis and serology reveal a new plant capillovirus in sweet cherry. J. Gen. Virol. 1995, 76 Pt 8, 2015–2024. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Xu, Y.; Candresse, T.; He, Z.; Li, S.; Ma, Y.; Lu, M. Further insight into genetic variation and haplotype diversity of Cherry virus A from China. PLoS ONE 2017, 12, e0186273. [Google Scholar] [CrossRef] [PubMed]
- Marais, A.; Svanella, D.L.; Barone, M.; Gentit, P.; Faure, C.; Charlot, G.; Ragozzino, A.; Candresse, T. Development of a polyvalent RT-PCR detection assay covering the genetic diversity of Cherry capillovirus A. Plant Pathol. 2012, 61, 195–204. [Google Scholar] [CrossRef]
- Kesanakurti, P.; Belton, M.; Saeed, H.; Rast, H.; Boyes, I.; Rott, M. Comparative analysis of cherry virus A genome sequences assembled from deep sequencing data. Arch. Virol. 2017, 162, 2821–2828. [Google Scholar] [CrossRef] [PubMed]
- Marais, A.; Faure, C.; Svanella-Dumas, L.; Candresse, T. First Report of Cherry virus A in Prunus mume in China. Plant Dis. 2008, 92, 1589. [Google Scholar] [CrossRef]
- Keim-Konrad, R.; Jelkmann, W. Genome analysis of the 3′-terminal part of the little cherry disease associated dsRNA reveals a monopartite clostero-like virus. Arch. Virol. 1996, 141, 1437–1451. [Google Scholar] [CrossRef] [PubMed]
- Matic, S.; Minafra, A.; Sánchez-Navarro, J.A.; Pallás, V.; Myrta, A.; Martelli, G.P. ‘Kwanzan Stunting’ syndrome: Detection and molecular characterization of an Italian isolate of Little cherry virus 1. Virus Res. 2009, 143, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Candresse, T.; Marais, A.; Faure, C.; Gentit, P. Association of Little cherry virus 1 (LChV1) with the Shirofugen Stunt Disease and Characterization of the Genome of a Divergent LChV1 Isolate. Phytopathology 2013, 103, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Glasa, M. First report of little cherry virus-1 in Slovakia. J. Plant Pathol. 2015, 97, 542. [Google Scholar]
- Sabanadzovic, S.; Aboughanem, N.; Rowhani, A.; Grant, J.A.; Uyemoto, J. Detection of Cherry virus A, Cherry necrotic rusty mottle virus and Little cherry virus 1 in California orchards. J. Plant Pathol. 2005, 87, 173. [Google Scholar]
- Komorowska, B.; Cieślińska, M. First Report of Cherry virus A and Little cherry virus-1 in Poland. Plant Dis. 2004, 88, 909. [Google Scholar] [CrossRef]
- Bajet, N.B.; Unruh, T.R.; Druffel, K.L.; Eastwell, K.C. Occurrence of Two Little Cherry Viruses in Sweet Cherry in Washington State. Plant Dis. 2008, 92, 234–238. [Google Scholar] [CrossRef]
- Katsiani, A.T.; Maliogka, V.I.; Amoutzias, G.D.; Efthimiou, K.E.; Katis, N.I. Insights into the genetic diversity and evolution of Little cherry virus 1. Plant Pathol. 2015, 64, 817–824. [Google Scholar] [CrossRef]
- Czotter, N.; Molnár, J.; Pesti, R.; Demián, E.; Baráth, D.; Varga, T.; Várallyay, É. Use of siRNAs for Diagnosis of Viruses Associated to Woody Plants in Nurseries and Stock Collections. In Viral Metagenomics: Methods and Protocols; Pantaleo, V., Chiumenti, M., Eds.; Springer: New York, NY, USA, 2018; pp. 115–130. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schaffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, T.; Candresse, T.; Ravelonandro, M.; Dunez, J. A polymerase chain reaction assay adapted to plum pox potyvirus detection. J. Virol. Methods 1991, 33, 355–365. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Wilks, J.M.; Welsh, M.F. Host range studies of the little cherry disease virus. Can. J. Plant Sci. 1961, 41, 544–548. [Google Scholar] [CrossRef]
- Jelkmann, W.; Fechtner, B.; Agranovsky, A.A. Complete genome structure and phylogenetic analysis of little cherry virus, a mealybug-transmissible closterovirus. J. Gen. Virol. 1997, 78, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of Next Generation Sequencing for Diagnostic Testing of Tree Fruit Viruses and Viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Islas, C.; Rowhani, A. Comparison of Next-Generation Sequencing Versus Biological Indexing for the Optimal Detection of Viral Pathogens in Grapevine. Phytopathology 2015, 105, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Massart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.-S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A Framework for the Evaluation of Biosecurity, Commercial, Regulatory, and Scientific Impacts of Plant Viruses and Viroids Identified by NGS Technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef] [PubMed]
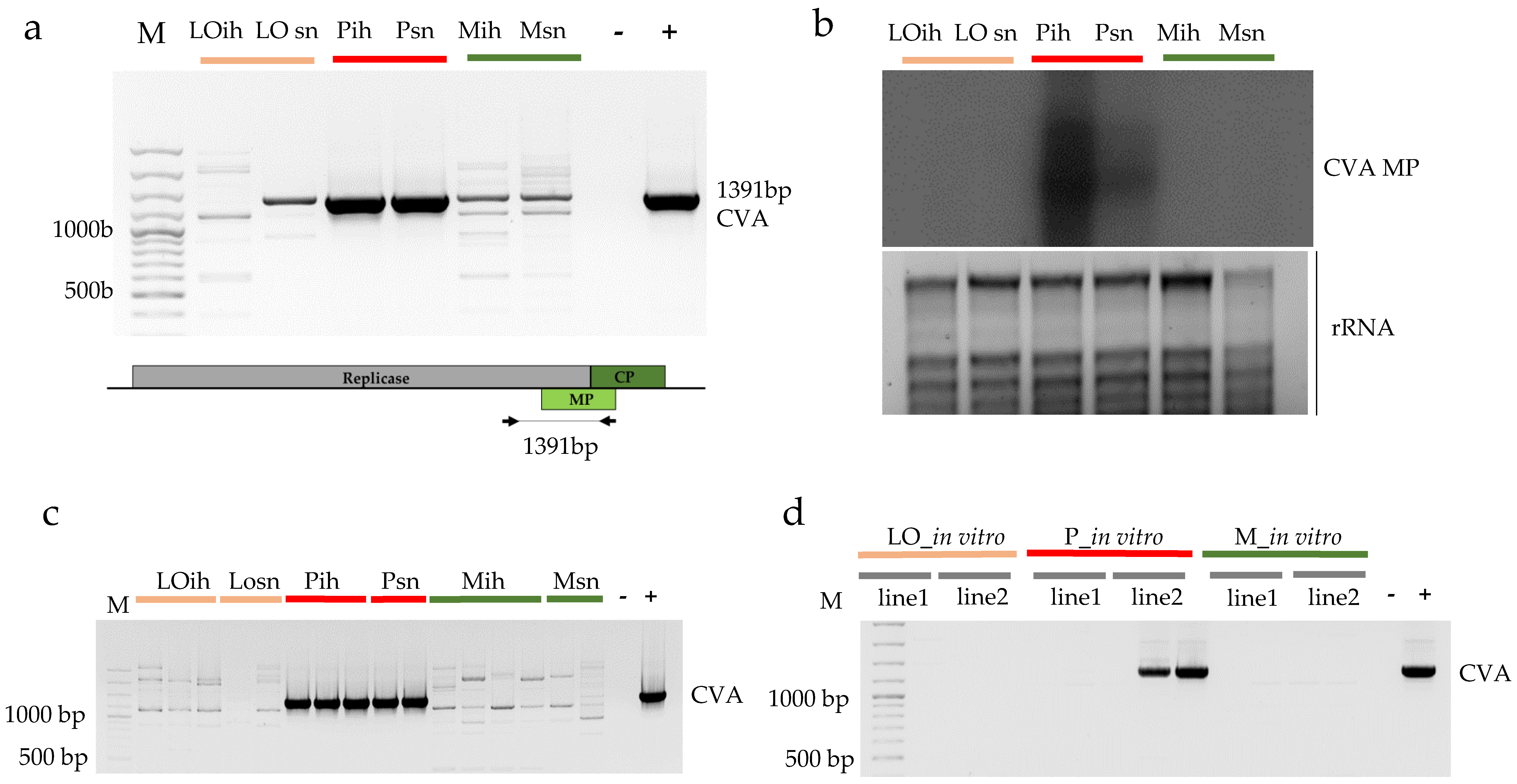

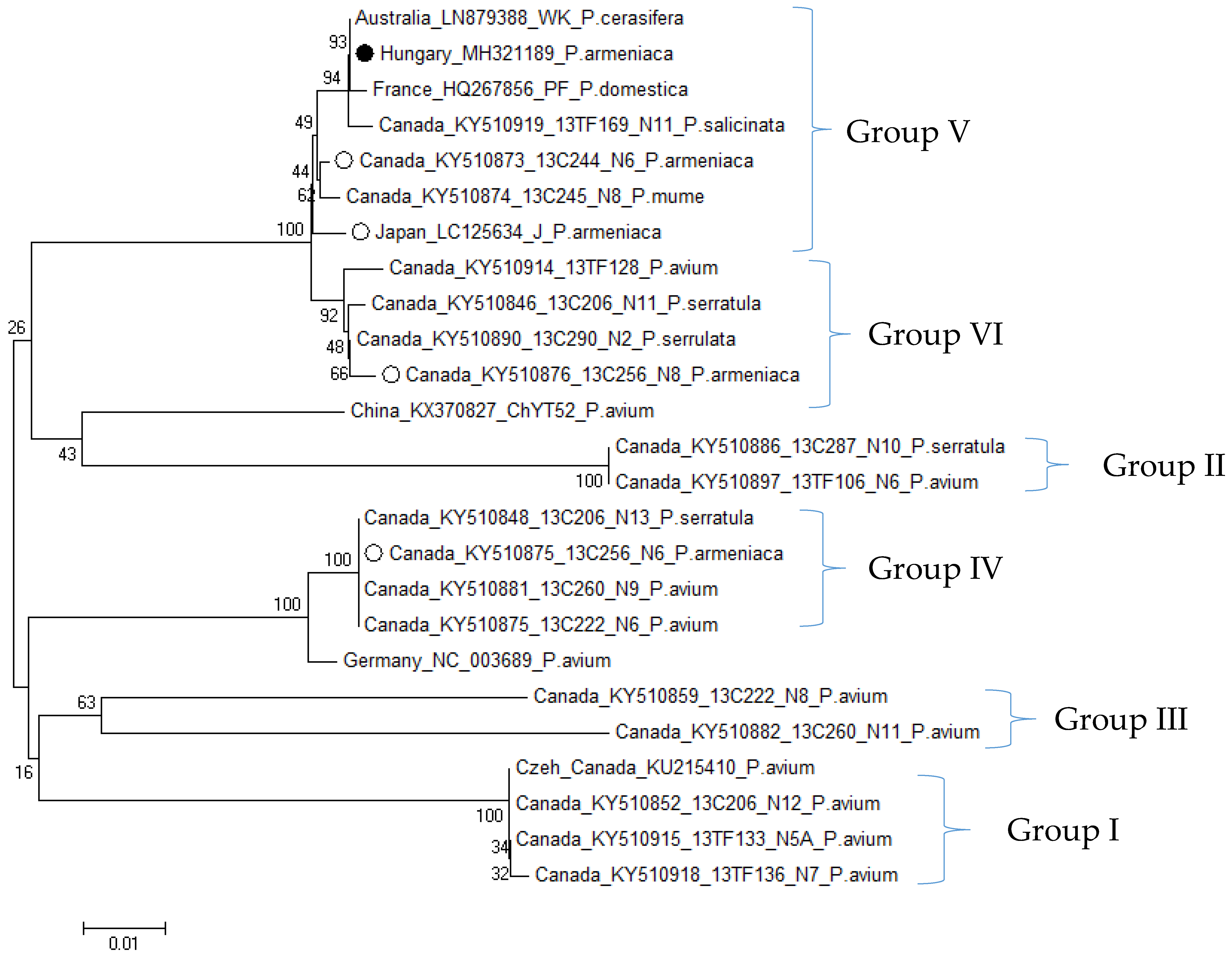
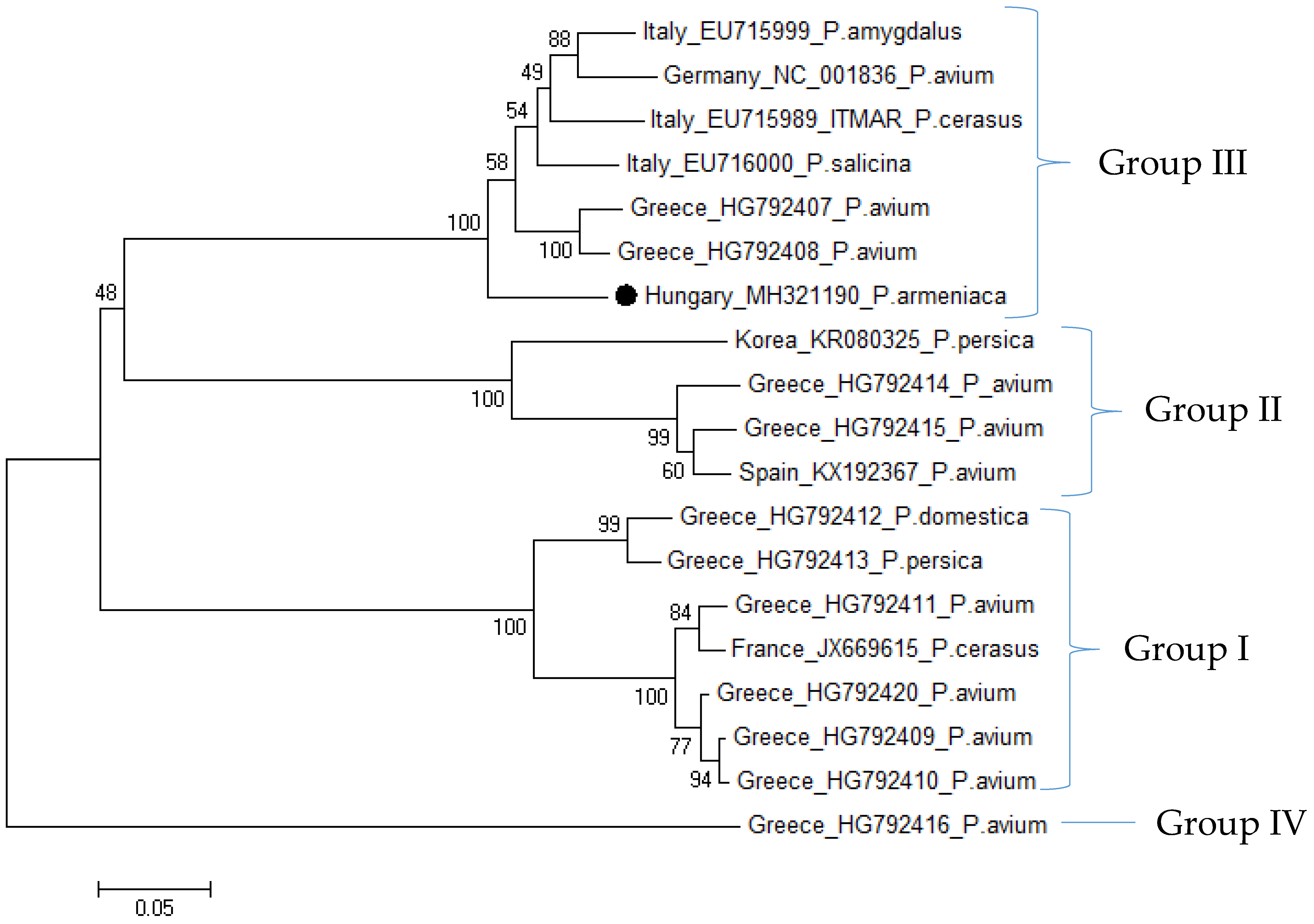
| Variety | Type of Analysis | Origin of Samples | ||
|---|---|---|---|---|
| In Vitro | Isolator House | Stock Nursery | ||
| Ligeti óriás | NGS virus hit | - | 0 | PPV |
| PCR * | 0 | 0 | 1/2 PPV | |
| Pannónia | NGS virus hit | - | CVA | CVA |
| PCR * | 2/4 CVA | 3/3 CVA | 2/2 CVA | |
| Magyar kajszi | NGS virus hit | - | 0 | PPV, LChV-1 |
| PCR * | 0 | 0 | 1/2 PPV, 1/2 LChV-1 | |
| Virus | Variety | Genebank Identifier | Genome Used as a Reference | Position on the Reference Genome | Function of the Amplified Part of the Genome | Identity on Nucleotide Level (%) | Identity on Amino Acid Level (%) |
|---|---|---|---|---|---|---|---|
| CVA | Pannónia kajszi/Pannonian apricot | MH321189 | NC_003689.1 | 5401–6791 | replicase (partial) | 1275/1392(92%) | 392/463(85%) |
| 5400–6791 | movement protein | 421/463(91%) | |||||
| LChV-1 | Magyar kajszi/Hungarian apricot | MH321190 | NC_001836.1 | 12,165–13,261 | coat protein | 1011/1097(92%) | 338/365(93%) |
| LChV-1 | Magyar kajszi/Hungarian apricot | MH321191 | NC_001836.1 | 9493–10,288 | HSP70h | 753/796(95%) | 251/265(95%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baráth, D.; Jaksa-Czotter, N.; Molnár, J.; Varga, T.; Balássy, J.; Szabó, L.K.; Kirilla, Z.; Tusnády, G.E.; Preininger, É.; Várallyay, É. Small RNA NGS Revealed the Presence of Cherry Virus A and Little Cherry Virus 1 on Apricots in Hungary. Viruses 2018, 10, 318. https://doi.org/10.3390/v10060318
Baráth D, Jaksa-Czotter N, Molnár J, Varga T, Balássy J, Szabó LK, Kirilla Z, Tusnády GE, Preininger É, Várallyay É. Small RNA NGS Revealed the Presence of Cherry Virus A and Little Cherry Virus 1 on Apricots in Hungary. Viruses. 2018; 10(6):318. https://doi.org/10.3390/v10060318
Chicago/Turabian StyleBaráth, Dániel, Nikoletta Jaksa-Czotter, János Molnár, Tünde Varga, Júlia Balássy, Luca Krisztina Szabó, Zoltán Kirilla, Gábor E. Tusnády, Éva Preininger, and Éva Várallyay. 2018. "Small RNA NGS Revealed the Presence of Cherry Virus A and Little Cherry Virus 1 on Apricots in Hungary" Viruses 10, no. 6: 318. https://doi.org/10.3390/v10060318
APA StyleBaráth, D., Jaksa-Czotter, N., Molnár, J., Varga, T., Balássy, J., Szabó, L. K., Kirilla, Z., Tusnády, G. E., Preininger, É., & Várallyay, É. (2018). Small RNA NGS Revealed the Presence of Cherry Virus A and Little Cherry Virus 1 on Apricots in Hungary. Viruses, 10(6), 318. https://doi.org/10.3390/v10060318






