Detection of Specific ZIKV IgM in Travelers Using a Multiplexed Flavivirus Microsphere Immunoassay
Abstract
1. Introduction
2. Materials and Methods
2.1. Bead Coupling to Monoclonal Antibody for FlaviMIA
2.2. Production of Purified Whole Virus Antigen Preparations for FlaviMIA
2.3. Binding of Flavivirus Preparations to Monoclonal Antibody-Coupled Microspheres for FlaviMIA
2.4. Reference Sera
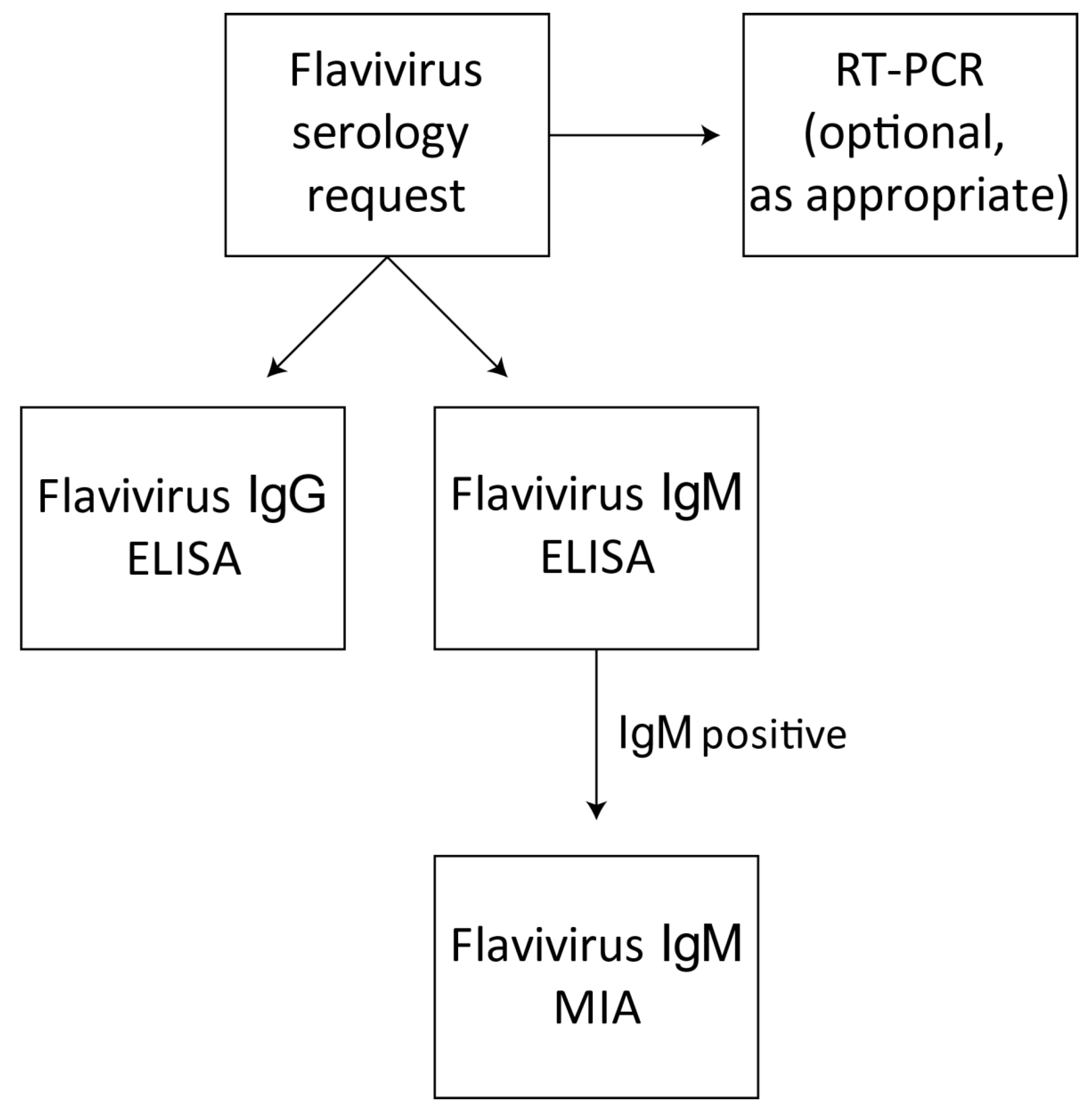
2.5. Flavivirus Screening Assays
2.6. Flavivirus-Specific IgM Serology—FlaviMIA
2.7. ZIKV PRNT
3. Results
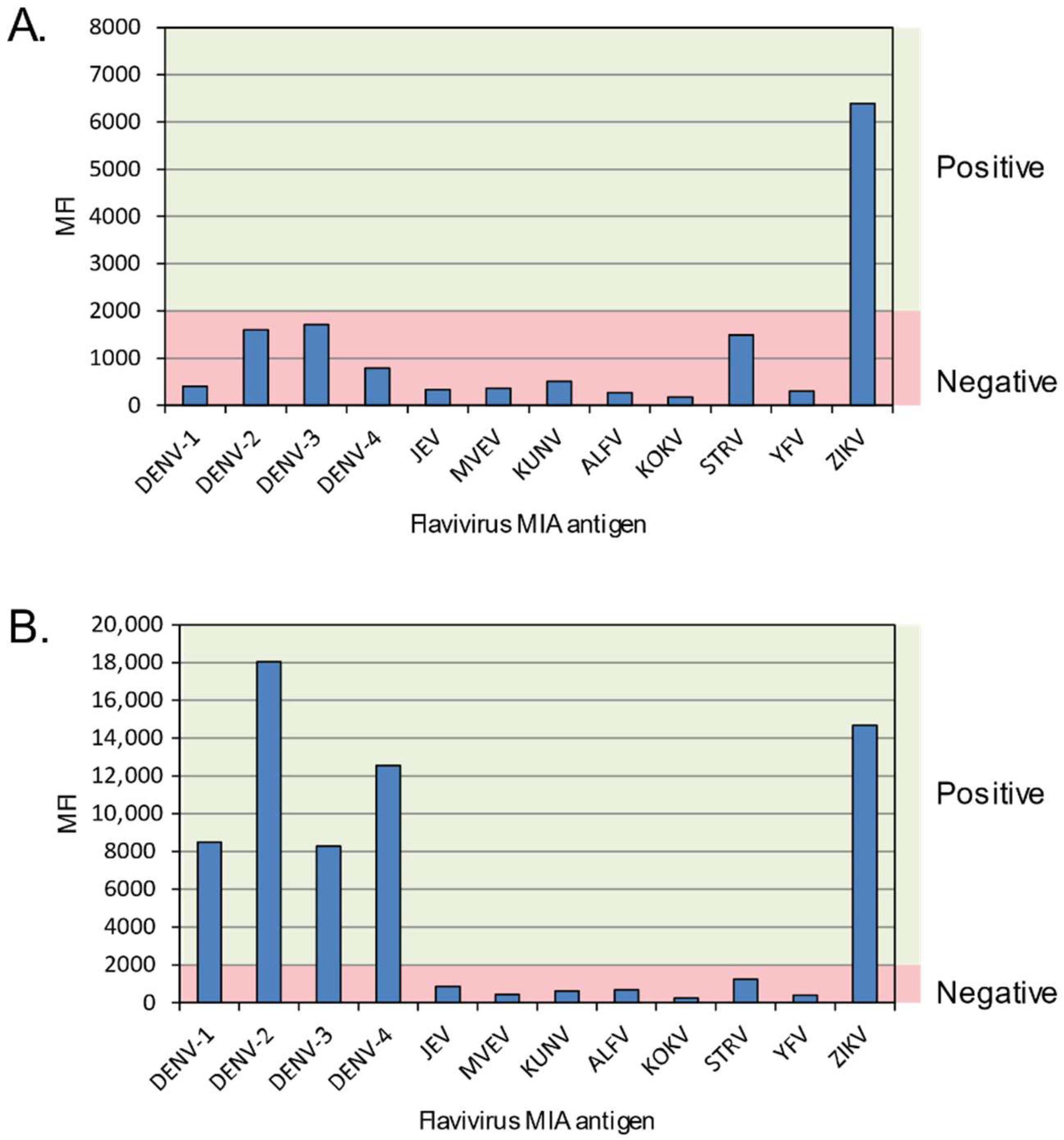
3.1. FlaviMIA Validation
3.2. Analysis of Patient Samples
3.3. Patients with Recent Primary ZIKV Infections
3.4. Patients with Recent Secondary ZIKV Infections
3.5. Patients with Past ZIKV Infection
3.6. Patients with ZIKV/DENV IgM
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika Virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika virus outbreak on Yap Island, federated states of Micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Besnard, M.; Eyrolle-Guignot, D.; Guillemette-Artur, P.; Lastere, S.; Bost-Bezeaud, F.; Marcelis, L.; Abadie, V.; Garel, C.; Moutard, M.L.; Jouannic, J.M.; et al. Congenital cerebral malformations and dysfunction in fetuses and newborns following the 2013 to 2014 Zika virus epidemic in French Polynesia. Eur. Commun. Dis. Bull. 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Zika Virus Disease Interim Case Definition. 12 February 2016. Available online: http://who.int/csr/disease/zika/case-definition/en/ (accessed on 5 July 2016).
- Musso, D.; Nilles, E.J.; Cao-Lormeau, V.M. Rapid spread of emerging Zika virus in the Pacific area. Clin. Microbiol. Infect. 2014, 20, 595–596. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Melo, A.S.; Malinger, G.; Ximenes, R.; Szejnfeld, P.O.; Alves Sampaio, S.; Bispo de Filippis, A.M. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: Tip of the iceberg? Ultrasound Obstet. Gynecol. 2016, 47, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Hubner, R. Zika virus—Brazil (16): (Pernambuco) Microcephaly Cause Determined. Available online: http://promedmail.org/post/20151118.3799192 (accessed on 18 November 2015).
- Pan American Health Organization. Epidemiological Update Neurological Syndrome, Congenital Anomalies, and Zika Virus Infection. 17 January 2016. Available online: http://www.paho.org/hq/index.php?option=com_docman&task=doc_view&Itemid=270&gid=32879&lang=en (accessed on 5 July 2016).
- Taylor, C.; Simmons, R.; Smith, I. Development of immunoglobulin M capture enzyme-linked immunosorbent assay to differentiate human flavivirus infections occurring in Australia. Clin. Diagn. Lab. Immunol. 2005, 12, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Pyke, A.T.; Daly, M.T.; Cameron, J.N.; Moore, P.R.; Taylor, C.T.; Hewitson, G.R.; Humphreys, J.L.; Gair, R. Imported Zika virus infection from the Cook Islands into Australia, 2014. PLoS Curr. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Roehrig, J.T.; Mathews, J.H.; Trent, D.W. Identification of epitopes on the E glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology 1983, 128, 118–126. [Google Scholar] [CrossRef]
- Dowd, K.A.; DeMaso, C.R.; Pelc, R.S.; Speer, S.D.; Smith, A.R.; Goo, L.; Platt, D.J.; Mascola, J.R.; Graham, B.S.; Mulligan, M.J.; et al. Broadly neutralizing activity of Zika virus-immune sera identifies a single viral serotype. Cell Rep. 2016, 16, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Basile, A.J.; Horiuchi, K.; Panella, A.J.; Laven, J.; Kosoy, O.; Lanciotti, R.S.; Venkateswaran, N.; Biggerstaff, B.J. Multiplex microsphere immunoassays for the detection of IgM and IgG to arboviral diseases. PLoS ONE 2013, 8, e75670. [Google Scholar] [CrossRef] [PubMed]
- Pyke, A.T.; Phillips, D.A.; Chuan, T.F.; Smith, G.A. Sucrose density gradient centrifugation and cross-flow filtration methods for the production of arbovirus antigens inactivated by binary ethylenimine. BMC Microbiol. 2004, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Pyke, A.T.; Moore, P.R.; Hall-Mendelin, S.; McMahon, J.L.; Harrower, B.J.; Constantino, T.R.; van den Hurk, A.F. Isolation of Zika virus imported from Tonga into Australia. PLoS Curr. 2016, 8. [Google Scholar] [CrossRef]
- Balmaseda, A.; Zambrana, J.V.; Collado, D.; Garcia, N.; Saborio, S.; Elizondo, D.; Mercado, J.C.; Gonzalez, K.; Cerpas, C.; Nunez, A.; et al. Comparison of four serological methods and two RT-PCR assays for diagnosis and surveillance of Zika. J. Clin. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mansuy, J.M.; Mengelle, C.; Pasquier, C.; Chapuy-Regaud, S.; Delobel, P.; Martin-Blondel, G.; Izopet, J. Zika virus infection and prolonged viremia in whole-blood specimens. Emerg. Infect. Dis. 2017, 23, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Lum, F.M.; Lin, C.; Susova, O.Y.; Teo, T.H.; Fong, S.W.; Mak, T.M.; Lee, L.K.; Chong, C.Y.; Lye, D.C.B.; Lin, R.T.P.; et al. A sensitive method for detecting Zika virus antigen in patients’ whole-blood specimens as an alternative diagnostic approach. J. Infect. Dis. 2017, 216, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.J.; Furuya, A.; Zou, J.; Xie, X.; Dupuis, A.P., 2nd.; Kramer, L.D.; Shi, P.Y. A multiplex microsphere immunoassay for Zika virus diagnosis. EBioMedicine 2017, 16, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Balmaseda, A.; Stettler, K.; Medialdea-Carrera, R.; Collado, D.; Jin, X.; Zambrana, J.V.; Jaconi, S.; Cameroni, E.; Saborio, S.; Rovida, F.; et al. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 8384–8389. [Google Scholar] [CrossRef] [PubMed]
- Steinhagen, K.; Probst, C.; Radzimski, C.; Schmidt-Chanasit, J.; Emmerich, P.; van Esbroeck, M.; Schinkel, J.; Grobusch, M.P.; Goorhuis, A.; Warnecke, J.M.; et al. Serodiagnosis of Zika virus (ZIKV) infections by a novel NS1-based ELISA devoid of cross-reactivity with dengue virus antibodies: A multicohort study of assay performance, 2015 to 2016. Eur. Commun. Dis. Bull. 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pinsky, B.A.; Ananta, J.S.; Zhao, S.; Arulkumar, S.; Wan, H.; Sahoo, M.K.; Abeynayake, J.; Waggoner, J.J.; Hopes, C.; et al. Diagnosis of Zika virus infection on a nanotechnology platform. Nat. Med. 2017, 23, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Ronnberg, B.; Gustafsson, A.; Vapalahti, O.; Emmerich, P.; Lundkvist, A.; Schmidt-Chanasit, J.; Blomberg, J. Compensating for cross-reactions using avidity and computation in a suspension multiplex immunoassay for serotyping of Zika versus other flavivirus infections. Med. Microbiol. Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Ortiz, D.A.; Yang, Y.; Wong, S.J.; Kramer, L.D.; Shi, P.Y.; Loeffelholz, M.J.; Ren, P. Evaluation of a novel reporter virus neutralization test for serological diagnosis of Zika and dengue virus infection. J. Clin. Microbiol. 2017, 55, 3028–3036. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, M.J.; Fischer, M.; Panella, A.J.; Kosoy, O.I.; Laven, J.J.; Lanciotti, R.S.; Staples, J.E. Zika virus disease in travelers returning to the United States, 2010–2014. Am. J. Trop. Med. Hyg. 2016, 95, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Ludolfs, D.; van An, L.; Schmitz, H. Laboratory diagnosis of primary and secondary dengue infection. J. Clin. Virol. 2004, 31, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Halstead, S.B.; Rojanasuphot, S.; Sangkawibha, N. Original antigenic sin in dengue. Am. J. Trop. Med. Hyg. 1983, 32, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Furuya-Kanamori, L.; Liang, S.; Milinovich, G.; Soares Magalhaes, R.J.; Clements, A.C.; Hu, W.; Brasil, P.; Frentiu, F.D.; Dunning, R.; Yakob, L. Co-distribution and co-infection of chikungunya and dengue viruses. BMC Infect. Dis. 2016, 16, 84. [Google Scholar] [CrossRef] [PubMed]
- Villamil-Gomez, W.E.; Rodriguez-Morales, A.J.; Uribe-Garcia, A.M.; Gonzalez-Arismendy, E.; Castellanos, J.E.; Calvo, E.P.; Alvarez-Mon, M.; Musso, D. Zika, dengue, and chikungunya co-infection in a pregnant woman from Colombia. Int. J. Infect. Dis. 2016, 51, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, J.J.; Gresh, L.; Vargas, M.J.; Ballesteros, G.; Tellez, Y.; Soda, K.J.; Sahoo, M.K.; Nunez, A.; Balmaseda, A.; Harris, E.; et al. Viremia and clinical presentation in nicaraguan patients infected with Zika virus, chikungunya virus, and dengue virus. Clin. Infect. Dis. 2016, 63, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Faccini-Martinez, A.A.; Botero-Garcia, C.A.; Benitez-Baracaldo, F.C.; Perez-Diaz, C.E. With regard about the case of dengue, chikungunya and Zika co-infection in a patient from Colombia. J. Infect. Public Health 2016, 9, 687–688. [Google Scholar] [CrossRef] [PubMed]
| Group Reference Number | Laboratory Reference No. | Sex | Age | Flavivirus IgG ELISA Result | Flavivirus IgG ELISA Results | FlaviMIA Result | FlaviMIA Result | SI | RT-PCR Result | PRNT Result | Year/Travel Region |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st Sample | 2nd Sample | 1st Sample | 2nd Sample | ||||||||
| Group 1: recent ZIKV infection—primary | |||||||||||
| #1 | 1 | M | 40 | Negative | Positive | Negative | Positive (ZIKV specific) | 19 | Not detected | Positive | 2013 Thailand |
| #2 | 2 | M | 34 | Negative | Positive | Negative | Positive (ZIKV specific) | 14 | Not detected | Positive | 2013 Vietnam |
| #3 | 3 | M | 30 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Insufficient | 2014 Cook Islands |
| #4 | 7 | M | 58 | Negative | Positive | Negative | Positive (ZIKV specific) | 6 | ZIKV RNA detected | Positive | 2014 Cook Islands |
| #5 | 9 | M | 32 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Negative | 2014 Cook Islands |
| #6 | 12 | M | 38 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Insufficient | 2014 Cook Islands |
| #7 | 13 | M | 79 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Insufficient | 2014 Cook Islands |
| #8 | 14 | M | 24 | Negative | Negative | Negative | Positive (ZIKV specific) | 4 | ZIKV RNA detected | Negative | 2014 Cook Islands |
| #9 | 15 | F | 7 | Negative | Positive | Negative | Positive (ZIKV specific) | 35 | Not detected | Positive | 2014 Country of travel not identified |
| #10 | 16 | F | 37 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 15 | Not detected | Positive | 2014 Country of travel not identified |
| #11 | 19 | F | 42 | Negative | Positive | Negative | Positive (cross-reactive) | 19 | ZIKV RNA detected | Positive | 2014 Vanuatu |
| #12 | 20 | F | 43 | Negative | Positive | Negative | Positive (ZIKV specific) | 16 | Not detected | Positive | 2015 Bali |
| #13 | 26 | M | 48 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Negative | 2015 Solomon Islands |
| #14 | 27 | M | 45 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Negative | 2015 Solomon Islands |
| #15 | 29 | M | 33 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 15 | ZIKV RNA detected | Positive | 2015 Solomon Islands |
| #16 | 30 | M | 52 | Negative | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2015 Solomon Islands |
| #17 | 32 | M | 29 | Negative | Positive | Negative | Positive (ZIKV specific) | 91 | Not detected | Positive | 2015 Solomon Islands |
| #18 | 33 | F | 22 | Negative | Positive | Negative | Positive (ZIKV specific) | 10 | ZIKV RNA detected | Positive | 2015 Vanuatu |
| #19 | 34 | F | 33 | Negative | nss | Positive (ZIKV specific) | nss | n/a | ZIKV RNA detected | Positive | 2015 Vanuatu |
| #20 | 47 | F | 30 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 14 | ZIKV RNA detected | Positive | 2016 Fiji |
| #21 | 52 | F | 22 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 17 | Not detected | Positive | 2016 Fiji |
| #22 | 53 | F | 62 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 16 | Not detected | Positive | 2016 Fiji |
| #23 | 57 | F | 17 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 24 | Not detected | Positive | 2016 Fiji/Samoa |
| #24 | 60 | M | 50 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Insufficient | 2016 Guyana |
| #25 | 61 | M | 30 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 26 | ZIKV RNA detected | Positive | 2016 Jamaica |
| #26 | 64 | F | 33 | Negative | nss | Positive (ZIKV specific) | nss | n/a | ZIKV RNA detected | Negative | 2016 Mexico |
| #27 | 65 | F | 33 | Negative | Positive | Negative | Positive (ZIKV specific) | 14 | Not detected | Positive | 2016 Mexico |
| #28 | 68 | M | 37 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 8 | Not detected | Positive | 2016 Mexico |
| #29 | 69 | F | 42 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 21 | Not detected | Positive | 2016 Mexico |
| #30 | 71 | F | 21 | Negative | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Nicaragua |
| #31 | 74 | F | 28 | Negative | Positive | Negative | Positive (ZIKV specific) | 16 | ZIKV RNA detected | Positive | 2016 Samoa |
| #32 | 76 | F | 2 | Negative | nss | Positive (ZIKV specific) | nss | n/a | ZIKV RNA detected | Positive | 2016 Samoa |
| #33 | 81 | F | 28 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 15 | Not detected | Positive | 2016 Solomon Is |
| #34 | 84 | F | 43 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 14 | Not detected | Positive | 2016 Thailand |
| #35 | 86 | F | 37 | Negative | nss | Negative | Positive (ZIKV specific) | 46 | ZIKV RNA detected | Positive | 2016 Tonga |
| #36 | 88 | F | 52 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Negative | 2016 Tonga |
| #37 | 89 | F | 23 | Negative | Positive | Positive (cross-reactive) | Positive (ZIKV specific) | 14 | Not detected | Positive | 2016 Tonga |
| #38 | 92 | M | 41 | Negative | Negative | Positive (ZIKV specific) | Positive (ZIKV specific) | 11 | Not detected | Positive | 2016 Tonga |
| #39 | 94 | M | 43 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 94 | ZIKV RNA detected | Positive | 2016 Vanuatu/Fiji |
| #40 | 95 | F | 56 | Negative | nss | Negative | nss | n/a | ZIKV RNA detected | Negative | 2016 Vietnam |
| #41 | 97 | F | 71 | Negative | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 12 | Not detected | Positive | 2017 Country of travel not identified |
| #42 | 99 | F | 49 | Negative | nss | Negative | nss | n/a | Not detected | Positive | 2017 Cuba |
| #43 | 100 | M | 33 | Negative | Positive | Negative | Positive (ZIKV specific) | 20 | ZIKV RNA detected | Positive | 2017 Cuba |
| Group 2: recent ZIKV infection—secondary | |||||||||||
| #1 | 4 | F | 42 | Positive | nss | Positive (cross-reactive) | nss | n/a | ZIKV RNA detected | Positive | 2014 Cook Islands |
| #2 | 5 | M | 30 | Positive | nss | Positive (ZIKV specific) | nss | n/a | ZIKV RNA detected | Negative | 2014 Cook Islands |
| #3 | 6 | F | 43 | Positive | nss | Positive (cross-reactive) | nss | n/a | ZIKV RNA detected | Insufficient | 2014 Cook Islands |
| #4 | 8 | F | 65 | Positive | Positive | Positive (DENV-4 specific) | Positive (ZIKV specific) | 23 | ZIKV RNA detected | Positive | 2014 Cook Islands |
| #5 | 10 | F | 38 | Positive | Positive | Negative | Positive (DENV-2 specific) | 20 | ZIKV RNA detected | Positive | 2014 Cook Islands |
| #6 | 11 | M | 31 | Positive | Positive | Positive (DENV-2 specific) | Positive (cross-reactive) | 23 | ZIKV RNA detected | Positive | 2014 Cook Islands |
| #7 | 23 | F | 51 | Positive | Positive | Negative | Positive (cross-reactive) | 30 | ZIKV RNA detected | Positive | 2015 El Salvador |
| #8 | 24 | M | 55 | Positive | Positive | Negative | Positive (ZIKV specific) | 25 | Not detected | Positive | 2015 Samoa |
| #9 | 25 | M | 46 | Positive | nss | Negative | nss | n/a | ZIKV RNA detected | Insufficient | 2015 Solomon Islands |
| #10 | 28 | F | 26 | Positive | nss | Negative | nss | n/a | ZIKV RNA detected | Positive | 2015 Solomon Islands |
| #11 | 36 | M | 54 | Positive | Positive | Negative | Positive (cross-reactive) | 13 | ZIKV RNA detected | Positive | 2016 Bali |
| #12 | 38 | M | 25 | Negative | nss | Positive (YFV specific) | nss | n/a | ZIKV RNA detected | Negative | 2016 Colombia |
| #13 | 45 | M | 42 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 15 | ZIKV RNA detected | Positive | 2016 El Salvador |
| #14 | 46 | F | 45 | Positive | Positive | Negative | Positive (ZIKV specific) | 13 | ZIKV RNA detected | Positive | 2016 Fiji |
| #15 | 62 | F | 25 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 14 | ZIKV RNA detected | Positive | 2016 Mexico |
| #16 | 63 | M | 34 | Positive | nss | Positive (ZIKV specific) | nss | n/a | ZIKV RNA detected | Negative | 2016 Mexico |
| #17 | 75 | M | 52 | Positive | nss | Positive (ZIKV specific) | nss | n/a | ZIKV RNA detected | Positive | 2016 Samoa |
| #18 | 82 | F | 60 | Positive | Positive | Negative | Positive (ZIKV specific) | 22 | ZIKV RNA detected | Positive | 2016 Solomon Islands |
| #19 | 83 | F | 30 | Negative | Positive | Positive (YFV specific) | Positive (ZIKV specific) | 18 | ZIKV RNA detected | Positive | 2016 Thailand |
| #20 | 85 | F | 71 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 13 | ZIKV RNA detected | Positive | 2016 Tonga |
| #21 | 87 | M | 34 | Positive | nss | Negative | nss | n/a | ZIKV RNA detected | Insufficient | 2016 Tonga |
| Group 3: patients with past ZIKV infection | |||||||||||
| #1 | 18 | M | 77 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 11 | Not detected | Positive | 2014 Papua New Guinea |
| #2 | 21 | M | 43 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 30 | Not tested | Positive | 2015 Brazil |
| #3 | 37 | F | 68 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 9 | Not tested | Positive | 2016 Caribbean |
| #4 | 39 | F | 23 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 26 | Not detected | Positive | 2016 Colombia |
| #5 | 40 | M | 35 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 22 | Not detected | Positive | 2016 Country of travel not identified |
| #6 | 41 | F | 39 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 14 | Not detected | Positive | 2016 Country of travel not identified |
| #7 | 43 | F | 61 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 17 | Not detected | Positive | 2016 Curacao |
| #8 | 51 | F | 23 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 17 | Not detected | Positive | 2016 Fiji |
| #9 | 67 | M | 48 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 28 | Not detected | Positive | 2016 Mexico |
| #10 | 72 | M | 88 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 9 | Not detected | Positive | 2016 No recent travel |
| #11 | 73 | M | 82 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 20 | Not detected | Positive | 2016 No recent travel |
| #12 | 80 | M | 73 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 36 | Not detected | Positive | 2016 Solomon Is |
| #13 | 90 | M | 27 | Positive | Positive | Positive (cross-reactive) | Positive (ZIKV specific) | 14 | Not detected | Positive | 2016 Tonga |
| #14 | 91 | F | 27 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 10 | Not detected | Positive | 2016 Tonga |
| Group 4: patients with ZIKV/DENV IgM | |||||||||||
| #1 | 17 | F | 43 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2014 Maldives |
| #2 | 22 | M | 78 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV/DENV) | 16 | Not tested | Positive | 2015 Burma |
| Group 5: patients with recent or past ZIKV infection | |||||||||||
| #1 | 31 | M | 37 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2015 Solomon Islands |
| #2 | 35 | F | 34 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not tested | Positive | 2015 Vanuatu |
| #3 | 42 | M | 31 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Country of travel not identified |
| #4 | 44 | F | 57 | Positive | Positive | Positive (ZIKV specific) | Positive (ZIKV specific) | 6 | Not detected | Positive | 2016 Dominican Republic |
| #5 | 48 | F | 25 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Fiji |
| #6 | 49 | M | 56 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Fiji |
| #7 | 50 | F | 41 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Fiji |
| #8 | 54 | M | 12 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Fiji |
| #9 | 55 | F | 31 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not tested | Positive | 2016 Fiji |
| #10 | 58 | M | 54 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Fiji/Tonga |
| #11 | 59 | F | 30 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not tested | Positive | 2016 Guatemala/Belize |
| #12 | 66 | F | 25 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Mexico |
| #13 | 70 | F | 36 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Mexico |
| #14 | 78 | M | 73 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Samoa |
| #15 | 79 | F | 27 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Solomon Is |
| #16 | 93 | F | 31 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Tonga |
| #17 | 96 | M | 32 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2016 Vietnam/Myanmar |
| #18 | 98 | F | 20 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Positive | 2017 Country of travel not identified |
| #19 | 101 | F | 36 | Positive | nss | Positive (ZIKV specific) | nss | n/a | Not tested | Positive | 2017 Fiji |
| Group 6: patients with unconfirmed ZIKV infection/false positives | |||||||||||
| #1 | 56 | M | 16 | Negative | Negative | Positive (ZIKV specific) | Positive (ZIKV specific) | 12 | Not detected | Negative | 2016 Fiji |
| #2 | 77 | F | 52 | Negative | nss | Positive (ZIKV specific) | nss | n/a | Not detected | Negative | 2016 Samoa |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, C.T.; Mackay, I.M.; McMahon, J.L.; Wheatley, S.L.; Moore, P.R.; Finger, M.J.; Hewitson, G.R.; Moore, F.A. Detection of Specific ZIKV IgM in Travelers Using a Multiplexed Flavivirus Microsphere Immunoassay. Viruses 2018, 10, 253. https://doi.org/10.3390/v10050253
Taylor CT, Mackay IM, McMahon JL, Wheatley SL, Moore PR, Finger MJ, Hewitson GR, Moore FA. Detection of Specific ZIKV IgM in Travelers Using a Multiplexed Flavivirus Microsphere Immunoassay. Viruses. 2018; 10(5):253. https://doi.org/10.3390/v10050253
Chicago/Turabian StyleTaylor, Carmel T., Ian M. Mackay, Jamie L. McMahon, Sarah L. Wheatley, Peter R. Moore, Mitchell J. Finger, Glen R. Hewitson, and Frederick A. Moore. 2018. "Detection of Specific ZIKV IgM in Travelers Using a Multiplexed Flavivirus Microsphere Immunoassay" Viruses 10, no. 5: 253. https://doi.org/10.3390/v10050253
APA StyleTaylor, C. T., Mackay, I. M., McMahon, J. L., Wheatley, S. L., Moore, P. R., Finger, M. J., Hewitson, G. R., & Moore, F. A. (2018). Detection of Specific ZIKV IgM in Travelers Using a Multiplexed Flavivirus Microsphere Immunoassay. Viruses, 10(5), 253. https://doi.org/10.3390/v10050253






