LiDAR-Based Regional Inventory of Tall Trees—Wellington, New Zealand
Abstract
1. Introduction
2. Materials and Methods
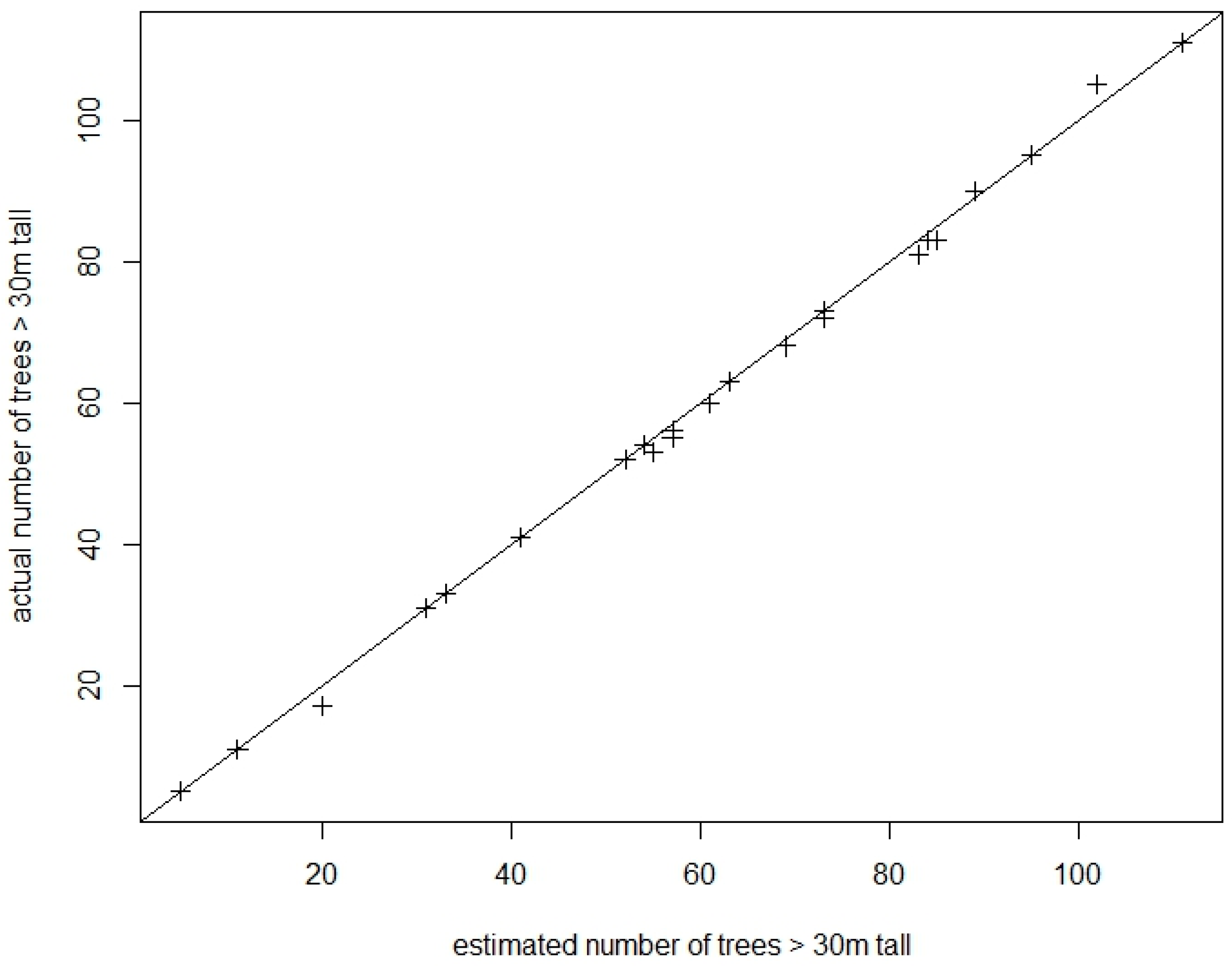
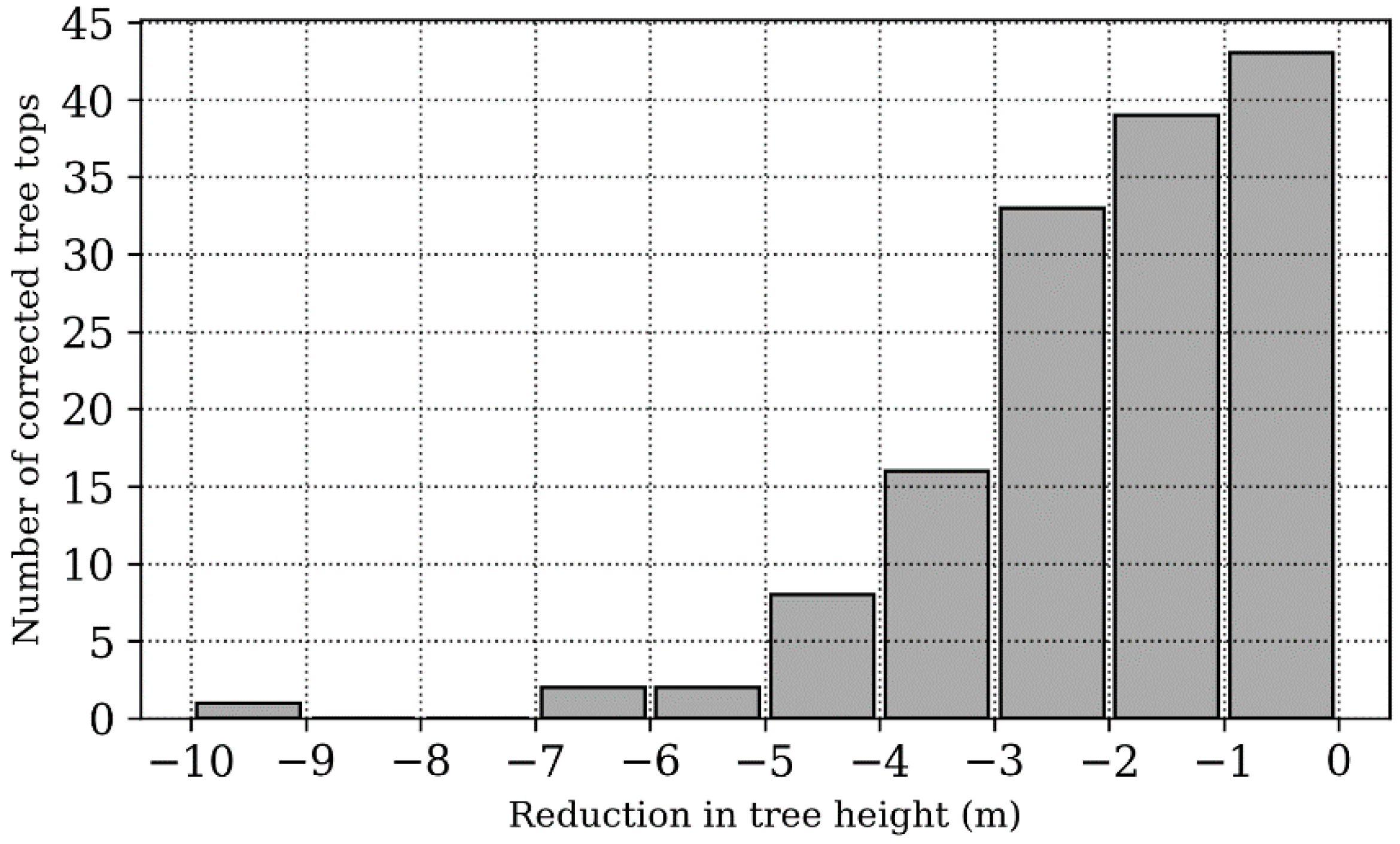
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dymond, J.R.; Shepherd, J.D.; Newsome, P.F.; Belliss, S. Estimating change in areas of indigenous vegetation cover in New Zealand from the New Zealand Land Cover Database (LCDB). N. Z. J. Ecol. 2017, 41. [Google Scholar] [CrossRef]
- Dymond, J.R.; Ausseil, A.-G.E.; Peltzer, D.A.; Herzig, A. Conditions and trends of ecosystem services in New Zealand—A synopsis. Solut. J. 2014, 5, 38–45. [Google Scholar]
- Wardle, P. Vegetation of New Zealand; Cambridge University Press: Cambridge, UK, 1991; ISBN 9780521258739. [Google Scholar]
- Díaz, I.A.; Sieving, K.E.; Peña-Foxon, M.E.; Larraín, J.; Armesto, J.J. Epiphyte diversity and biomass loads of canopy emergent trees in Chilean temperate rain forests: A neglected functional component. For. Ecol. Manag. 2010, 259, 1490–1501. [Google Scholar] [CrossRef]
- Dislich, R.; Mantovani, W. Vascular epiphyte assemblages in a Brazilian Atlantic Forest fragment: Investigating the effect of host tree features. Plant Ecol. 2016, 217. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.; Shi, X.-M.; Liu, W.-Y.; Song, L.; Lu, H.-Z.; Chen, X.; Wu, C.-S. Forest Type and Tree Characteristics Determine the Vertical Distribution of Epiphytic Lichen Biomass in Subtropical Forests. Forests 2017, 8, 436. [Google Scholar] [CrossRef]
- Lesak, A.A.; Radeloff, V.C.; Hawbaker, T.J.; Pidgeon, A.M.; Gobakken, T.; Contrucci, K. Modeling forest songbird species richness using LiDAR-derived measures of forest structure. Remote Sens. Environ. 2011, 115, 2823–2835. [Google Scholar] [CrossRef]
- Flaspohler, D.J.; Giardina, C.P.; Asner, G.P.; Hart, P.; Price, J.; Lyons, C.K.; Castaneda, X. Long-term effects of fragmentation and fragment properties on bird species richness in Hawaiian forests. Biol. Conserv. 2010, 143, 280–288. [Google Scholar] [CrossRef]
- Greene, D.F.; Johnson, E.A. A Model of Wind Dispersal of Winged or Plumed Seeds. Ecology 1989, 70, 339–347. [Google Scholar] [CrossRef]
- Loehle, C. Strategy Space and the Disturbance Spectrum: A Life-History Model for Tree Species Coexistence. Am. Nat. 2000, 156, 14–33. [Google Scholar] [CrossRef] [PubMed]
- McGlone, M.S.; Richardson, S.J.; Jordan, G.J. Comparative biogeography of New Zealand trees: Species richness, height, leaf traits and range sizes. N. Z. J. Ecol. 2010, 34, 137–151. [Google Scholar]
- Coomes, D.A.; Šafka, D.; Shepherd, J.; Dalponte, M.; Holdaway, R. Airborne laser scanning of natural forests in New Zealand reveals the influences of wind on forest carbon. For. Ecosyst. 2018, 5, 10. [Google Scholar] [CrossRef]
- Koukoulas, S.; Blackburn, G.A. Mapping individual tree location, height and species in broadleaved deciduous forest using airborne LIDAR and multi-spectral remotely sensed data. Int. J. Remote Sens. 2005, 26, 431–455. [Google Scholar] [CrossRef]
- Latifi, H.; Nothdurft, A.; Koch, B. Non-parametric prediction and mapping of standing timber volume and biomass in a temperate forest: Application of multiple optical/LiDAR-derived predictors. Forestry 2010, 83, 395–407. [Google Scholar] [CrossRef]
- O’Neil-Dunne, J.; MacFaden, S.; Royar, A. A Versatile, Production-Oriented Approach to High-Resolution Tree-Canopy Mapping in Urban and Suburban Landscapes Using GEOBIA and Data Fusion. Remote Sens. 2014, 6, 12837–12865. [Google Scholar] [CrossRef]
- McPherson, E.G.; Simpson, J.R.; Xiao, Q.; Wu, C. Million trees Los Angeles canopy cover and benefit assessment. Landsc. Urban Plan. 2011, 99, 40–50. [Google Scholar] [CrossRef]
- Crowther, T.W.; Glick, H.B.; Covey, K.R.; Bettigole, C.; Maynard, D.S.; Thomas, S.M.; Smith, J.R.; Hintler, G.; Duguid, M.C.; Amatulli, G.; et al. Mapping tree density at a global scale. Nature 2015, 525, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Lefsky, M.A.; Cohen, W.B.; Acker, S.A.; Parker, G.G.; Spies, T.A.; Harding, D. Lidar Remote Sensing of the Canopy Structure and Biophysical Properties of Douglas-Fir Western Hemlock Forests. Remote Sens. Environ. 1999, 70, 339–361. [Google Scholar] [CrossRef]
- Hyyppä, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.; Zhang, L. Trends in Automatic Individual Tree Crown Detection and Delineation—Evolution of LiDAR Data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Ayrey, E.; Fraver, S.; Kershaw, J.A.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer Stacking: A Novel Algorithm for Individual Forest Tree Segmentation from LiDAR Point Clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Hamraz, H.; Contreras, M.A.; Zhang, J. A robust approach for tree segmentation in deciduous forests using small-footprint airborne LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 532–541. [Google Scholar] [CrossRef]
- Duncanson, L.I.; Cook, B.D.; Hurtt, G.C.; Dubayah, R.O. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Bunting, P.; Lucas, R. The delineation of tree crowns in Australian mixed species forests using hyperspectral Compact Airborne Spectrographic Imager (CASI) data. Remote Sens. Environ. 2006, 101, 230–248. [Google Scholar] [CrossRef]
- Wang, Y.; Hyyppa, J.; Liang, X.; Kaartinen, H.; Yu, X.; Lindberg, E.; Holmgren, J.; Qin, Y.; Mallet, C.; Ferraz, A.; et al. International Benchmarking of the Individual Tree Detection Methods for Modeling 3-D Canopy Structure for Silviculture and Forest Ecology Using Airborne Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2016, 54, 5011–5027. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A New Method for Segmenting Individual Trees from the Lidar Point Cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef]
- Pirotti, F.; Kobal, M.; Roussel, J.R. A comparison of tree segmentation methods using very high density airborne laser scanner data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, 42, 285–290. [Google Scholar] [CrossRef]
- Dalponte, M.; Coomes, D.A. Tree-centric mapping of forest carbon density from airborne laser scanning and hyperspectral data. Methods Ecol. Evol. 2016, 7, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Fonstad, M.A.; Dietrich, J.T.; Courville, B.C.; Jensen, J.L.; Carbonneau, P.E. Topographic structure from motion: A new development in photogrammetric measurement. Earth Surf. Process. Landf. 2013, 38, 421–430. [Google Scholar] [CrossRef]
- Baltsavias, E.; Gruen, A.; Eisenbeiss, H.; Zhang, L.; Waser, L.T. High-quality image matching and automated generation of 3D tree models. Int. J. Remote Sens. 2008, 29, 1243–1259. [Google Scholar] [CrossRef]
- White, J.; Wulder, M.; Vastaranta, M.; Coops, N.; Pitt, D.; Woods, M. The Utility of Image-Based Point Clouds for Forest Inventory: A Comparison with Airborne Laser Scanning. Forests 2013, 4, 518–536. [Google Scholar] [CrossRef]
- St-Onge, B.; Jumelet, J.; Cobello, M.; Véga, C. Measuring individual tree height using a combination of stereophotogrammetry and lidar. Can. J. For. Res. 2004, 34, 2122–2130. [Google Scholar] [CrossRef]
- Dandois, J.P.; Ellis, E.C. Remote Sensing of Vegetation Structure Using Computer Vision. Remote Sens. 2010, 2, 1157–1176. [Google Scholar] [CrossRef]
- Westoby, M.J.; Brasington, J.; Glasser, N.F.; Hambrey, M.J.; Reynolds, J.M. ‘Structure-from-Motion’ photogrammetry: A low-cost, effective tool for geoscience applications. Geomorphology 2012, 179, 300–314. [Google Scholar] [CrossRef]
- Brandtberg, T.; Warner, T.A.; Landenberger, R.E.; McGraw, J.B. Detection and analysis of individual leaf-off tree crowns in small footprint, high sampling density lidar data from the eastern deciduous forest in North America. Remote Sens. Environ. 2003, 85, 290–303. [Google Scholar] [CrossRef]
- Hirata, Y. The effects of footprint size and sampling density in airborne laser scanning to extract individual trees in mountainous terrain. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 36, 102–107. [Google Scholar]
- Hansen, E.; Ene, L.; Gobakken, T.; Ørka, H.; Bollandsås, O.; Næsset, E. Countering Negative Effects of Terrain Slope on Airborne Laser Scanner Data Using Procrustean Transformation and Histogram Matching. Forests 2017, 8, 401. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Wang, T.; Isenburg, M.; Khoshelham, K. Effect of slope on treetop detection using a LiDAR Canopy Height Model. ISPRS J. Photogramm. Remote Sens. 2015, 104, 44–52. [Google Scholar] [CrossRef]
- Duan, Z.; Zhao, D.; Zeng, Y.; Zhao, Y.; Wu, B.; Zhu, J. Assessing and Correcting Topographic Effects on Forest Canopy Height Retrieval Using Airborne LiDAR Data. Sensors 2015, 15, 12133–12155. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.; Korstjens, A.H.; Hill, R.A. Influence of micro-topography and crown characteristics on tree height estimations in tropical forests based on LiDAR canopy height models. Int. J. Appl. Earth Obs. Geoinf. 2018, 65, 105–113. [Google Scholar] [CrossRef]
- Regional High-Resolution DEM Now on LDS, Wellington Regional Government GIS Group (2016). Available online: http://mapping.gw.govt.nz/News06.htm (accessed on 3 October 2018).
- Coomes, D.A.; Dalponte, M.; Jucker, T.; Asner, G.P.; Banin, L.F.; Burslem, D.F.R.P.; Lewis, S.L.; Nilus, R.; Phillips, O.L.; Phua, M.-H.; et al. Area-based vs tree-centric approaches to mapping forest carbon in Southeast Asian forests from airborne laser scanning data. Remote Sens. Environ. 2017, 194, 77–88. [Google Scholar] [CrossRef]
- Kandare, K.; Dalponte, M.; Ørka, H.; Frizzera, L.; Næsset, E. Prediction of Species-Specific Volume Using Different Inventory Approaches by Fusing Airborne Laser Scanning and Hyperspectral Data. Remote Sens. 2017, 9, 400. [Google Scholar] [CrossRef]
- Allen, R.B.; Bellingham, P.J.; Holdaway, R.J.; Wiser, S.K. New Zealand’s indigenous forests and shrublands. In Ecosystem Services in New Zealand—Conditions and Trends; Dymond, J.R., Ed.; Manaaki Whenua Press: Lincoln, New Zealand, 2013; pp. 34–48. ISBN 9780478347364. [Google Scholar]
- Müller, M.U.; Shepherd, J.D.; Dymond, J.R. Support vector machine classification of woody patches in New Zealand from synthetic aperture radar and optical data, with LiDAR training. J. Appl. Remote Sens. 2015, 9, 095984. [Google Scholar] [CrossRef]
- Bunting, P.; Armston, J.; Clewley, D.; Lucas, R. The sorted pulse data software library (SPDLib): Open source tools for processing LiDAR data. In Proceedings of the SilviLaser 2011, 11th International Conference on LiDAR Applications for Assessing Forest Ecosystems, University of Tasmania, Hobart, Australia, 16–20 October 2011; pp. 1–11. [Google Scholar]
- Zhang, K.; Chen, S.; Whitman, D.; Shyu, M.; Yan, J.; Zhang, C. A progressive morphological filter for removing nonground measurements from airborne LiDAR data. IEEE Trans. Geosci. Remote Sens. 2003, 41, 872–882. [Google Scholar] [CrossRef]
- Evans, J.S.; Hudak, A.T. A Multiscale Curvature Algorithm for Classifying Discrete Return LiDAR in Forested Environments. IEEE Trans. Geosci. Remote Sens. 2007, 45, 1029–1038. [Google Scholar] [CrossRef]
- Dalponte, M. itcSegment: Individual Tree Crowns Segmentation. R Package Version 0.8. Available online: https://CRAN.R-project.org/package=itcSegment (accessed on 3 October 2018).
- Zörner, J.; Dymond, J.; Shepherd, J.; Jolly, B. PyCrown-Fast Raster-Based Individual Tree Segmentation for LiDAR Data; Landcare Research Ltd.: Lincoln, New Zealand, 2018. [Google Scholar] [CrossRef]
- Roussel, J.-R.; Auty, D. lidR: Airborne LiDAR Data Manipulation and Visualization for Forestry Applications. R Package Version 1.5.1. Available online: https://CRAN.R-project.org/package=lidR (accessed on 3 October 2018).
- NeSI Pan Cluster, Centre for eResearch. Available online: https://wiki.auckland.ac.nz/display/CER/NeSI+Pan+Cluster (accessed on 3 October 2018).
- Wiser, S.; Thomson, F.; de Cáceres, M. Expanding an existing classification of New Zealand vegetation to include non-forested vegetation. N. Z. J. Ecol. 2016, 40, 160–178. [Google Scholar] [CrossRef]
- Wiser, S.K.; de Cáceres, M. New Zealand’s plot-based classification of vegetation. Phytocoenologia 2018, 48, 153–161. [Google Scholar] [CrossRef]
- Shepherd, J.D.; Ausseil, A.-G.; Dymond, J.R. EcoSat Forests: A 1:750,000 Scale Map of Indigenous Forest Classes in New Zealand; Manaaki Whenua Press: Lincoln, New Zealand, 2005; Available online: https://lris.scinfo.org.nz/ (accessed on 3 October 2018).
- Erdas Imagine, Hexagon Geospatial. Available online: https://www.hexagongeospatial.com (accessed on 3 October 2018).
- Dymond, J.R.; Derose, R.C.; Harmsworth, G.R. Automated mapping of land components from digital elevation data. Earth Surf. Process. Landf. 1995, 20, 131–137. [Google Scholar] [CrossRef]
- Bellingham, P.; Wiser, S.; Coomes, D.; Dunningham, A. Review of Permanent Plots for Long-Term Monitoring of New Zealand’s Indigenous Forests; Science for Conservation; Department of Conservation: Wellington, New Zealand, 2000; ISBN 9780478219586.
- Leathwick, J.; Wilson, G.; Rutledge, D.; Wardle, P.; Morgan, F.; Johnston, K.; McLeod, M.; Kirkpatrick, R. Land Environments of New Zealand; David Bateman Ltd.: Auckland, New Zealand, 2003; ISBN 9781869535223. [Google Scholar]
- Wardle, P. Facets of the distribution of forest vegetation in New Zealand. N. Z. J. Bot. 1964, 2, 352–366. [Google Scholar] [CrossRef]
- Ministry for Culture and Heritage. Te Ara: The Encyclopaedia of New Zealand. Available online: https://teara.govt.nz/en/logging-native-forests/page-1 (accessed on 3 October 2018).
- Dymond, J.R.; Shepherd, J.D.; Newsome, P.F.; Gapare, N.; Burgess, D.W.; Watt, P. Remote sensing of land-use change for Kyoto Protocol reporting: The New Zealand case. Environ. Sci. Policy 2012, 16, 1–8. [Google Scholar] [CrossRef]
- Gatziolis, D.; Fried, J.S.; Monleon, V.S. Challenges to estimating tree height via LiDAR in closed-canopy forests: A parable from Western Oregon. For. Sci. 2010, 56, 139–155. [Google Scholar]
- Vierling, K.T.; Vierling, L.A.; Gould, W.A.; Martinuzzi, S.; Clawges, R.M. Lidar: Shedding new light on habitat characterization and modeling. Front. Ecol. Environ. 2008, 6, 90–98. [Google Scholar] [CrossRef]
- Turner, A.; Colby, J.; Csontos, R.; Batten, M. Flood Modeling Using a Synthesis of Multi-Platform LiDAR Data. Water 2013, 5, 1533–1560. [Google Scholar] [CrossRef]
- New Zealand National Aerial LiDAR Base Specification, Version 1.1. 2018. Available online: https://www.linz.govt.nz/system/files_force/media/doc/loci_nz-national-aerial-lidar-base-specification_20180629.pdf (accessed on 3 October 2018).
- The New Zealand Land Resource Inventory. Available online: https://lris.scinfo.org.nz/layer/48055-nzlri-vegetation/ (accessed on 3 October 2018).
- Asner, G.P.; Martin, R.E.; Keith, L.M.; Heller, W.P.; Hughes, M.A.; Vaughn, N.R.; Hughes, R.F.; Balzotti, C. A Spectral Mapping Signature for the Rapid Ohia Death (ROD) Pathogen in Hawaiian Forests. Remote Sens. 2018, 10, 404. [Google Scholar] [CrossRef]
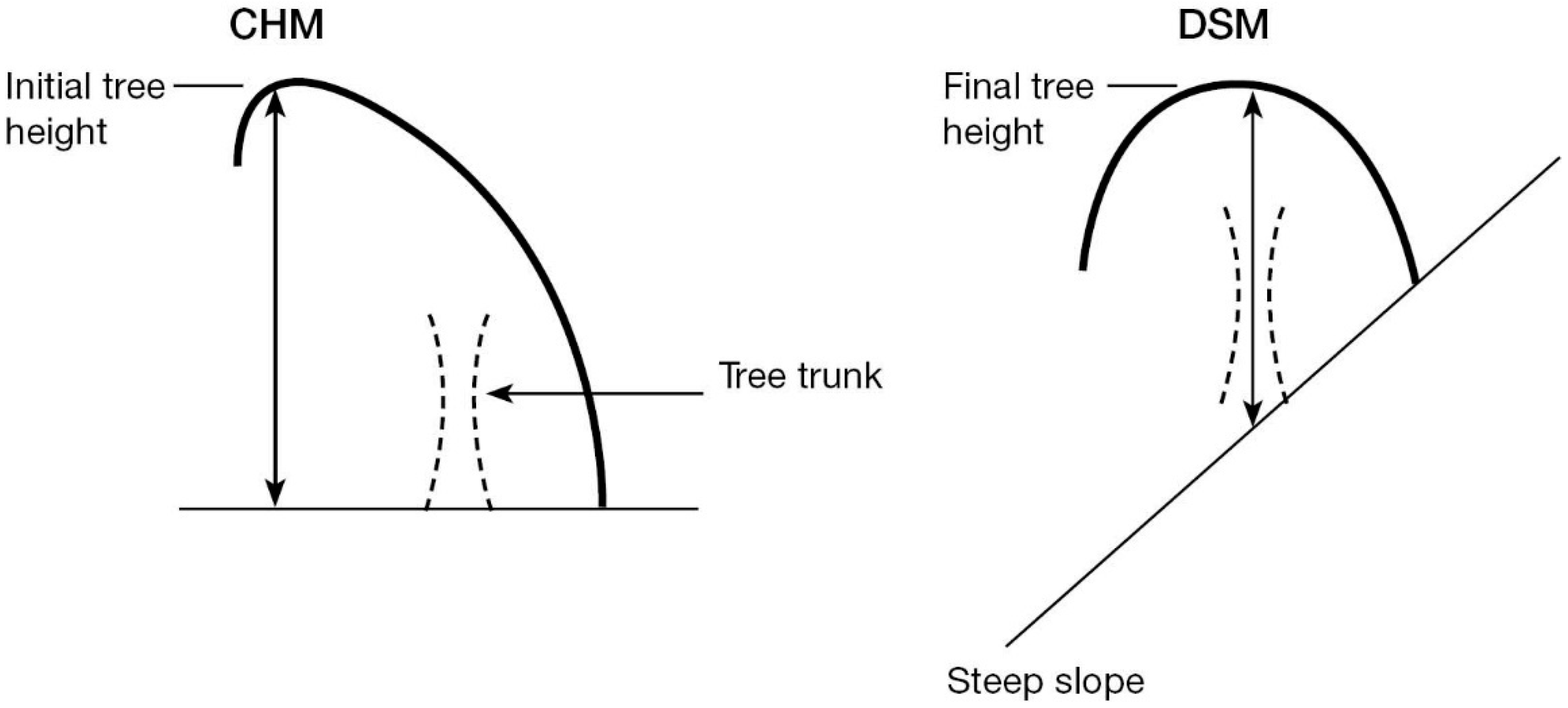
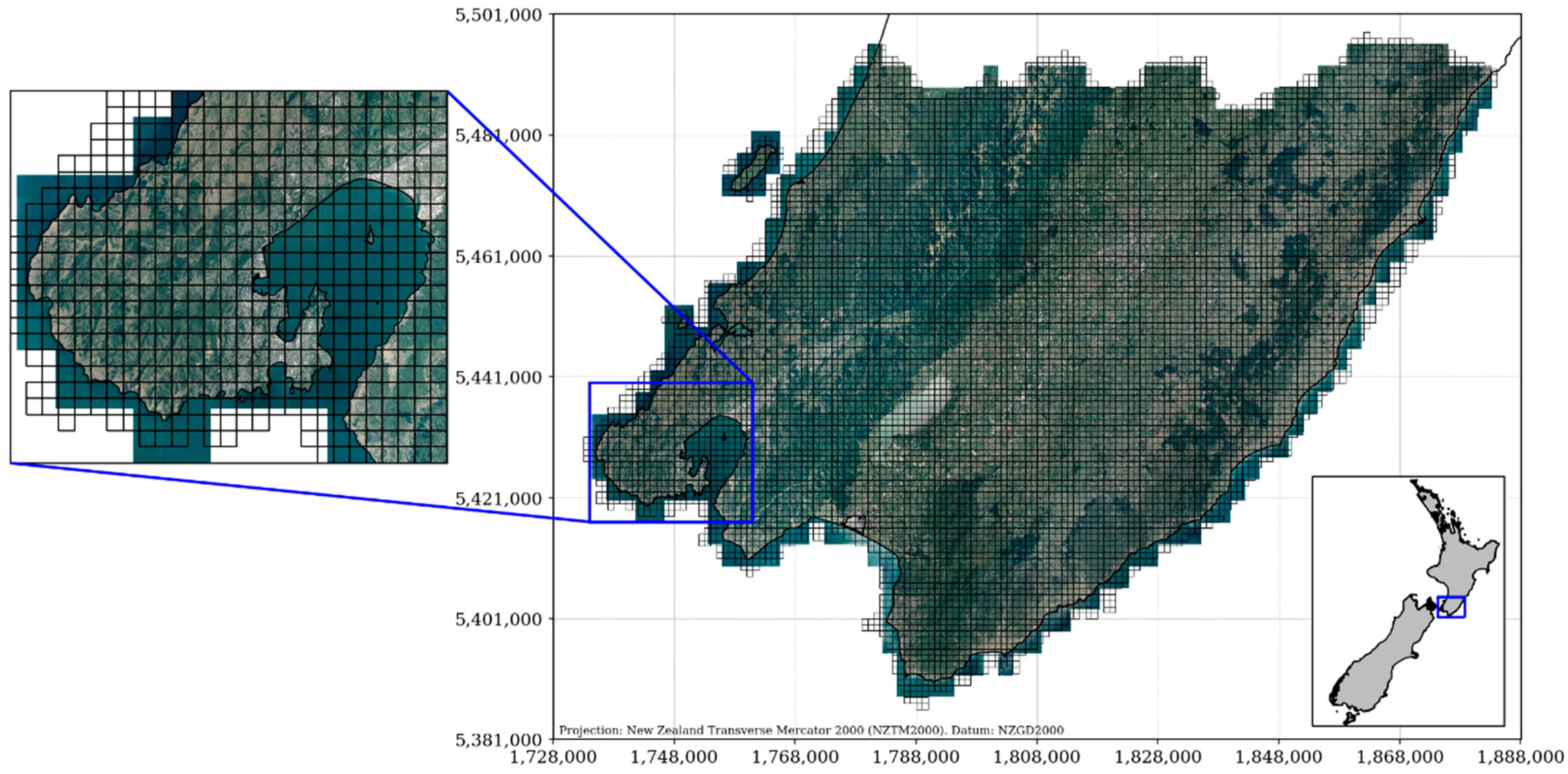
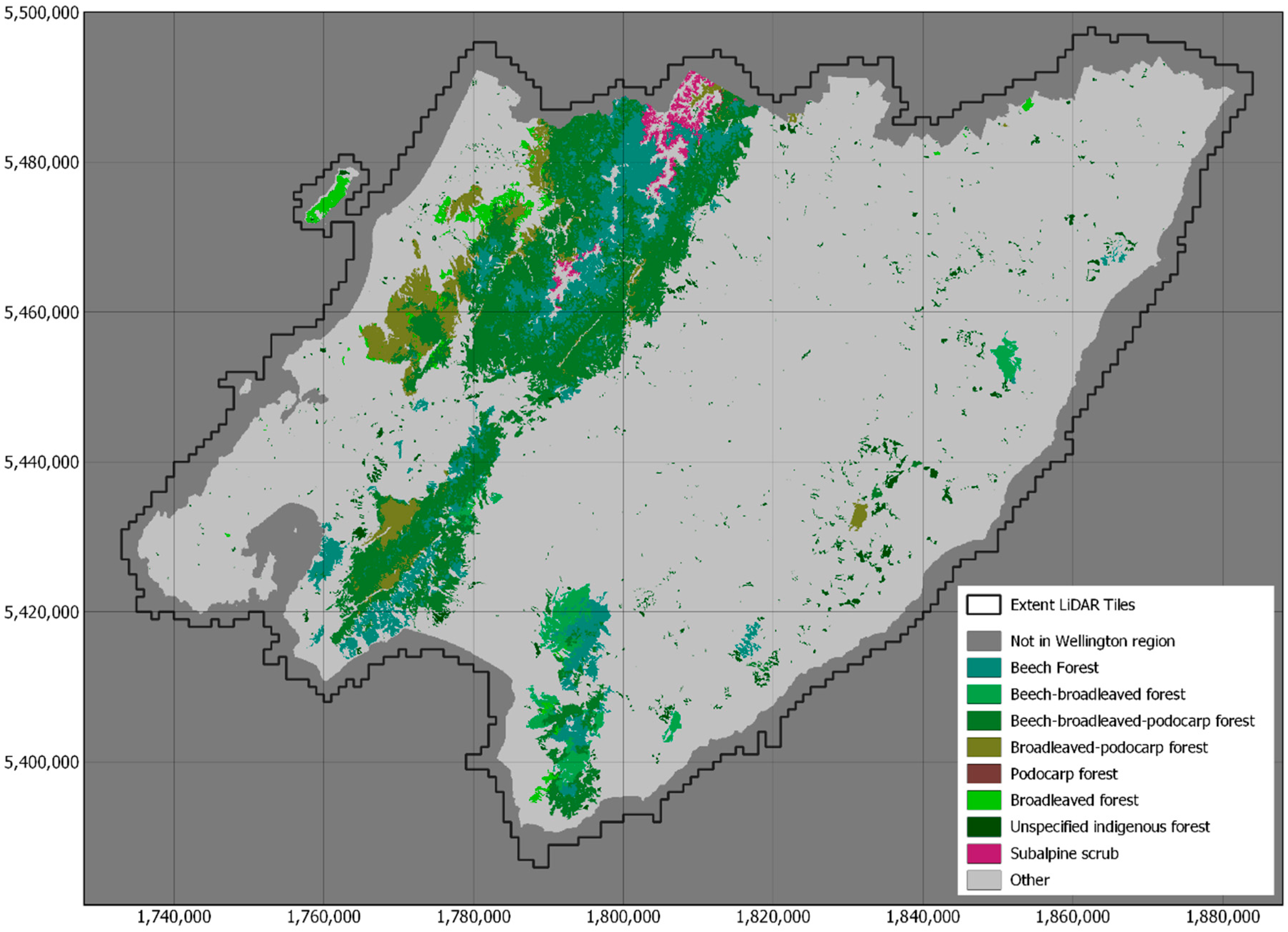
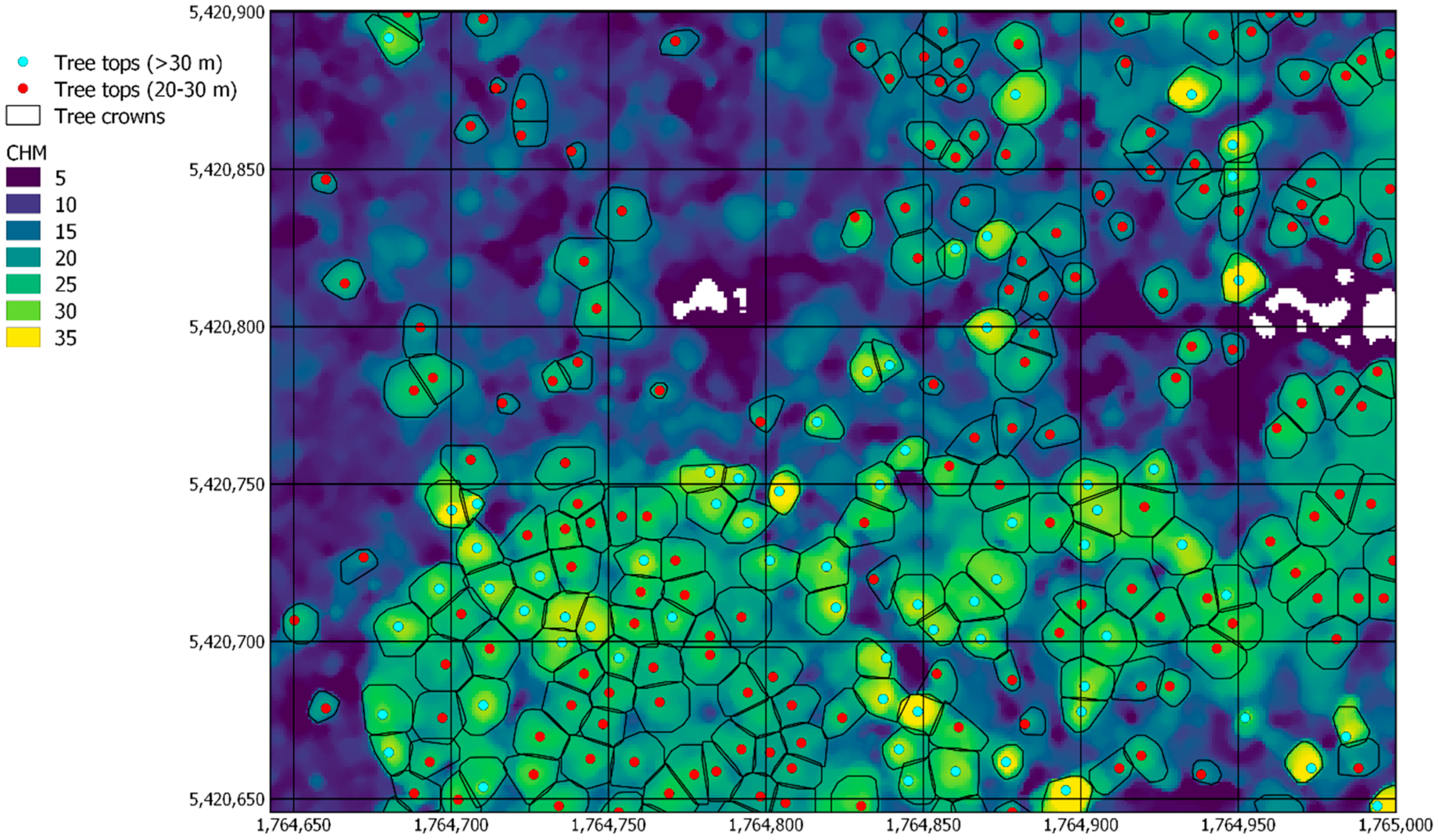
| Forest Alliance Group | EcoSat Forests Classes |
|---|---|
| Beech forest | Beech forest |
| Beech-broadleaved forest | Beech-broadleaved forest |
| Beech-broadleaved podocarp forest | Podocarp-broadleaved/Beech forest Beech/Podocarp-broadleaved forest |
| Broadleaved-podocarp forest | Podocarp-broadleaved forest Kauri forest |
| Podocarp forest | Podocarp forest |
| Broadleaved forest | Broadleaved forest Coastal forest |
| Unspecified indigenous forest | Unspecified indigenous forest |
| Forest Alliance Group | Wellington | North Island | South Island | New Zealand |
|---|---|---|---|---|
| Beech forest | 37,402 | 360,104 | 1,826,940 | 2,187,044 |
| Beech-broadleaved forest | 7946 | 28,703 | 69,606 | 98,309 |
| Beech-broadleaved podocarp forest | 81,155 | 685,952 | 1,148,242 | 1,834,194 |
| Broadleaved-podocarp forest | 17,341 | 891,306 | 447,823 | 1,339,129 |
| Podocarp forest | 71 | 7939 | 57,362 | 65,301 |
| Broadleaved forest | 5172 | 241,297 | 112,423 | 353,720 |
| Unspecified indigenous forest | 9956 | 317,247 | 184,225 | 501,472 |
| Total indigenous forest | 159,043 | 2,532,548 | 3,846,621 | 6,379,169 |
| Total land area | 811,727 | 11,442,900 | 15,286,900 | 26,729,800 |
| Forest Alliance Group | Area (ha) | >30 m | >35 m | >40 m | >45 m | >50 m |
|---|---|---|---|---|---|---|
| Beech forest | 37,402 | 47,646 | 4911 | 410 | 44 | 0 |
| Beech-broadleaved forest | 7946 | 4911 | 598 | 108 | 9 | 0 |
| Beech-broadleaved podocarp forest | 81,155 | 176,518 | 28,903 | 3202 | 293 | 0 |
| Broadleaved-podocarp forest | 17,341 | 33,493 | 11,268 | 2103 | 226 | 0 |
| Podocarp forest | 71 | 129 | 4 | 0 | 0 | 0 |
| Broadleaved forest | 5172 | 3689 | 1357 | 302 | 43 | 0 |
| Unspecified indigenous forest | 9956 | 19,202 | 5944 | 1210 | 170 | 0 |
| Subalpine shrubland | 3841 | 453 | 44 | 5 | 0 | 0 |
| Total | 162,884 | 286,041 | 53,029 | 7340 | 785 | 0 |
| Forest Alliance Group | >30 m | >35 m | >40 m | >45 m |
|---|---|---|---|---|
| Beech forest | 1.27 | 0.13 | 0.01 | 0.001 |
| Beech-broadleaved forest | 0.62 | 0.08 | 0.01 | 0.001 |
| Beech-broadleaved podocarp forest | 2.18 | 0.36 | 0.04 | 0.004 |
| Broadleaved-podocarp forest | 1.93 | 0.65 | 0.12 | 0.013 |
| Podocarp forest | 1.82 | 0.06 | 0.00 | 0.000 |
| Broadleaved forest | 0.71 | 0.26 | 0.06 | 0.008 |
| Unspecified indigenous forest | 1.93 | 0.60 | 0.12 | 0.017 |
| Subalpine shrubland | 0.12 | 0.01 | 0.00 | 0.000 |
| Forest Alliance Group | 30–35 m | 35–40 m | 40–45 m | 45–50 m |
|---|---|---|---|---|
| Beech forest | 638 | 565 | 478 | 332 |
| Beech-broadleaved forest | 385 | 338 | 224 | 181 |
| Beech-broadleaved podocarp forest | 470 | 418 | 373 | 332 |
| Broadleaved-podocarp forest | 332 | 307 | 291 | 286 |
| Podocarp forest | 461 | 464 | ||
| Broadleaved forest | 278 | 245 | 218 | 217 |
| Unspecified indigenous forest | 191 | 191 | 172 | 154 |
| Subalpine shrubland | 731 | 742 |
| CHM Median Filter | ||||
|---|---|---|---|---|
| ws = 3 | ws = 5 | ws = 7 | ||
| Local Maximum Filter | ws = 3 | 1081 | 768 | 546 |
| ws = 5 | 882 | 711 | 511 | |
| ws = 7 | 777 | 646 | 492 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zörner, J.; Dymond, J.R.; Shepherd, J.D.; Wiser, S.K.; Jolly, B. LiDAR-Based Regional Inventory of Tall Trees—Wellington, New Zealand. Forests 2018, 9, 702. https://doi.org/10.3390/f9110702
Zörner J, Dymond JR, Shepherd JD, Wiser SK, Jolly B. LiDAR-Based Regional Inventory of Tall Trees—Wellington, New Zealand. Forests. 2018; 9(11):702. https://doi.org/10.3390/f9110702
Chicago/Turabian StyleZörner, Jan, John R. Dymond, James D. Shepherd, Susan K. Wiser, and Ben Jolly. 2018. "LiDAR-Based Regional Inventory of Tall Trees—Wellington, New Zealand" Forests 9, no. 11: 702. https://doi.org/10.3390/f9110702
APA StyleZörner, J., Dymond, J. R., Shepherd, J. D., Wiser, S. K., & Jolly, B. (2018). LiDAR-Based Regional Inventory of Tall Trees—Wellington, New Zealand. Forests, 9(11), 702. https://doi.org/10.3390/f9110702






