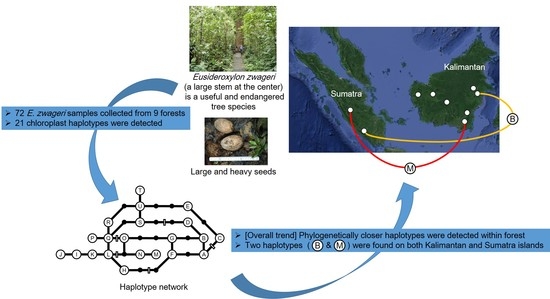
Genetic Structure of the Tropical Tree Eusideroxylon zwageri in Indonesia Revealed by Chloroplast DNA Phylogeography
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Chloroplast DNA Sequencing
2.3. Data Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Partomihardjo, T. The ulin wood which is threatened to extinction. Duta Rimba 1987, 13, 3–15. [Google Scholar]
- Ohtani, M.; Kondo, T.; Tani, N.; Ueno, S.; Lee, L.S.; Ng, K.K.; Muhammad, N.; Finkeldey, R.; Na’iem, M.; Indrioko, S.; et al. Nuclear and chloroplast DNA phylogeography reveals pleistocene divergence and subsequent secondary contact of two genetic lineages of the tropical rainforest tree Species Shorea leprosula (Dipterocarpaceae) in South-East Asia. Mol. Ecol. 2013, 22, 2264–2279. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Kremer, A.; Wagner, D.B. Geographic structure of chloroplast DNA polymorphisms in European oaks. Theor. Appl. Genet. 1993, 87, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Hampe, A. Some evolutionary consequences of being a tree. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 187–214. [Google Scholar] [CrossRef]
- Bossdorf, O.; Auge, H.; Lafuma, L.; Rogers, W.E.; Siemann, E.; Prati, D. Phenotypic and genetic Differentiation between native and introduced plant populations. Oecologia 2005, 144, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, D.M.; Ramakrishnan, A.P.; Cruzan, M.B. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol. Ecol. 2008, 17, 4657–4669. [Google Scholar] [CrossRef] [PubMed]
- Pairon, M.; Petitpierre, B.; Campbell, M.; Guisan, A.; Broennimann, O.; Baret, P.V.; Jacquemart, A.L.; Besnard, G. Multiple introductions boosted genetic diversity in the invasive range of black cherry (Prunus serotina; Rosaceae). Ann. Bot. 2010, 105, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Kurokochi, H.; Saito, Y.; Chuman, M.; Ide, Y. Low Chloroplast Diversity despite Phylogenetically Divergent Haplotypes in Japanese Populations of Ailanthus altissima (Simaroubaceae). Botany 2012, 91, 148–154. [Google Scholar] [CrossRef]
- Kurokochi, H.; Nurtjahjaningsih, I.L.G.; Tan, E.; Asakawa, S.; Saito, Y.; Ide, Y. Development of polymorphic chloroplast DNA markers for the endangered tree Eusideroxylon zwageri through chloroplast isolation and next-generation sequencing. Conserv. Genet. Resour. 2015, 7, 845–850. [Google Scholar] [CrossRef]
- Sugiura, N.; Tang, D.; Kurokochi, H.; Saito, Y.; Ide, Y. Genetic structure of Quercus gilva Blume in Japan as revealed by chloroplast DNA sequences. Botany 2015, 93, 873–880. [Google Scholar] [CrossRef]
- Hall, T. BioEdit Sequence Alignment Editor; Version 7.0.1.; Ibis Therapeutics: Carlsbad, CA, USA, 2004. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistics analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA-sequence data. 3. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Tnah, L.H.; Lee, S.L.; Ng, K.K.; Tani, N.; Bhassu, S.; Othman, R.Y. Geographical traceability of an important tropical timber (Neobalanocarpus heimii) inferred from chloroplast DNA. For. Ecol. Manag. 2009, 258, 1918–1923. [Google Scholar] [CrossRef]
- Kamiya, K.; Nanami, S.; Kenzo, T.; Yoneda, R.; Diway, B.; Chong, L.; Azani, M.A.; Majid, N.M.; Lum, S.K.; Wong, K.M.; Harada, K. Demographic history of Shorea curtisii (Dipterocarpaceae) inferred from chloroplast DNA sequence variations. Biotropica 2012, 44, 577–585. [Google Scholar] [CrossRef]
- Iwasaki, T.; Aoki, K.; Seo, A.; Murakami, N. Comparative phylogeography of four component species of deciduous broad-leaved forests in Japan based on chloroplast DNA variation. J. Plant Res. 2012, 125, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Gaskin, J.F.; Schaal, B.A. Hybrid tamarix widespread in US invasion and undetected in native Asian range. Proc. Natl. Acad. Sci. USA 2002, 99, 11256–11259. [Google Scholar] [CrossRef] [PubMed]
- Kolbe, J.J.; Glor, R.E.; Schettino, L.R.; Lara, A.C.; Larson, A.; Losos, J.B. Genetic variation increases during biological invasion by a Cuban lizard. Nature 2004, 431, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Besnard, G.; Henry, P.; Wille, L.; Cooke, D.; Chapuis, E. On the origin of the invasive olives (Olea europaea L., Oleaceae). Heredity 2007, 99, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Henry, P.; Le Lay, G.; Goudet, J.; Guisan, A.; Jahodová, Š.; Besnard, G. Reduced genetic diversity, increased isolation and multiple introductions of invasive giant hogweed in the western Swiss Alps. Mol. Ecol. 2009, 18, 2819–2831. [Google Scholar] [CrossRef] [PubMed]
- Fineschi, S.; Taurchini, D.; Villani, F.I.; Vendramin, G.G. Chloroplast DNA polymorphism reveals little geographical structure in Castanea sativa Mill. (Fagaceae) throughout southern European countries. Mol. Ecol. 2000, 9, 1495–1503. [Google Scholar] [CrossRef] [PubMed]
- Irawan, B. Soil Properties and the abundance of ironwood (Eusideroxylon zwageri Teijsm. & Binn.) varieties in Jambi, Indonesia. J. Manaj. Hutan Trop. 2015, 21, 155–161. [Google Scholar]

| Region | Population | Longitude | Latitude | Number of Detected Haplotypes | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B * | C | D | E | F | G | H | I | J | K | L * | M * | N | O * | P | Q * | R * | S | T | U | ||||
| Kalimantan | Kutai | 117°35′29″ E | 0°33′53″ N | 1 | 6 | 1 | ||||||||||||||||||
| Kalimantan | Berau | 117°11′52″ E | 0°35′03″ N | 1 | 1 | 1 | 1 | 3 | 1 | |||||||||||||||
| Kalimantan | Bukit Soeharto | 116°59′50″ E | 0°35′41″ S | 2 | 3 | 2 | 1 | 2 | 1 | |||||||||||||||
| Kalimantan | Kintap | 115°09′05″ E | 3°42′17″ S | 7 | 1 | |||||||||||||||||||
| Kalimantan | Busang | 113°55′43″ E | 0°14′23″ S | 1 | 1 | 6 | ||||||||||||||||||
| Kalimantan | Kapuas Hulu | 111°59′23″ E | 1°03′35″ N | 6 | 2 | |||||||||||||||||||
| Kalimantan | Gunung Palung | 109°59′11″ E | 0°56′42″ N | 1 | 1 | 3 | ||||||||||||||||||
| Sumatra | Musi Rawas | 103°17′00′′ E | 2°59′39″ S | 2 | 1 | 5 | ||||||||||||||||||
| Sumatra | Jambi | 102°53′17″ E | 1°47′01″ S | 8 | ||||||||||||||||||||
| a | Ez_Cp_01 | Ez_Cp_05 | Ez_Cp_07 | Ez_Cp_09 | Ez_Cp_11 | Ez_Cp_12 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| b | 530 | 648 | 490 | 467 | 505 | 497 | |||||||
| c | 63 | 264 | 56 | 164 | 166 | 560 | 468 | 171–175 | 45 | 137 | 28 | 431 | |
| Haplotype | d | SNP | SNP | SNP | SNP | SNP | SNP | SNP | SI | SNP | SNP | SNP | SNP |
| A | C | A | A | A | A | G | C | CTACT | T | A | G | A | |
| B | C | A | A | A | A | G | A | CTACT | T | A | G | A | |
| C | C | A | A | A | A | G | C | AGTAG | T | A | G | A | |
| D | C | A | A | A | A | G | A | CTACT | T | G | G | A | |
| E | C | A | A | A | A | G | C | AGTAG | G | G | G | A | |
| F | T | C | A | A | A | G | C | CTACT | T | A | G | A | |
| G | T | C | A | A | A | G | A | CTACT | T | A | G | A | |
| H | T | C | A | G | A | G | C | AGTAG | T | A | T | A | |
| I | T | C | A | G | C | A | C | AGTAG | T | G | T | A | |
| J | T | C | A | G | C | A | A | AGTAG | T | G | T | A | |
| K | T | C | A | G | C | G | C | AGTAG | T | G | T | A | |
| L | T | C | A | G | A | G | C | AGTAG | T | G | T | A | |
| M | T | C | G | G | A | G | C | CTACT | T | G | T | C | |
| N | T | C | A | G | A | G | C | CTACT | T | G | T | C | |
| O | T | C | A | G | A | G | A | CTACT | T | G | T | A | |
| P | T | C | G | G | A | G | A | AGTAG | T | G | T | A | |
| Q | T | C | A | G | A | G | A | AGTAG | T | G | T | A | |
| R | T | C | A | G | A | G | A | AGTAG | G | G | T | A | |
| S | C | A | A | G | A | G | A | AGTAG | T | G | T | A | |
| T | C | A | A | G | A | G | A | AGTAG | G | G | T | C | |
| U | C | A | A | G | A | G | A | AGTAG | G | G | T | A | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nurtjahjaningsih, I.L.G.; Sukartiningsih; Kurokochi, H.; Saito, Y.; Ide, Y. Genetic Structure of the Tropical Tree Eusideroxylon zwageri in Indonesia Revealed by Chloroplast DNA Phylogeography. Forests 2017, 8, 229. https://doi.org/10.3390/f8070229
Nurtjahjaningsih ILG, Sukartiningsih, Kurokochi H, Saito Y, Ide Y. Genetic Structure of the Tropical Tree Eusideroxylon zwageri in Indonesia Revealed by Chloroplast DNA Phylogeography. Forests. 2017; 8(7):229. https://doi.org/10.3390/f8070229
Chicago/Turabian StyleNurtjahjaningsih, I.L.G., Sukartiningsih, Hiroyuki Kurokochi, Yoko Saito, and Yuji Ide. 2017. "Genetic Structure of the Tropical Tree Eusideroxylon zwageri in Indonesia Revealed by Chloroplast DNA Phylogeography" Forests 8, no. 7: 229. https://doi.org/10.3390/f8070229
APA StyleNurtjahjaningsih, I. L. G., Sukartiningsih, Kurokochi, H., Saito, Y., & Ide, Y. (2017). Genetic Structure of the Tropical Tree Eusideroxylon zwageri in Indonesia Revealed by Chloroplast DNA Phylogeography. Forests, 8(7), 229. https://doi.org/10.3390/f8070229