Abstract
The Białowieża Primeval Forest (BPF) is Europe’s last primeval forest and an irreplaceable area for biodiversity conservation due to its size, protection status, and substantially undisturbed nature. There is no other forest in Europe with such a large surface representing highly-advanced natural succession. This article reports on the first analysis of the genetic variability and demographic structure of a self-renewed Pinus sylvestris population located in BPF, using both chloroplast and mitochondrial DNA markers. The analysis of molecular variance (AMOVA) for chloroplast simple sequence repeats (cpSSRs) revealed a significant genetic differentiation among age classes that accounted for about 2% of the total variance, comparable to those reported among different populations of Scots pine. None of the 117 detected chloroplast haplotypes were common to all age classes. Haplotype diversity ranged from 0.370 to 0.415 for cpSSRs and from 0.320 to 0.455 for mitochondrial markers. The genetic variation of the studied age classes—represented by mitochondrial markers—strongly depicts the maternal genetic structure, indicating limited seed dispersal. Temporal genetic substructuring is maintained within a self-renewed population of Scots pine from the BPF.
1. Introduction
The Białowieża Primeval Forest (BPF; 1.470 km2, 52°30/−53° N, 23°30/−24°15 E) is located on the border between Poland and Belarus, and is Europe’s last remaining lowland broad-leaved and mixed forest that has not been exploited by forestry or influenced by other human activities for nearly one hundred years [1]. The forest was protected by successive Polish monarchs and Russian Tsars as a hunting reserve. The strictly protected part of the forest on the Polish side—the Białowieża National Park (BNP), established in 1921—is the oldest park in Poland and among the oldest parks throughout Europe, and comprises exceptional stands of primeval forest [2]. The BNP was added to the World Heritage List in 1979. It is estimated that over 4500 plant species, about 3000 fungal species, and nearly 10,000 animal species have been preserved in the Białowieża Primeval Forest [2]. There is no other forest in Europe with such a large surface representing well-advanced natural succession [3]. The BPF is considered as a “flagship ecosystem in European nature conservation” [3]. The BFP is an invaluable reference area for scientists studying the natural characteristics of European forests.
Nowadays, most central and northern European forests are intensely managed, and almost no natural primary forests remain in Europe [4]. It is difficult to study the dynamics of natural temperate zone forests in Europe due to the necessity of operating over a suitable timescale, and also because the suitable stands are so few in number and of too small a size [5]. The genetic diversity of tree species is a key component for the functioning of a forest ecosystem [6]. Regeneration is the basic process that maintains the dynamics of a forest ecosystem, and as such, is a key aspect of any sustainable forest management system [7]. A self-renewed natural population involves individuals that often belong to genetically different age classes along with a differentiated sensitivity of these classes for selection, which may cause fluctuations in genetic variation [8].
Selection occurring upon germination and the following stages may act differently than during the adult stage. For example, early-stage shade tolerance for seedlings may be favored in dense populations, whereas light tolerance may prove to be important at later stages for the same tree [9]. At the population level, selection for light will favor fast-growing and vigorous seedlings in dense stands. The age-dependent climate sensitivity of French oaks [10], Picea glauca [11], and Pinus sylvestris [12] was reported to possibly have been caused by differences in the physiological processes of differently-aged trees. Chen et al. [13] observed temporal trends in the growth and net biomass change in the boreal forests of Canada. The relationships between the genetic diversity and demographic structure of a population is of fundamental importance for understanding the adaptive strategies of forest trees [14]. Studies of genetic variation in the naturally regenerated population of P. sylvestris from the BPF (performed using nuclear microsatellites and isoenzyme markers) revealed a high level of intra-population diversity between generations. The significant subdivision of genetic variation (FST) detected across the age classes was comparable to those found between different populations of this species [15].
The Scots pine (P. sylvestris) dominates the boreal forests of Northern and Eastern Europe, but is also an important species in Europe’s managed temperate forests. In the Białowieża Primeval Forest, it occupies more than 27% of the total area [16].
We combine genetic data (chloroplast and mitochondrial DNA markers) to analyze the consequences of demographic events on genetic diversity and structuring in a self-renewed population of Scots pine. The combination of data from multiple polymorphic sites in the non-recombining, uniparentally-inherited haploid chloroplast and mitochondrial genome offers a robust and highly-informative assay for the analysis of diversity in natural populations of plant species.
2. Materials and Methods
2.1. Study Site and Sampling
We collected samples for genetic analysis in the naturally regenerated tree stands of P. sylvestris in the “Sitki” reserve of the Białowieża Primeval Forest (compartment 667B and 668A of the Hajnówka Forest District). The “Sitki” reserve (34.09 ha) was created in 1979 to preserve forms of oligotrophic pine forests in their natural state, which do not occur in any of the existing reserves of the BPF. The removal of trees that had toppled over and had been infested by bark beetles was the only recently applied treatment in this reserve [17]. The age of each individual tree was estimated by measuring its diameter at breast height and by counting its growth rings (using cores). The distance between sampled trees was between 10 m and 100 m. Fresh needles were collected from 125 individuals, representing four age classes of Scots pines (Table 1). Additionally, cones were collected from 40 adult fruiting trees (age class M and O)—one cone per tree. Seeds from these cones were mixed together in equal proportions, as a pooled sample, which was then germinated in laboratory conditions to form the youngest age class E, comprising 31 individuals (Table 1).

Table 1.
Number of alleles and haplotype genetic diversity (H) of the analyzed cp and mt loci in age classes of Pinus sylvestris population.
2.2. DNA Extraction and Amplification
DNA was extracted from needles and two-week-old seedlings (100 mg of fresh material) by means of the modified cetyltrimethylammonium bromide (CTAB) protocol [18]. The quality and quantity of the extracted the DNA was measured using a Nanodrop™ ND-1000 spectrophotometer (ThermoScientific, Waltham, MA, USA).
The ten variable chloroplast microsatellite loci provided a high-quality amplification product and were used for the analysis of all samples. These loci consisted of: PCP102652, PCP1289, PCP41131, PCP87314 [19], Pt15169, Pt26081, Pt30204, Pt36480, Pt45002, and Pt71936 [20]. PCR amplification was carried out in a total volume of 25 µL, containing about 20 ng of template DNA, 2.5 mM MgCl2, 100 µM of each dNTP, 0.2 µM of each primer, and 1U HiFiTaq Polymerase (Novazym, Poznań, Poland), with the respective 1 × PCR buffer in accordance with Celiński et al. [21]. The PCR conditions were as follows: initial denaturation for 3 min at 94 °C, followed by 35 cycles of 15 s denaturation at 95 °C, 1 min annealing at 59 °C, 1 min incubation at 72 °C, and final extension at 72 °C for 10 min. The amplification products were separated using the 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) capillary electrophoresis system with GeneScan™ 600 LIZ™ as an internal size standard. The reverse primer of each primer pair was labeled with fluorescent 6FAM, PET, and VIC dyes. Individuals were analyzed and genotyped using GeneMapper version 3.7 software (Applied Biosystems). In order to amplify a known polymorphic indel 1 in nad1 intron (exon B/C), the diagnostic primers nad1 H and nad1 I were used according to Soranzo et al. [22]. For the nad7 intron 1, sequences from the GeneBank [23] were used to design primers flanking the deletion of 5 bp and 32 bp (DQ665913–DQ665915). Thus, the PCR products would result in the following fragments: 300 bp (a mitotype), 295 (b mitotype), and 268 (c mitotype). The total volume of PCR reaction for nad1 and nad7 was 25 µL, containing 20 ng of template DNA, 2.5 mM MgCl2, 100 µM of each dNTP, 0.2 µM of each primer, and 1U HiFiTaq Polymerase (Novazym, Poznań, Poland), with the respective 1 × PCR buffer. The indel of 31-bp in nad1 intron (exon B/C) was scored by an electrophoresis of the PCR product on 1.5% agarose gel containing ethidium bromide. The amplification protocol for nad1 was 4 min at 94 °C, followed by 35 cycles of 40 s at 94 °C, annealing for 1 min at 53 °C, 1 min at 72 °C, and finally 10 min at 72 °C. The PCR conditions for nad7 consisted of 2 min at 95 °C, followed by 25 cycles of 30 s at 94 °C, annealing for 45 s at 60 °C, 1 min at 72 °C, and finally 10 min at 72 °C.
The PCR products were separated by capillary electrophoresis in a Genetic Analyser (Applied Biosystems). Genotyping of individuals was made with GeneMapper version 3.7 software (Applied Biosystems).
2.3. Data Analysis
The level of polymorphism of both the entire population and within each age class was estimated using the percentage of polymorphic loci (P%), allele frequencies at each locus, the number of alleles per locus (A), effective number of alleles (Ne), number of haplotypes (H), and haplotype diversity [24] using GenAlex 6.5 [25]. Fisher’s exact test [26] calculated by GenePop v.4.0 [27,28] was used to estimate the pairwise p values for allele distribution between generations.
The level of genetic differentiation between generations was estimated using PhiPT-statistics for haploid data (analogous to F-statistics for diploid data) calculated via the analysis of molecular variance (AMOVA) option following Merimans [29]. The genetic distance between pairs of subpopulations was computed according to Nei [30]. Genetic relationships between populations were evaluated via principal coordinate analysis (PCoA) based on the genetic distance between each age classes.
3. Results
3.1. Chloroplast simple sequence repeats (cpSSRs)
The cpSSR loci were highly polymorphic among the Scots pine age classes: from 80% (class E) to 100% (class O). Loci Pt15169 and Pt 30204 (eight alleles) appeared to be the most polymorphic, whereas loci Pt36480 and PCP102652 (three alleles) appeared to be the least polymorphic (Table S1). A total of 48 alleles from the ten chloroplast microsatellite loci were found, ranging from 31 (class E) to 36 (classes S, Y, M) (Table 1).
The lowest mean number of alleles per locus (Na), 3.1, was noted in the class of Embryos, while the highest, 3.6, was observed in the classes of Seedlings, Young trees, and Middle-age trees (Table 2). The mean effective number of alleles per locus (Ne) was lower, and ranged from 2.0 to 2.2. A total of 12 private alleles were found: one in the Oldest trees, five in the Middle-age trees, and two alleles in the other classes. A considerable variance in the allele frequencies between particular age classes was observed, especially in respect to loci Pt15169, Pt30204, and Pt45002. Fisher’s exact probability test showed that allele frequencies in locus Pt30204 of the Oldest trees class were significantly different from classes E, Y, and M (p values were 0.013, 0.035, and 0.011, respectively).

Table 2.
Genetic characteristics of the analyzed Scots pine age classes.
A joint analysis of the alleles of the ten chloroplast loci identified 117 different haplotypes among the 156 individuals, from 23 (class M) to 32 (class O). The majority of these haplotypes (77%) were detected only once (unique haplotypes), and the rest were observed in two-to-six individuals. None of the detected haplotypes were common amongst all of the age classes of the studied population. The majority of specimens in an age class demonstrated a private haplotype: from 13 (M class) to 22 (E class) private haplotypes, with a mean of 79% (Table 2).
Haplotype diversity differed considerably between the chloroplast microsatellite loci, from 0.025 in PCP102652 to 0.734 in Pt15169, with the mean over the entire population amounting to 0.393 (Table 1). The lowest values were observed in the Embryos and the Oldest trees classes (0.381 and 0.370, respectively), while the highest value was observed amongst the Young trees (0.415) (Figure 1).
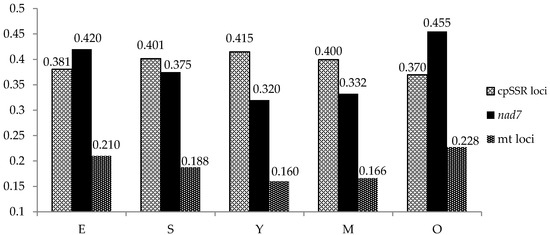
Figure 1.
Haplotype diversity among age classes for chloroplast and mitochondrial markers. E—embryos germinated from seeds; S—seedlings 1–3 years old; Y—young trees 10–20 years old; M—middle-aged trees 40–80 years old; O—old trees above 100 years.
An analysis of molecular variance (AMOVA) using chloroplast SSRs revealed a significant (probability 0.027) variation among the age classes, accounting for about 2% of the total variance (Table 3). The highest differentiation occurred in loci Pt30204 (6.8%, probability 0.009) and Pt45002 (4.4%, probability 0.035) (Table 4). The pairwise PhiPT value varied among the age classes, from 0 to 0.084 (Table 5).

Table 3.
Analysis of molecular variance (AMOVA) in Scots pine age classes for cpSSRs markers.

Table 4.
Genetic differentiation (AMOVA PhiPT) across five age classes of Scots pine.

Table 5.
Pairwise age class PhiPT values (below diagonal) and Nei’s genetic distance (above diagonal) for cpSSRs markers.
The lowest Nei’s [30] genetic distance was observed between the class of Embryos and the Young trees (0.011), and the greatest distance was observed between the Oldest trees and the Young trees (0.029) (Table 5).
PCoA analysis based on the genetic distance between each age class revealed the genetic distinctiveness of the Oldest tree group (Figure 2). The first two factors of the PCoA analysis explained more than 84% of the variation found in the genetic distance matrix.

Figure 2.
Principal coordinates analysis (PCoA) based on the genetic distances among the age classes of Scots pine.
3.2. Mitochondrial Markers
All analyzed individuals had a single a mitotype (217 bp) for the locus nad1. At the locus nad7, two mitotypes were observed in all age classes: universal a of 300 bp (65–80%; overall 74%) and northern b of 295 bp (20–36%; overall 26%) (Table 1 and Table 2). Haplotype diversity H (calculated for polymorphic nad7 locus) ranged from 0.320 (class Y) to 0.455 (class O) and 0.420 (class E), indicating that the greatest values occurred in the oldest and the youngest age classes. The distribution of H values in different age classes showed the opposite tendency compared to the chloroplast loci (Figure 1). An AMOVA analysis conducted on the allele frequencies did not indicate any significant differentiation between the age classes (Table 4), and the entire genetic variation was found within the age classes. Nei’s genetic distances among particular generations were very low: from 0 to 0.013.
4. Discussion
This article reports on the first analysis of the temporal genetic diversity across five age classes of a naturally renewing P. sylvestris population located in the BPF using both chloroplast and mitochondrial DNA markers. The ten chloroplast polymorphic microsatellite loci indicated a high genetic variation within the particular generations in terms of the number of alleles and haplotypes (31–36 and 23–32, respectively). Provan et al. [19] observed lower values for the number of haplotypes (from 10 to 18) by analyzing the loci of 17 cpSSRs in populations of the naturally-occurring range of Scots pine in Europe. A very high number of haplotypes (36–43) was found across three Estonian populations of this species [31]. In total, 152 haplotypes were identified of four cpSSR loci in 38 populations of P. sylvestris in Asia and Eastern Europe [32]. The alleles of six cpSSR loci were combined into 32 different haplotypes, all found in the set of 235 individuals from the 12 different populations of P. pinaster [33]. Parducci et al. [34] identified from 11 to 22 different haplotypes among the 169 individuals of four Abies species. The nine cpSSR loci were examined in P. resinosa, giving a total of 23 chloroplast haplotypes among the 159 individuals [35]. Schmidt et al. [36] observed 43 haplotypes in A. sibirica, 49 in A. sachalinensis, and 31 in A. nephrolepis in two chloroplast microsatellite loci. We detected a high percentage of unique haplotypes in every age class (from 56% to 76%), whereby 59% of the analyzed individuals could be individually genotyped. A similar percentage of unique chloroplast haplotypes (55%) was found in 13 Scots pine populations from the Iberian Peninsula [37]. A study by Naydenov et al. [38] revealed as much as 90% of unique haplotypes in a study of 12 populations of P. sylvestris in Bulgaria.
An analysis of molecular variance (AMOVA) using chloroplast SSRs revealed that the significant variation among age classes of P. sylvestris population from the BPF accounted for about 2% of the total variance. The PhiPT values for pairwise age classes were much higher, showing a significant genetic differentiation (0–6.2%). These values were comparable to those found between the different natural populations of this species through an analysis of chloroplast SSRs: 2.1% between populations from Asia and Eastern Europe [39], and 1.5% between three bog populations from Estonia [31]. Previous studies on P. sylvestris [40,41,42] have shown that inter-population variation accounted for between 1.7% and 7.5% of the total observed variation. Such pronounced genetic differentiation within a single population may be a consequence of a temporal variation in population reproductive episodes, such as the different participation of trees in pollen generation and pollination (wind and flowering period) or pollen inflow from the outside. Paternally-inherited cpDNA is dispersed by the movement of both seeds and pollen. In landscapes dominated by forests, the pollen source area may have a radius of 50–100 m [43]. In mixed woodlands, such as in the BPF, the presence of different tree species in the canopy may restrict pollen movement and cause partial reproductive isolation [44], and thus a higher genetic differentiation between age classes. These factors result in differences of allele frequencies among generations of seeds produced. The distribution of diversity at nuclear SSRs and allozyme loci appeared to follow a similar pattern, as revealed by our previous study on the same Scots pine population [15].
A significant subdivision of genetic variation at the level of 5% was particularly evident in two loci: Pt 30204 and Pt 45002. The same loci revealed significant differences (Fisher’s exact probability test) in allele frequencies for age class pairs. Locus Pt30204 is the intergenic space between the ATP-dependent protease proteolytic subunit (clpP) gene and ribosomal protein S 12 (rpsl2) gene [45]. SSR markers are assumed to be neutral, but there is some evidence that certain microsatellite loci show functional significance and may be closely linked to the target of natural selection or may even be directly under selection themselves [46,47]. Recent studies indicated that polymorphism in organelle genomes may be important for adaptation processes in plants [48]. Rodriguez et al. [49] investigated the associations between cpSSR groups and phenotypes in Phaseolus coccineus. The existence of a cytonuclear disequilibrium between allozymes and cpDNA in a natural population of P. ponderosa was shown by Latta et al. [50].
The mitochondrial genome of the Pinaceae family does not recombinate and is passed through the maternal line over generations. In conifers, mtDNA shows a low level of variation through its exons and introns [51,52].
Among the age classes of the studied population from the BPF, only mitotype a (217 bp) of the nad1 intron B/C was present and shared in common. It predominates in the Scots pine populations of Central and Northern Europe [22,23]. Locus nad7 was represented by two mitotypes occurring in all of the analyzed generations, with mitotype a and northern mitotype b being more frequent. Both frequencies of alleles as well as haplotype diversity values did not differ between generations, which confirmed the AMOVA analysis. In conifers, variation in the mitochondrial genome represents the gene flow mediated by seeds, while variation in the chloroplast genome describes gene flow attributable to both pollen and seeds [53]. Because of the ease of movement, pollen dispersion is considered to be generally more important in the gene flow of forest trees than the dispersion of seeds, especially for wind-pollinated species [54,55]. Forest gaps and the surrounding crown canopies may locally influence the microenvironmental factors, thus affecting the seeds and pollen migration. As a result, genetic variation is limited by the low distance of seed dispersion, and the population structure of mtDNA variability within species remains conservative over the long term [56]. We can conclude that the genetic variation of the studied age classes—represented by mitochondrial markers—strongly depicts the maternal genetic structure of the overlying canopy, indicating limited seed dispersal. The same pattern of genetic variation was found in other conifer species; e.g., P. ponderosa [57] and Picea abies [58]. The results indicate that temporal genetic substructuring is more or less maintained within this self-renewed population of Scots pine from the Białowieża Primeval Forest.
5. Conclusions
The populations of long-lived forest tree species have a structure with overlapping generations, whose dynamic changes inevitably involve genetic differentiation at the spatial as well as temporal scales. The genetic study of successive generations allows for the inference of long-term demographic processes, and is a powerful tool for the analysis of processes that underlie the observed genetic structure [59]. The cpSSRs indicated that a natural population of Scots pine from the BPF appears to maintain a high level of genetic diversity between generations, while the mitochondrial genome markers represent a stable level of variation. The genetic structure of the studied self-renewed P. sylvestris population is naturally dynamic, calling for research into its fluctuations over time to better understand how they contribute to the adaptive potential of the population. Our study provides information on the genetic dynamics of a natural population of a keystone long-lived tree species, and is significant for the fields of conservation and adaptation. Finally, our analysis highlights the need to incorporate age profiles in analyses of the genetic variation of forest tree populations.
Supplementary Materials
The following is available online at www.mdpi.com/1999-4907/8/7/227/s1, Table S1: Allele size of 10 chloroplast loci and 2 mitochondrial loci in the studied individuals from five age classes of Pinus sylvestris.
Acknowledgments
This study was financed by the Ministry of Science and Higher Education in Poland (grant number 3P04F 007 25) and by the National Science Centre in Poland (grant number NN304169740). The authors are grateful to Adolf Korczyk, Witold Wachowiak, and Łukasz Myczko for their support in plant material collection in the Białowieża Primeval Forest.
Author Contributions
A.W.-P. conceived and designed the experiments, analyzed the data and wrote the manuscript; K.C. performed the experiments and analyzed the data; E.C. analyzed the data and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Baker, A.G.; Zimny, M.; Keczyński, A.; Bhagwat, S.A.; Willis, K.J.; Latałowa, M. Pollen productivity estimates from old-growth forest strongly differ from those obtained in cultural landscapes: Evidence from the Białowieża National Park, Poland. Holocene 2016, 26, 80–92. [Google Scholar] [CrossRef]
- Faliński, J.B. Vegetation Dynamics in Temperate Lowland Primeval Forests: Ecological Studies in Białowieża Forest; Junk Publishers: Dordrecht, The Netherlands, 1986. [Google Scholar]
- Bobiec, A. Białowieża Primeval Forest.The largest area of natural deciduous lowland forest in Europe. Int. J. Wilderness 2002, 8, 33–37. [Google Scholar]
- Lorenz, M.; Fischer, R.; Mues, V. Forest Resources in Europe and Their Condition; Arbora Publishers: Zvolen, Slovakia, 2005. [Google Scholar]
- Koop, H. Forest Dynamics. SILVI-STAR: A Comprehensive Monitoring System; Springer: Berlin, Germany, 1989. [Google Scholar]
- Ratnam, W.; Rajora, O.P.; Finkeldey, R.; Aravanopoulos, F.; Bouvet, J.M.; Vaillancourtf, R.E.; Kanashiro, M.; Fady, B.; Tomita, M.; Vinson, C. Genetic effects of forest management practices: Global synthesis and perspectives. For. Ecol. Manag. 2014, 333, 52–65. [Google Scholar] [CrossRef]
- Ackzell, L. Comparison of planting, sowing and natural regeneration for Pinus sylvestris (L.) in Boreal Sweden. For. Ecol. Manag. 1993, 61, 229–245. [Google Scholar] [CrossRef]
- Chomicz, E. Factor shaping genetic diversity of forest trees. Kosmos 2013, 62, 597–605, (In Polish with English summary). [Google Scholar]
- Poorter, L.; Bongers, F.; Sterck, F.J.; Woll, H. Beyond the regeneration phase: Differentiation of height-light trajectories among tropical tree species. J. Ecol. 2005, 93, 256–267. [Google Scholar] [CrossRef]
- Grey, B.M. Comment on transfer functions. In Climate from Tree Ring; Huges, M.K., Kelly, P.M., Pilcher, J.M., La Marche, V.C., Eds.; Cambridge University Press: Cambridge, UK, 1982; pp. 56–58. [Google Scholar]
- Szeicz, J.M.; MacDonald, G.M. Age-dependent tree ring growth on subarctic white spruce to climate. Can. J. For. Res. 1994, 24, 120–132. [Google Scholar] [CrossRef]
- Linderholm, H.W.; Linderholm, K. Age-dependent climate sensitivity of Pinus sylvestris L. in the central Scandinavian Mountains. Boreal Environ. Res. 2004, 9, 307–317. [Google Scholar]
- Chen, H.Y.H.; Luo, Y.; Reich, P.B.; Searle, E.B.; Biswas, S.R. Climate change-associated trends in net biomass change are age dependent in western boreal forests of Canada. Ecol. Lett. 2016, 19, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Korczyk, A. Protection and conservation of gene resources of forest trees in the Białowieża Forest. In Protection of Forest Ecosystems Biodiversity of Białowieża Primeval Forest; Paschalis, P., Rykowski, K., Zajączkowski, S., Eds.; Pol Forest Biodiversity Protection Project; Warsaw, Poland, 1995; pp. 95–102. [Google Scholar]
- Wojnicka-Półtorak, A.; Celiński, K.; Chudzińska, E. Genetic differentiation between generations of Pinus sylvestris natural population: A case study from the last European primeval forest. Austrian J. For. Sci. 2017, in press. [Google Scholar]
- Sokołowski, A. Lasy Puszczy Białowieskiej; Centrum Informacyjne Lasów Państwowych: Warszawa, Poland, 2004. (In Polish) [Google Scholar]
- Korczyk, A.F. Identification of Genetic Diversity. Part I. Demographic Structure of Natural Populations of Pinus sylvestris L. and Picea abies Karst in the Białowieża Forest; Report GEF 05/21655 Pol part 3.04.; Global Environment Facility: Warsaw, Poland, 1994; (In Polish with English abstract). [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 1, 13–15. [Google Scholar]
- Provan, J.; Soranzo, N.; Wilson, N.J.; McNicol, J.W.; Forrest, G.I.; Cottrell, J.; Powell, W. Gene-pool variation in Caledonian and European Scots pine (Pinus sylvestris L.) revealed by chloroplast simple-sequence repeats. Proc. R. Soc. B Biol. Sci. 1998, 265, 1697–1705. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, G.G.; Lelli, L.; Rossi, P.; Morgante, M. A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Mol. Ecol. 1996, 5, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Celiński, K.; Pawlaczyk, E.M.; Wojnicka-Półtorak, A.; Chudzińska, E.; Prus-Głowacki, W. Cross-species amplification and characterization of microsatellite loci in Pinus mugo Turra. Biologia 2013, 68, 621–626. [Google Scholar] [CrossRef]
- Soranzo, N.; Alia, R.; Provan, J.; Powell, W. Patterns of variation at a mitochondrial sequence-tagged-site locus provides new insights into the postglacial history of European Pinus sylvestris populations. Mol. Ecol. 2000, 9, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Naydenov, K.; Senneville, S.; Beaulieu, J.; Tremblay, F.; Bosquet, J. Glacial vicariance in Eurasia: Mitochondrial DNA evidence from Scots pine for a complex heritage involving genetically distinct refugia at mid-northern latitudes and in Asia Minor. BMC Evol. Biol. 2007, 7, 233. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-un update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. An exact test for population differentiation. Evolution 1995, 49, 1280–1283. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. GenePop (version 1.2): A population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Res. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 2006, 60, 2399–2402. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Pazouki, L.; Shanjani, P.S.; Fields, P.D.; Martins, K.; Suhhorutsenko, M.; Viinalass, H.; Niinemets, U. Large within-population genetic diversity of the widespread conifer Pinus sylvestris at its soil fertility limit characterized by nuclear and chloroplast microsatellite markers. Eur. J. For. Res. 2016, 135, 161–177. [Google Scholar] [CrossRef]
- Semerikov, V.L.; Semerikova, S.A.; Dymshakova, O.S.; Zatsepina, K.G.; Tarakanov, V.V.; Tikhonova, I.V.; Ekart, A.K.; Vidyakin, A.I.; Jamiyansuren, S.; Rogovtsev, R.V.; et al. Microsatellite loci polymorphism of chloroplast DNA of the pine tree (Pinus sylvestris L.) in Asia and Eastern Europe. Genetika 2014, 50, 660–669. [Google Scholar] [PubMed]
- Ribeiro, M.M.; Plomion, P.; Petit, R.; Vendramin, G.G.; Szmidt, A.E. Variation in chloroplast single-sequence repeats in Portuguese maritime pine (Pinus pinaster Ait.). Theor. Appl. Genet. 2001, 102, 97–103. [Google Scholar] [CrossRef]
- Parducci, R.; Szmidt, A.E.; Madaghiele, M.; Anzidei, M.; Vendramin, G.G. Genetic variation at chloroplast microsatellites (cpSSRs) in Abies nebrodensis (Lojac.) Mattei and three neighboring Abies species. Theor. Appl. Genet. 2001, 102, 733–740. [Google Scholar] [CrossRef]
- Echt, C.S.; DeVerno, L.L.; Anzidei, M.; Vendramin, G.G. Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Mol. Ecol. 1998, 7, 307–316. [Google Scholar] [CrossRef]
- Schmidt, A.; Semerikova, S.A.; Semerikov, V.L. The diversity of chloroplast microsatellite loci in Siberian fir (Abies sibirica Ledeb.) and two Far East fir species A. nephrolepis (Trautv.) Maxim and A. sachalinensis Fr. Schmidt. Russ. J. Genet. 2007, 43, 1373–1381. [Google Scholar]
- Robledo-Arnuncio, J.J.; Alia, R.; Gil, L. High levels of genetic diversity in a long-term European glacial refugium of Pinus sylvestris L. For. Genet. 2004, 11, 239–248. [Google Scholar]
- Naydenov, K.D.; Tremblay, F.M.; Alexandrov, A.; Fenton, N.J. Structure of Pinus sylvestris L. populations in Bulgaria revealed by chloroplast microsatellites and terpenes analysis: Provenance tests. Biochem. Syst. Ecol. 2005, 33, 1226–1245. [Google Scholar] [CrossRef]
- Semerikov, V.L.; Semerikova, S.A.; Dymshakova, O.S.; Zatsepina, K.G.; Tarakanov, V.V.; Tikhonova, I.V.; Ekart, A.K.; Vidyakin, A.I.; Jamiyansuren, S.; Rogovtsev, R.V.; et al. Microsatellite loci polymorphism of chloroplast DNA of Scots pine (Pinus sylvestris L.) in Asia and Eastern Europe. Russ. J. Genet. 2014, 50, 577–585. [Google Scholar] [CrossRef]
- Wang, X.-R.; Szmidt, A.E.; Lindgren, D.A.G. Allozyme differentiation among populations of Pinus sylvestris (L.) from Sweden and China. Hereditas 1991, 114, 219–226. [Google Scholar] [CrossRef]
- Goncharenko, G.G.; Silin, A.E.; Padutov, V.E. Allozyme variation in natural populations of Eurasian pines.3. Population structure, diversity, differentiation and gene flow in central and isolated populations of Pinus sylvestris L. in Eastern-Europe and Siberia. Silvae Genet. 1994, 43, 119–132. [Google Scholar]
- Shigapov, Z.K.H.; Bakhtiyarova, R.M.; Yanbaev, Y.A. Genetic variation and differentiation in natural populations of the Scots Pine, Pinus silvestris L. Russ. J. Genet. 1995, 31, 1180–1186. [Google Scholar]
- Sugita, S. Pollen representation of vegetation in Quaternary sediments: Theory and method in patchy vegetation. J. Ecol. 1994, 82, 881–897. [Google Scholar] [CrossRef]
- Bacilieri, R.; Labbe, T.; Kremer, R. Intraspecific genetic structure in a mixed population of Quercus petraea (Matt.) Liebl and Q. robur L. Heredity 1994, 73, 130–141. [Google Scholar] [CrossRef]
- Joung, Y.H.I.; Roh, M.S. Mapping characterization of Pinus sylvestris var. silvestriformis based on chloroplast DNA microsatellite markers. For. Genet. 2005, 12, 89–97. [Google Scholar]
- Acheré, V.; Favre, J.M.; Besnard, S.; Jeandroz, S. Genomic organization of molecular differentiation in Norway spruce (Picea abies). Mol. Ecol. 2005, 14, 3191–3201. [Google Scholar] [CrossRef] [PubMed]
- Lind-Riehl, J.F.; Sullivan, A.R.; Gailing, O. Evidence for selection on a CONSTANS-like gene between two red oak species. Ann. Bot. 2014, 113, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Bock, D.G.; Andrew, R.L.; Seberg, L.H. On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 2014, 23, 4899–4911. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Rau, D.; Angioi, S.A.; Bellucci, E.; Bitocchi, E.; Nanni, L.; Knüpffer, H.; Negri, V.; Papa, R.; Attene, G. European Phaseolus coccineus L. landraces: Population structure and adaptation, as revealed by cpSSRs and phenotypic analyses. PLoS ONE 2013, 8, e57337. [Google Scholar] [CrossRef] [PubMed]
- Latta, R.G.; Linhart, Y.B.; Mitton, J.B. Cytonuclear disequilibrium and genetic drift in a natural population of Ponderosa pine. Genetics 2001, 158, 843–850. [Google Scholar] [PubMed]
- Laroche, J.; Li, P.; Maggia, L.; Bousquet, J. Molecular evolution of angiosperm mitochondrial introns and exons. Proc. Natl. Acad. Sci. USA 1997, 94, 5722–5727. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo-Correa, J.P.; Bousquet, J.; Beaulieu, J.; Isabel, N.; Perron, M.; Bouille, M. Cross-species amplification of mitochondrial DNA sequence-tagged-site markers in conifers: The nature of polymorphism and variation within and among species in Picea. Theor. Appl. Genet. 2003, 106, 1353–1367. [Google Scholar] [CrossRef] [PubMed]
- Burban, C.; Petit, R.J. Phylogeography of maritime pine inferred with organelle markers having contrasted inheritance. Mol. Ecol. 2003, 12, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Burczyk, J.; DiFazio, S.P.; Adams, W.T. Gene flow in forest trees: How far do genes really travel? For. Genet. 2004, 1, 179–192. [Google Scholar]
- Vidyakin, A.I.; Semerikov, V.L.; Polezhaeva, M.A.; Dymshakova, O.S. Spread of mitochondrial DNA haplotypes in population of Scots pine (Pinus sylvestris L.) in northern European Russia. Russ. J. Genet. 2012, 48, 1267–1271. [Google Scholar] [CrossRef]
- Latta, R.G.; Linhart, Y.B.; Fleck, D.; Elliot, M. Direct and indirect estimates of seed versus pollen movement within a population of Ponderosa pine. Evolution 1998, 52, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Wojnicka-Półtorak, A.; Celiński, K.; Chudzińska, E. Temporal dynamics in the genetic structure of a natural population of Picea abies. Biologia 2016, 71, 875–884. [Google Scholar] [CrossRef]
- Chung, M.Y.; Epperson, B.K.; Chung, M.G. Genetic structure of age classes in Camellia japonica (Theaceae). Evolution 2003, 57, 62–73. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).