Abstract
Quercus variabilis is a tree species of ecological and economic value that is widely distributed in China. To effectively evaluate, use, and conserve resources, we applied 25 pairs of simple sequence repeat (SSR) primers to study its genetic diversity and genetic structure in 19 natural forest or natural secondary forest populations of Q. variabilis (a total of 879 samples). A total of 277 alleles were detected. Overall, the average expected heterozygosity (He) was 0.707 and average allelic richness (AR) was 7.79. Q. variabilis manifested a loss of heterozygosity, and the mean of inbreeding coefficient (FIS) was 0.044. Less differentiation among populations was observed, and the genetic differentiation coefficient (FST) was 0.063. Bayesian clustering analysis indicated that the 19 studied populations could be divided into three groups based on their genetic makeup, namely, the Southwest group, Central group, and Northeastern group. The Central group, compared to the populations of the Southwest and Northeast group, showed higher genetic diversities and lower genetic differentiations. As a widely distributed species, the historical migration of Q. variabilis contributed to its genetic differentiation.
1. Introduction
Fagaceae is one of the most important components of northern sub-tropical forests and temperate forests. Quercus spp. consists of about 450 different species [1] and is the largest Fagaceae genus. Moreover, these have important ecological and economic values. Also known as Chinese cork oak, Quercus variabilis Bl. is a species of oak in the section Quercus Sect. Cerris [2] and native to a wide area of China [3], as well as the Korean peninsula and the southwestern Japanese archipelago. In China, the Qinling and Dabie mountain land areas are considered the geographical distribution centers of Q. variabilis [4]. Having one of the widest amplitudes of distribution and most complex climate types in worldwide geographic distribution [5], Q. variabilis forests have formed many stable forest ecosystems in warm temperate and northern subtropical regions in China and are playing a significant role in the conservation and improvement of water and soil [6]. It is a precious timber and economic tree species, and also considered as an important resource for natural raw materials in cork production due to its most unique feature, i.e., its bark [7,8]. In recent years, with the development of cork, tannin, and other industries in China, the phenomenon of over harvesting of Q. variabilis has become a prominent agricultural problem that has resulted in extensive destruction and waste of natural resources [9]. Currently, the main distribution areas of Q. variabilis in China are mostly natural secondary forests [4]. The original forest has become extremely rare; scattered forests are found in Guizhou, Yunnan, and Fujian (on-site investigations). The destruction of the natural populations has been strongly associated with climate change, soil erosion, and a series of ecological problems [10], hence a shortage in natural resources for Q. variabilis has caused a sharp decline in raw material production.
Genetic diversity in forests is determined by gene flow, genetic drift, selection, mutation, and other factors [11,12,13], and the levels of genetic variation and structures are the combined effects of evolutionary history, distribution area, life-styles, and breeding forms [14]. For a tree with a relatively long life cycle, genetic diversity determines its ability to adapt to changing environments, which in turn serves as the basis for maintaining long-term stability of forest ecosystems. The evaluation of genetic variability, especially for forests with wide distribution ranges and high ecological and economic values, is of great significance for forest ecosystem conservation and management of genetic resources [15]. Therefore, investigating the evolutionary history and distribution, even including the development of protection strategies and measures based on forest diversity levels and genetic structures, is imperative.
Studies on Q. variabilis have mainly concentrated on population structure and classification, biodiversity characteristics, population dynamics, and ecological function [16,17,18]. In addition, various features of reproduction, seedling cultivation, and planting techniques have also been studied [19,20,21]. Various reports on the population genetics of Quercus such as Q. acutissima [22], Q. rubra and Q. ellipsoidalis [23], Q. mongolica [24], and Q. infectoria [25] have been published in recent years. There are relatively few genetic investigations on Q. variabilis, but Xu [6] was the first to conduct a preliminary study on its genetic diversity of Chinese five natural populations by using simple sequence repeat (SSR) markers. Recent studies have focused on Q. variabilis pedigree geography such as Chen [26], who used chloroplast simple sequence repeat (cpSSR) to relatively and comprehensively analyze the historical evolution and its geographical distribution. However, this study was limited by the number of samples, and may therefore not fully represent the genetic variation of existing populations. Therefore, based on the natural resource distribution of all existing populations, the establishment of a suitable means of detection, a sufficient number of markers, and more comprehensive sample size is necessary to study the genetic structure and mating system.
China is considered as the global distribution center for Q. variabilis distribution. Furthermore, research studies on its genetic structure based on a broader sampling strategy involving its main distribution area, are of high significance to better understand the distribution pattern and formation of existing populations. Furthermore, such studies may provide important molecular bases for the protection and exploitation of the genetic resources.
2. Materials and Methods
2.1. Population Sample Information
The tender leaves of mature trees were collected from 19 different sub-locations in nine provinces (Figure 1), which is representative of the entire distribution area covered from the Henan township Neixiang, Henan (33°50′ N 111°18′ E) to Leye, Guangxi (24°47′ N 106°57′ E). Annual temperature varies from 8.3 °C to 18.4 °C, spanning two climatic zones: subtropical monsoon and Temperate monsoon. More details on the number of samples and geographical features of each group are listed in Table 1. The studied forests are typical natural secondary forests, and the plants of each population were separated by at least 50 m. The collected samples were brought back to the lab, dried by silica, and stored at room temperature until analysis.
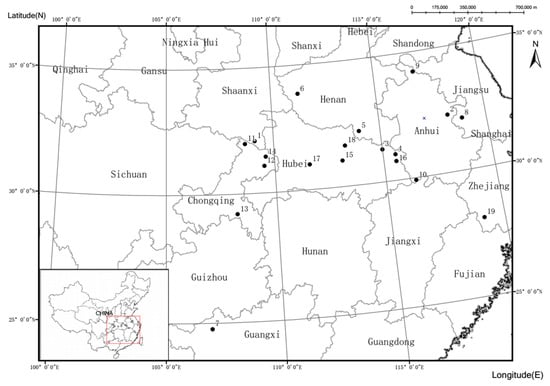
Figure 1.
Locations of the 19 investigated populations of Q. variabilis. Numbers correspond to the population numbers in Table 1.

Table 1.
Geographic locations and mean values of climate parameter in 19 populations of Q. variabilis.
2.2. Experimental Methods
Total DNA was extracted from the samples by using a modified cetyl trimethylammonium bromide (CTAB) lysis-silica beads adsorption method [27]. The SSR primers used in population identification are shown in Table 2. The total volum e of each single locus polymerase chain reaction (PCR) was 20 μL, which consisted of 20 ng of DNA, 50 mM KCl, 20 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 0.2 mM dNTP, 0.5 μM of each primer (forward and reverse), and 1 U of Taq enzyme (Qiagen Inc., Hilden, Germany) PCR thermal cycling was performed on a GeneAmp PCR System 9700 (Applied Biosystems Inc., Carlsbad, CA, USA) using the following conditions: an initial denaturation step of 94 °C for 4 min; followed by 30 cycles of denaturation at 94 °C for l min, annealing at N °C for 2 min (N is the specific annealing temperature for each pair of primers), and extension at 72 °C for 1 min; and a final extension at 72 °C for 8 min and final storage at 4 °C. The PCR products were detected by using capillary electrophoresis (Bioptic100-automated nucleic acid analyzer, BiOptic Inc., Tucson, AZ, USA). Electrophoresis results were interpreted using Q-editor software [28].

Table 2.
Source, repeat motif, and polymorphism information for the 25 microsatellite loci analyzed in the 879 Q. variabilis trees.
2.3. Data Analysis
Genetic parameters, calculated by the Fstat software, were: average number of alleles: Na; observed heterozygosity: Ho; expected heterozygosity or gene diversity, He [32]; allelic richness, AR [33]; the fixation index (FIS: the inbreeding coefficient) of every point and group level; the differentiation index between pairwise populations (FST, GST, RST) and the matrix of FST between various groups on which F-statistics were based [34,35]; Hardy-Weinberg equilibrium testing and estimation of allele frequency were calculated [36]. The polymorphic information index (PIC) and Linkage disequilibrium were calculated by using the PowerMarker v3.23 software (Bioinformatics Research Center; North Carolina State University, Raleigh, NC, USA) [37]. P values were adjusted by using a Bonferroni correction. Private alleles were calculated by using the GenAlEx software [38,39]. Gene flow (Nm) was calculated using the formula: Nm = (1 − FST)/4FST [40]. To investigate genetic diversity and geography-space-climate relationships [41], the average AR value was calculated using a total of 25 loci, whereas the relationships between He value and each geographic population (latitude and longitude), annual average temperature, and annual precipitation in climate parameters (obtained through DIVA-GIS software [42]) were calculated by using the Kendall rank-correlation test as provided in the SAS v9.1 software (SAS Institute 2001, Wallisellen, Switzerland). Detection of bottleneck effects was calculated by using BOTTLENECK version 1.2 software [43]. A two-phase mutation model (TPM) and a stepwise mutation model (SMM) were used for Wilcoxon signed-rank tests. The parameter settings included a 90% SMM and 10% TPM with a variance of 12%, and 1000 repeats [44].
For genetic relationships at the population level, a factorial correspondence analysis (FCA) was performed by using the Genetix 4.05 software (CNRSUMR 5000; Universite Montpellier II, Montpellier, France) [45]. Population genetic structure was analyzed based on Bayesian clustering using the STRUCTURE software [46]. For clustering from K = 1 to K = 20 (populations + 1), an admixture ancestry model and correlated allele frequency model were used to perform a Markov chain Monte Carlo simulation algorithm (MCMC) [47]. The length of the burn-in period at start time was set as 100,000; MCMC after the length of the burn-in period was set as 100,000, and for each of K value, simulation calculation was repeated 10 times. The method from Evanno was used to determine the optimal K value [48]. The relative proportions of the geographical distribution diagram were plotted by using the Arc-GIS software (Environmental Systems Research Institute, Inc., Redlands, CA, USA). Molecular variance analysis (AMOVA) for population genetic structures [49] and a Mantel test to detect the presence of geographic segregation between populations by means of logarithmic normalization of geospatial distance and genetic distance [50] were calculated by using the Arlequin software (CMPG, Institute of Ecology and Evolution; University of Berne, Berne, Switzerland).
3. Results
3.1. Genetic Diversity
A total of 277 alleles were amplified from 25 SSR primers. On average, each microsatellite loci could be amplified 11 times, and the number of alleles ranged from a minimum of 4 (G11, EE856) to a maximum of 26 (Q16). The Ho of each microsatellite loci ranged from 0.157 (PIE040) to 0.704 (Q16), and the average observed heterozygosity was 0.401. For each microsatellite loci, He was higher than Ho, and ranged from 0.418 (EE856) to 0.875 (Q16), and the average He was 0.707. AR ranged from 3.50 (EE856) to 21.12 (Q16), and the average AR was 9.51. The PIC of SSR microsatellite loci ranged from 0.423 to 0.911, and the average PIC was 0.760 (Table 2). After detecting significance (P < 0.05), we did not find the existence of linkage disequilibrium between any two points.
Based on statistics of population genetic diversities of 25 microsatellite loci from 19 geographic populations (Table 3), the average number of alleles in each population was 8.01; the population with the highest number of alleles was PZ (8.76), and the lowest was XX (7.04). The average number of effective alleles was 4.52; the highest was LY, and the lowest was FJ. The overall average AR was 7.79; the highest one (8.56) was distributed in XS, whereas the lowest was in XX (6.90). Q. variabilis manifested a significant loss of heterozygosity, and the mean of inbreeding coefficient was 0.044. The average He was 0.707; the highest one was 0.745 with a distribution in CK, whereas the lowest one was 0.623 in FJ. BOTTLENECK analysis showed that in the 19 analyzed populations, three groups could be identified: CZ (SMM, P < 0.05), PS (SMM, P < 0.05), and FJ (TPM, SMM, P < 0.05), which may have recently undergone a severe decline in population size, i.e., the bottleneck effect had occurred.

Table 3.
Mean values of genetic diversity statistics for 25 microsatellite loci in 19 Q. variabilis populations.
Kendall rank-correlation analysis of genetic diversity of geographic populations with latitude/longitude and climatic parameters showed that in general, the AR of Q. variabilis geographic populations has a significant positive correlation with higher average annual temperatures (r = 0.507, P < 0.01), whereas no correlations among He, geographic latitude, and climate were observed.
3.2. Genetic Differentiation and Genetic Structure
The overall population differentiation degree among geographical populations of Q. variabilis: FST = 0.063, GST = 0.060 (P < 0.001) have been reported in Table 3. Gene flow: Nm = 3.648. AMOVA analysis [52] showed that 6.3% (P < 0.001) of the genetic variations was among populations. The greatest percentage of variation was contained within populations. The FST matrix between every two populations is shown in Table 4; the differentiation coefficient between populations CK and SZ was the smallest, (FST = 0.028); that between XX and FJ was the largest (FST = 0.135).

Table 4.
The pairwise FST for all populations of Q. variabilis.
A FCA on genetic structure differences of pattern detection at the population level showed that the populations could be divided into three groups based on geographic location: (a) the Southwest populations: LY, PS, FJ, and WX; (b) the Central populations (the most resource-rich center of Q. variabilis distribution in China): AK, MC, XS, PZ, CK, JS, YS, YA, SZ, and LQ; and (c) the Northeast populations: NJ, CZ, ZM, NX, and XX. The Central populations were adjacent to the Southwest populations and Northeast populations in the FCA clustering groups, whereas the Southwest populations were distantly located from the Northeast populations in the FCA clustering groups (see Figure 2a,c). Small genetic differences between the Central populations and the other two populations were observed, whereas larger genetic differences were detected between the Southwest populations and the Northeast populations.
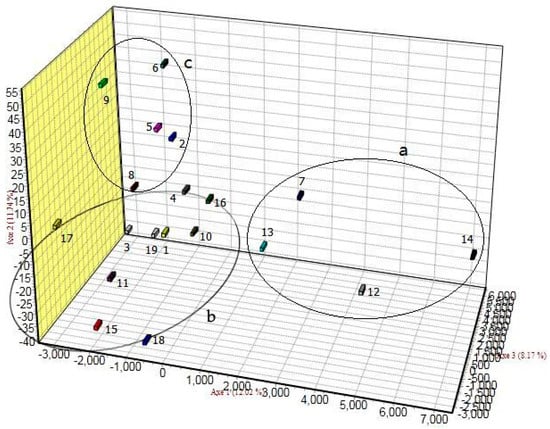
Figure 2.
Results of factorial correspondence analysis at the population level based on simple sequence repeat markers. The population codes are as in Table 1.
For a more detailed understanding of the genetic structures, using a ΔK value that Evanno proposed to determine a reasonable K, the ΔK value reached a maximum when K = 3 (Figure 3). The 19 geographic populations of Q. variabilis could then be divided into three genetically distinct groups (Figure 4). In addition, we used the relative proportions of the geographical distribution diagram to visually compare the distribution ratio of each group within the population (Figure 5). The 19 populations as a whole were divided into three groups based on geographic distribution (Figure 4 and Figure 5); the first group was the Southwest group (pop1–4), the second group was the Central group (pop5–14), and the third group was the Northeast group (pop15–19). Average distribution ratios [53] were 76%, 72% and 73%, respectively. The Central group had the lowest distribution ratio, i.e., the highest mixing degree among groups. XX (Northeast group, ratio: 87.9%), JS (Central group, ratio: 83.4%), and WX (Southwest group, ratio: 88.9%) were the source populations in each group, which had the highest distribution ratio.
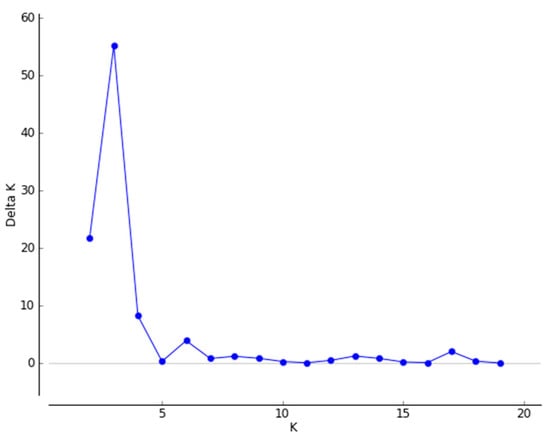
Figure 3.
Relationships between the number of clusters (K) and the corresponding ΔK statistics calculated according to ΔK based on STRUCTURE analysis [46].
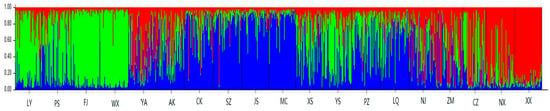
Figure 4.
Results of the structure analysis of Q. variabilis populations when K = 3. Each individual is represented by a single vertical bar, which is partitioned into three different colors. Each color represents a genetic cluster and the colored segments shows the individual’s estimated ancestry proportion to each of the genetic clusters. The population codes are as in Table 1.
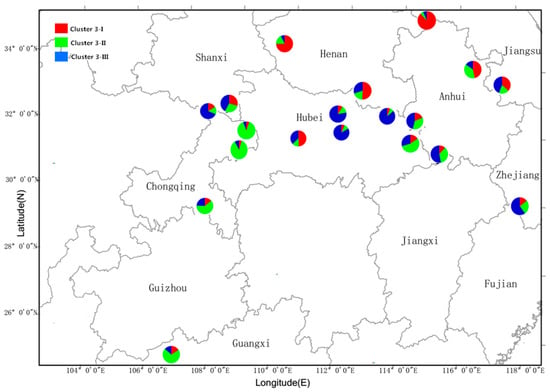
Figure 5.
Mean proportions of cluster memberships of analyzed individuals in each of the 19 Q. variabilis populations, based on STRUCTURE analysis at K = 3.
The results of a FCA and STRUCTUE analysis indicated that the population as a whole could be divided into three genetic groups. Furthermore, AMOVA analysis showed 1.75% variation between groups (P < 0.001), 5.59% variation within groups (P < 0.001), and 92.66% variation within populations (P < 0.001). All genetic groups were ordered according to the calculated FST value: Northeast group (FST = 0.070) > Southwest group (FST = 0.039) > Central Group (FST = 0.027). The standard genetic distance and geographic distance of FST/(1 − FST) were calculated using the Mantel test, which revealed that the changes in the FST values did not follow the geographical distance separating mode (R = 0.0013, P = 0.371). The geographical distances between populations AK, CK and FJ, WX were very small, but FCA and Bayesian analyses did not cluster those into the same groups, and AMOVA analysis showed that most of the variations that existed within groups resulted in significant genetic structural differences in the populations AK, CK and FJ, WX.
4. Discussion
4.1. Genetic Diversity
The level of species genetic diversity is often associated with specific characteristics such as the length of life cycle, mating system and reproduction, the size of the geographic range, and genetic exchange [11,54,55,56]. The He (0.707) of Q. variabilis in our study was slightly lower than that of Xu’s previous study (0.806) [6]. The main reason for this difference is that only five Chinese populations of Q. variabilis were studied in Xu’s study, whereas we examined representative populations within the distribution range from the South to the North. Therefore, the population genetic variation of the species reflected in the present study was more comprehensive. Compared to the overall genetic variation level of Q. variabilis with other Fagaceae Quercus genera species such as the summer oak (Q. robur) (He = 0.764) [29] and Liaodong oak (Q. liaotungensis) (He = 0.754) [30], the levels of diversity were about the same yet slightly higher than that of the Sawtooth oak (Q. acutissima) (He = 0.660) [22] and Mongolian oak (Q. mongolica) (He = 0.630) [24]. Compared to the other endangered species or narrow domains that were left over from the Quaternary Ice Age in China such as Ginkgo biloba (He = 0.241) [57], the overall genetic diversity level of Q. variabilis was high, which was mainly due to the extensive distribution of the species distributed from 19° N to 42° N across the temperate forests. Because of huge differences in climate and habitat conditions among populations in the distribution area, Q. variabilis may have differentiated adapted ecological and genetic types, thereby resulting in a wide range of genetic variations. In addition, affected by the fact that the populations were sampled from natural secondary forest, heterozygote deficits were shown (FIS = 0.044).
He and AR of each population by Kendall rank-correlation analysis revealed that neither followed the geographical space separation mode. However, changes in AR clearly showed significant correlations with changes in the average annual temperature, indicating that temperature has a significant impact on Q. variabilis. Among the 19 populations studied, the genetic diversity of the central populations (AR = 8.06, He = 0.718) was higher than that of the Southwest populations (AR = 7.55, He = 0.683) and the Northeast populations (AR = 7.45, He = 0.704). The two populations with the highest genetic diversity—XS (AR = 8.56) and CK (He = 0.745)—both belonged to the Central group; these results supported the conclusions of Chen [26] in the populations of Q. variabilis in the Central region, which experiences an average annual temperature of 15 °C or above and has a higher level of genetic diversities than other areas. Bottleneck detection revealed that the Southwest populations FJ and PS, and Northeast population CZ experienced a genetic bottleneck, which may be the main reason for the low level of genetic diversity in all three populations [14,43,58].
Mayr proposed a “core–periphery” hypothesis that states that compared to the core groups in general [59], the genetic diversity of peripheral groups decreased and genetic differentiation increased under the pressures of the bottleneck effect, genetic drift, and selection pressures [60]. Based on the genetic diversity of five Q. variabilis populations, Xu concluded that the genetic diversity of the central groups was higher than that of the peripheral groups [6], but because of the limitations of the sample population scales, these findings could only be used as a reference. In our study, the samples of 19 populations were collected from a representative distribution in the main distribution areas. For the populations located in the geographical centers of existing distribution areas (the Central group), overall genetic diversities were higher than those in the peripheral regions (the Southwest group and the Northeast group). In addition, private alleles were only found in the southwestern population, such as LY; this indicated that there was genetic variation in the peripheral population in adaptation to the climate and environment. Accordingly, the diversity distribution of Q. variabilis was in line with the “core–periphery” hypothesis.
4.2. Genetic Differentiation
The genetic differentiation level of Q. variabilis was moderately low (FST = 0.063), which was in agreement with the results of AMOVA analysis (among populations variation = 6.3%). The genetic differentiation level of Q. variabilis is similar to that of the summer oak (Q. robur) (FST = 0.080) [29] and Mongolian oak (Q. mongolica) (FST = 0.077) [24]. Its genetic differentiation level is close to that reported by Hamrick [61], who investigated diversity in widespread species, including long-lived woody perennials and wind-pollinated outcrossing species. As a perennial tree, Q. variabilis is a typical wind-pollinated and outcrossing species, which undoubtedly increases gene flow between populations and reduces differentiation among populations.
Gene flow determines the genetic structure and survival potential of future populations of a species [12]. The present study showed that Q. variabilis populations are characterized by a relatively large gene flow (Nm = 3.648) that may inhibit genetic drifting and prevent population genetic differentiation [11]. The large gene flow is mainly determined by its biological means of spreading via pollen and seeds. Xu [6] reported a higher gene flow (Nm = 5.239) than this study, which objectively reflected a decreasing gene flow for Q. variabilis in the last decade; the original natural forest of Q. variabilis has degenerated into a natural secondary forest, which may have caused a reduction in gene flow.
The clustering results of STRUCTURE Bayesian clustering showed that the 19 populations could be divided into three groups: the Southwest group, the Central group, and the Northeast group. These findings were in agreement with the results of a FCA, wherein the mix in the population genetic structure of the Southwest group and the Northeast group was relatively low, and there was a large difference in the genetic composition between the two groups. On the other hand, the Central populations were the largest; they showed a high mixing degree and the lowest differentiation. In general, the population that is capable of maintaining its level of genetic diversity is often the largest population group [62]. Therefore, as expected, the Central group showed the highest capacity to maintain its level of genetic diversity. The Mantel test showed that correlations of genetic and geographic distances between the groups were not significant (R = 0.0013, P = 0.371), which showed that geographical distance is not the main reason for genetic differentiation among populations.
As a widely distributed species and perennial tree, the genetic structure formation of Q. variabilis should not be analyzed from a single geographic distribution and distance. During the Quaternary Ice Age, which was affected by the global glacial climate, Q. variabilis in northern China was unable to adapt and eventually disappeared. Furthermore, the South of Qinling and Dabie Mountains had become barriers that blocked the Quaternary glaciers, with their complex landforms and diverse ecological environments rendering them sources of plant heterogeneous differentiations [63]. The area thus served as a sanctuary for Q. variabilis during Quaternary glaciation [64] and eventually became the geographical distribution center for Q. variabilis [4]. At the end of the Ice Age and with the recovery of temperature as well as other natural conditions, it is assumed that some populations in the distribution center differentiated into varying types which adapted to a cold, dry climate and began to migrate to high latitudes in the northeast, while some others adapted to warm and moist types and began to spread southwestward, and gradually formed the geographical distribution pattern of Q. variabilis (in the present study, there were significant correlations between AR and the average annual temperature). This evolutionary geographic history has been verified by simple haplotypes in peripheral populations, and populations in the central region represented almost all haplotypes of Q. variabilis [26]. It is in this complex historical dynamic migration process that the genetic structure of the existing distribution of Q. variabilis was formed.
In addition, human activities may also have an impact on the genetic structure of various species [65,66,67]. However, the results of the present study showed that among the 19 Q. variabilis populations, the population in the center of the distribution did not experience a bottleneck effect, whereas three populations at the periphery (FJ, PS, CZ) did, and three genetic lineages (cluster I,II and III) obtained from STRUCTURE showed distributions in every population (Figure 4 and Figure 5). Based on these results, we inferred that Q. variabilis has been a widely distributed species for several historical periods, and the history of its population dynamics and genetic variations stretches back further than the time of human activities [68], so human activity is not the major factor that has affected its current genetic structure.
5. Conclusions
This represents the first research carried out on the genetic diversity and structure of Q. variabilis based on a broader sampling strategy involving its main distribution area. Q. variabilis, in general, was in line with the “core–periphery” hypothesis. The overall genetic diversity level of Q. variabilis was relatively high; the genetic diversity level of populations in the geographical distribution center was higher than that of peripheral populations. The accessions from southwest, center and northeast areas clustered into three separate groups, and the genetic structure of Q. variabilis was mainly affected by the preferable adaptability to the climate and environment in the complex historical dynamic migration process. Due to the development of cork, tannin, and other industries, the distribution scale of Q. variabilis has gradually reduced; we should establish conservation measures to prevent population decline; peripheral populations FJ, PS and CZ, in particular, suffered the bottleneck effect. Additionally central populations with abundant genetic variation could be used as the preferred germplasm resource in plantations. Furthermore, private alleles were found only in peripheral population LY; they are of great research value to the maintenance and evolution of species in disadvantageous habitats.
Acknowledgments
This research was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author Contributions
This study was carried out with collaboration among all authors. Li-an Xu conceived and designed the experiments; Xiaomeng Shi and Xin Guo performed the experiments; Qiang Wen carryed out data correction and manuscript revision; Xiaomeng Shi and Mu Cao provided the experimental data; Xiaomeng Shi wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Johnson, P.S.; Shifley, S.R. The Ecology and Silviculture of Oaks; CABI Publishing: New York, NY, USA, 2001; pp. 9–10. ISBN 9780851995700. [Google Scholar]
- Kremer, A.; Petit, R.J. Gene diversity in natural Population of oak species. Ann. For. Sci. 1993, 50, 186–202. [Google Scholar] [CrossRef]
- eFlora. Org., Flora of China, Quercus variabilis Blume. Available online: http://www.efloras.org/ (accessed on 10 October 2017).
- Wang, J.; Wang, S.B.; Kang, H.Z.; Xin, Z.J.; Qian, Z.H.; Liu, C.J. Distribution Pattern of Oriental Oak (Quercus variabilis Blume) and the Characteristics of Climate of Distribution Area in Eastern Asia. J. Shanghai Jiaotong Univ. 2009, 27, 235–241. [Google Scholar]
- Wei, L. Preliminary investigation on the distribution of the cork oak. Sci. Silvae Sin. 1960, 6, 70–71. [Google Scholar]
- Xu, X.L.; Xu, L.A.; Huan, M.R.; Wang, Z.R. Genetic Diversity of Microsatellites (SSRs) of Natural Populations of Quercus variabilis. Hereditas 2004, 26, 683–688. [Google Scholar] [PubMed]
- Zhao, G.; Duan, X.F.; Guan, T.; Huang, L.H. Situation of Cork Utilization in the World and the Development Countermeasure of the China's Cork Industry. World For. Res. 2004, 17, 25–28. [Google Scholar]
- Flores, M.; Rosa, M.E. Properties and uses of consolidated cork dust. J. Mater. Sci. 1992, 27, 5629–5634. [Google Scholar] [CrossRef]
- Zhang, W.H.; Lu, Z.J. A study on the biological and ecological property and geographical distribution of Quercus variabilis population. Acta Bot. Boreali-occident. Sin. 2002, 22, 1093–1101. [Google Scholar]
- Lei, J.P.; Xiao, W.F.; Liu, J.F. Distribution of Quercus variabilis Blume and Its Ecological Research in China. World For. Res. 2013, 26, 57–62. [Google Scholar]
- Hamrick, J.L.; Godt, M.J.W. Allozyme diversity in plant species. In Plant Population Genetics, Breeding, and Genetic Resources; Brown, A.D.H., Clegg, M.T., Kahler, A.L., Weit, B.S., Eds.; Sinauer Associates: Sunderland, MA, USA, 1990; pp. 43–63. ISBN 9780878931170. [Google Scholar]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Heuertz, M.; Hausman, J.F.; Hardy, O.J.; Vendramin, G.G.; Frascarialacoste, N. Nuclear microsatellites reveal contrasting patterns of genetic structure between western and southeastern European populations of the common ash (Fraxinus excelsior L.). Evolution 2004, 58, 976–988. [Google Scholar] [PubMed]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2004; Volume 190, pp. 385–386. ISBN 9780521702713. [Google Scholar]
- Sutherland, W.J.; Albon, S.D.; Allison, H.S.; Armstrong, B.; Bailey, M.J.; Brereton, T.; Boyd, I.L.; Carey, P.; Edwards, J.; Gill, M.; et al. The identification of priority policy options for UK nature conservation. J. Appl. Ecol. 2010, 47, 955–965. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.W.; Fang, Y.M. Predicting the impact of global warming on the geographical distribution pattern of Quercus variabilis in China. Chin. J. Appl. Ecol. 2014, 25, 3381–3389. [Google Scholar]
- Zhou, J.Y.; Lin, J.; He, J.F.; Zhang, W.H. Review and Perspective on Quercus variabilis Research. J. Northwest For. Univ. 2010, 25, 43–49. [Google Scholar]
- Yuan, Z.L.; Wang, T.; Zhu, X.L.; Sha, Y.Y.; Ye, Y.Z. Patterns of spatial distribution of Quercus variabilis in deciduous broadleaf forests in Baotianman Nature Reserve. Biodivers. Sci. 2011, 19, 224–231. [Google Scholar]
- Hu, X.J.; Zhang, W.H.; He, J.F.; Yin, Y.N. Architectural analysis of crown geometry of Quercus variablis BL. natural regenerative seedlings in different habitats. Acta Ecol. Sin. 2014, 35, 789–795. [Google Scholar]
- Li, Z.P.; Zhang, W.H.; Cui, Y.C. Effects of NaCl and Na2CO3 stresses on seed germination and seedling growth of Quercus variabilis. Acta Ecol. Sin. 2015, 35, 742–751. [Google Scholar]
- Ran, R.; Zhang, W.H.; He, J.F.; Zhou, J.Y. Effects of thinning intensities on population regeneration of natural Quercus variabilis forest on the south slope of Qinling Mountains. Chin. J. Appl. Ecol. 2014, 25, 695–701. [Google Scholar]
- Zhang, Y.Y.; Fang, Y.M.; Yu, M.K.; Li, X.X.; Xia, T. Molecular characterization and genetic structure of Quercus acutissima germplasm in China using microsatellites. Mol. Biol. Rep. 2013, 40, 4083–4090. [Google Scholar] [CrossRef] [PubMed]
- Jennifer, F.L.; Oliver, G. Genetic structure of Quercus rubra L. and Quercus ellipsoidalis E.J. Hill populations at gene-based EST-SSR and nuclear SSR markers. Tree Genet. Genomes 2013, 9, 707–722. [Google Scholar]
- Saneyoshi, U.; Yoshihiko, T. Development of ten microsatellite markers for Quercus mongolica var. crispula by database mining. Conserv. Genet. 2008, 9, 1083–1085. [Google Scholar]
- Neophytou, C.H.; Dounavi, A.; Aravanopoulos, F.A. Conservation of Nuclear SSR Loci Reveals High Affinity of Quercus infectoria ssp. veneris A. Kern (Fagaceae) to Section Robur. Plant Mol. Biol. Rep. 2008, 26, 133–141. [Google Scholar] [CrossRef]
- Chen, D.M.; Zhang, X.X.; Kang, H.Z.; Sun, X.; Yin, S.; Du, H.M.; Yamanaka, N.; Gapare, W.; Wu, H.X.; Liu, C.J. Phylogeography of Quercus variabilis Based on Chloroplast DNA Sequence in East Asia: Multiple Glacial Refugia and Mainland-Migrated Island Populations. PLoS ONE 2013, 7, e47268. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Pan, H.X.; Zhu, G.Q.; Yin, T.M.; Zou, H.Y.; Huang, M.R. Analysis of Genetic Structure of Natural Populations of Castanopsis fargesii by RAPDs. Acta Bot. Sin. 2002, 44, 1321–1326. [Google Scholar]
- BiOptic Inc. Available online: http://www.bioptic.com.tw (accessed on 5 July 2016).
- Alexis, R.S.; Jennifer, F.L.; Tim, S.M.; Jeanne, R.S.; Oliver, G. Development and Characterization of Genomic and Gene-Based Microsatellite Markers in North American Red Oak Species. Plant Mol. Biol. Rep. 2012, 31, 231–239. [Google Scholar]
- Qin, Y.Y.; Han, H.R.; Kang, F.F.; Zhao, Q. Genetic diversity in natural populations of Quercus liaotungensis in Shanxi Province based on nuclear SSR markers. J. Beijing For. Univ. 2012, 34, 61–65. [Google Scholar]
- Durand, J.; Bodenes, C.; Chancerel, E.; Frigerio, J.M.; Vendramin, G.; Sebastiani, F.; Buonamici, A.; Gailing, O.; Koelewijn, H.P.; Villani, F.; et al. A fast and cost-effective approach to develop and map EST-SSR markers: Oak as a case study. BMC Genom. 2010, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; pp. 73–95. ISBN 9780231063210. [Google Scholar]
- Mousadik, E.L.; Petit, R.J. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels endemic to Morocco. Theor. Appl. Genet. 1996, 92, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT Version 2.9.3, A Program to Estimate and Test Gene Diversities and Fixation Indices. 2002. Available online: http://www.unil.ch/izea/softwares/fstat.html (accessed on 5 July 2016).
- Liu, J. POWERMARKER: A Powerful Software For Marker Data Analysis; North Carolina State University, Bioinformatics Research Center: Raleigh, NC, USA, 2002. [Google Scholar]
- Peakall, R.; Smouse, P.E. GENEALEX: Genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Curtu, A.L.; Gailing, O.; Leinemann, L.; Finkeldey, R. Genetic variation and differentiation within a natural community of five oak species (Quercus spp.). Plant Biol. 2007, 9, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M.; Barton, N.H. Methods for estimating geneflow. Evolution 1989, 43, 1349–1368. [Google Scholar] [CrossRef] [PubMed]
- Iwaizumi, M.G.; Tsuda, Y.; Ohtani, M.; Ohtani, M.; Tsumura, Y.; Takahashi, M. Recent distribution changes affect geographic clines in genetic diversity and structure of Pinus densiflora natural populations in Japan. For. Ecol. Manag. 2013, 304, 407–416. [Google Scholar] [CrossRef]
- Tian, C.; Mu, N.; Zhu, Z.Y.; Wang, C.J. Method for the Rapid Obtaining of Climate Data Based on DIVA-GIS. J. Agric. 2015, 5, 109–113. [Google Scholar]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Luikart, G.; Cornuet, J.M. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv. Biol. 1998, 12, 228–237. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX (ver.4.02): Logiciel sous Windows TM pour la Genetique des Populations, Laboratoire Genome, Populations, Interactions; CNRSUMR 5000; Universite Montpellier II: Montpellier, France, 2004. [Google Scholar]
- Pritchard, J.K.; Wen, X.; Falush, D. STRUCTURE Version 2.3.1. 2009. Available online: http://pritch.bsd.uchicago.edu/structure (accessed on 5 July 2016).
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. ARLEQIN (ver.3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2007, 1, 47–50. [Google Scholar] [PubMed]
- Excoffier, L.; Lischer, H. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 1463. [Google Scholar]
- Meirmans, P.G. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 2010, 60, 2399–2402. [Google Scholar] [CrossRef]
- Yan, B.Q.; Wang, T.; Hu, L.L. Population genetic diversity and structure of Schisandra sphenanthera, a medicinal plant in China. Chin. J. Ecol. 2009, 28, 811–819. [Google Scholar]
- Gaudeul, M.; Taberlet, P.; Till, B.I. Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from a mplified fragment length polymorphism markers. Mol. Ecol. 2000, 9, 1625–1637. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Jin, Z.X.; Lou, W.Y.; LI, J.M. Genetic diversity of Lithocarpus harlandii populations in three forest communities with different succession stage. Chin. J. Ecol. 2007, 26, 509–514. [Google Scholar] [CrossRef]
- Hellmanna, J.J.; Pineda, K.M. Constraints and reinforcement on adaptation under climate change: Selection of genetically correlated traits. Biol. Conserv. 2007, 13, 599–609. [Google Scholar] [CrossRef]
- Ge, Y.Q.; Qiu, Y.X.; Ding, B.Y.; Fu, C.X. An ISSR analysis on population genetic diversity of the relict plant Ginkgo biloba. Chin. Biodivers. 2003, 4, 276–287. [Google Scholar]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [PubMed]
- Mayr, E. Animal Species and Evolution; Harvard University Press: Cambridge, UK, 1963; pp. 425–457. ISBN 9780674037502. [Google Scholar]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Hamrick, J.L. IsoZymes ananysis of genetic structure of plant population. In Isozymes in Plant Biology; Soltis, D., Soltis, P., Eds.; Dioscorids Press: Washington, DC, USA, 1989; pp. 87–105. ISBN 9780412365003. [Google Scholar]
- Ohsawa, T.; Tsuda, Y.; Saito, Y.; Ide, Y. The genetic structure of Quercus crispula in northeastern Japan as revealed by nuclear simple sequence repeat loci. J. Plant Res. 2011, 124, 645–654. [Google Scholar] [CrossRef] [PubMed]
- He, C.R.; Chen, F.Q. The endemic genera of Chinese seed plants distributed in Three Gorges Reservoir Area of Changjiang River. Guihaia 1999, 19, 43–46. [Google Scholar]
- Jin, J.H.; Liao, W.B.; Wang, B.S.; Peng, S.L. Global change in Cenozoic and evolution of flora in China. Guihaia 2003, 23, 217–225. [Google Scholar]
- Knowles, P.; Perry, D.J.; Foster, H.A. Spatial genetic structure in two tamarack (Larix laricina) populations with differing establish menthi stories. Evolution 1992, 46, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Young, A.C.; Merriam, H.G. Effect of forest fragmentation on the spatial genetic structure of Acer saccharum Marsh. (sugar maple) populations. Heredity 1994, 72, 201–208. [Google Scholar] [CrossRef]
- Aldrich, P.R.; Hamrick, J.L.; Chavarriaga, P. Microsatellite analysis of de mographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Mol. Ecol. 1998, 7, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.K. Origian, Phylogeny and Disperal of Quercus from China. Acta Bot. Yunnanica 1992, 14, 227–236. [Google Scholar]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).