Applying Effective Population Size Estimates of Kandelia obovata Sheue, Liu and Yong to Conservation and Restoration Management
Abstract
:1. Introduction
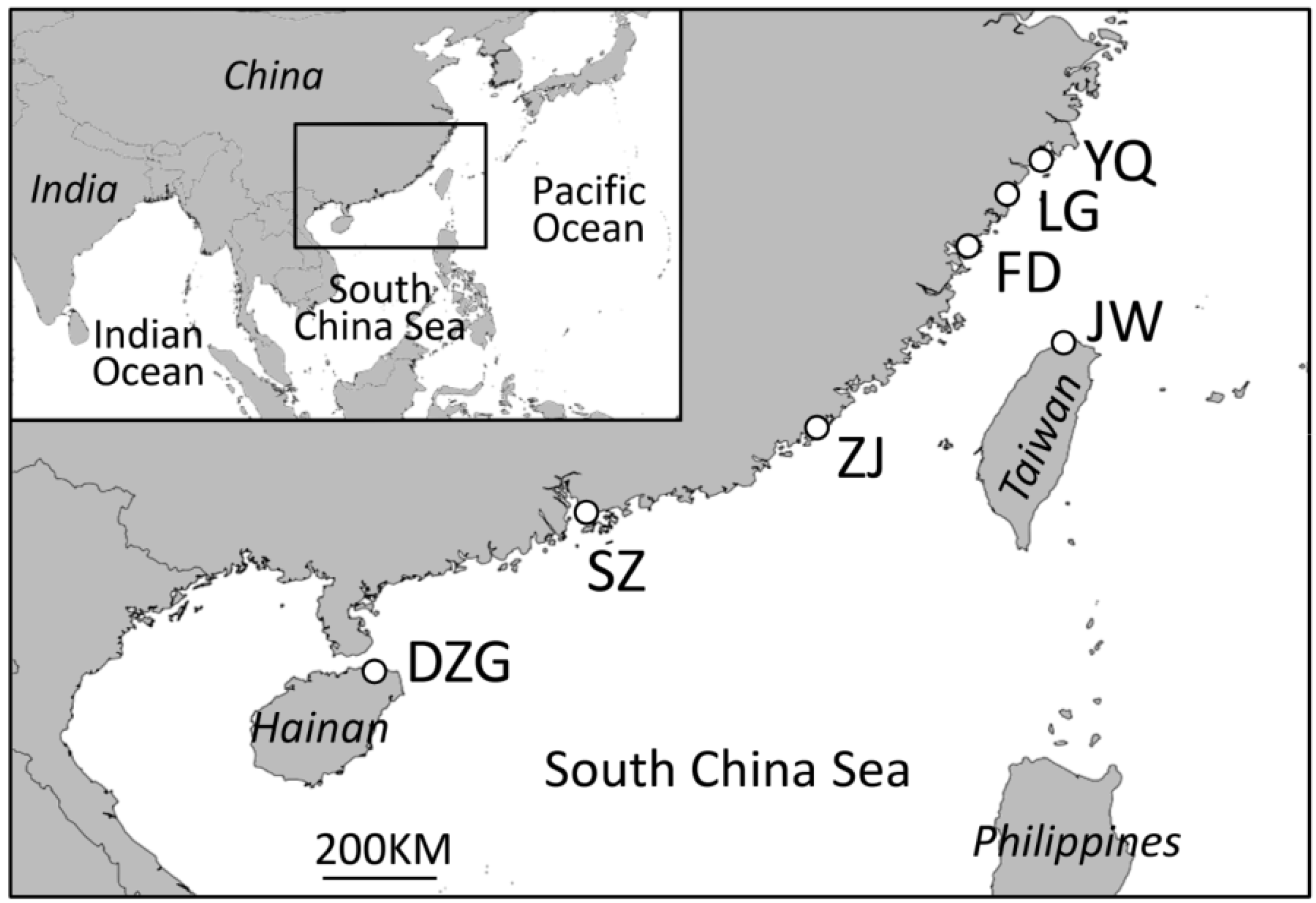
2. Experimental Section
2.1. Data Collection
2.2. Data Analyses
3. Results and Discussion
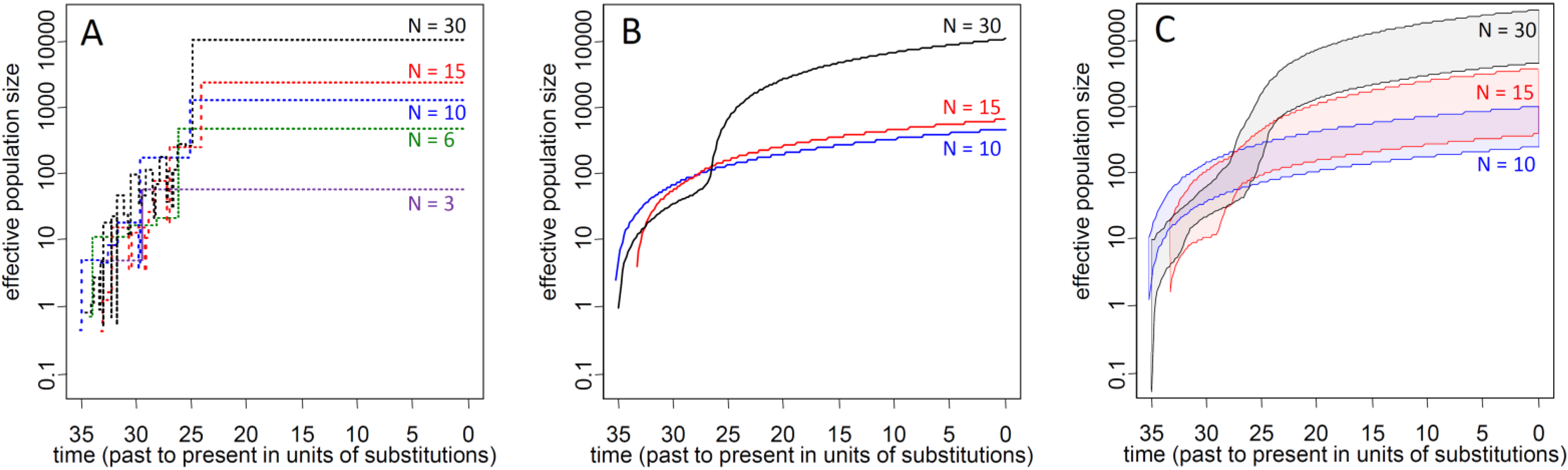
3.1. Influence of Demographic Inference on Adaptive Loci
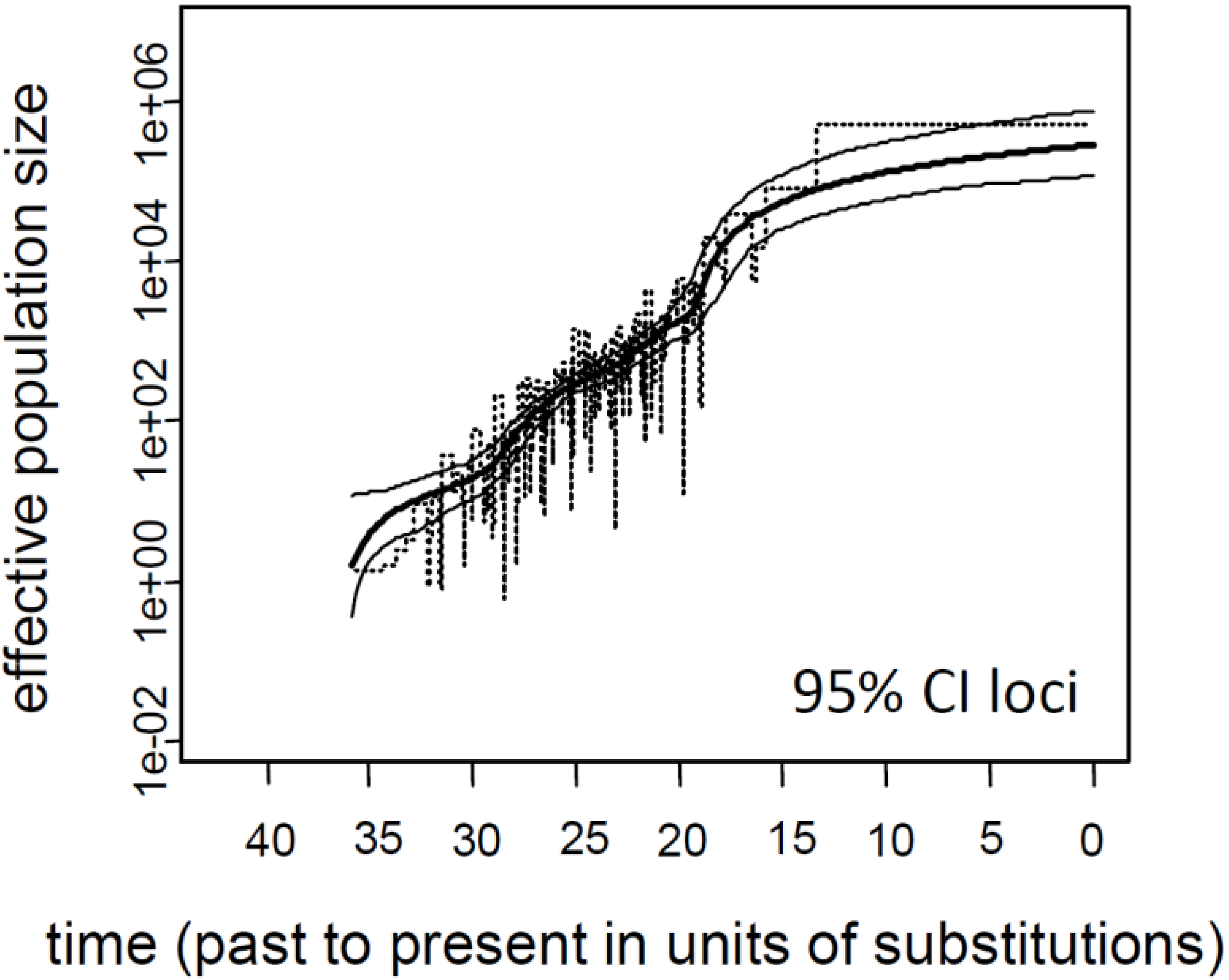
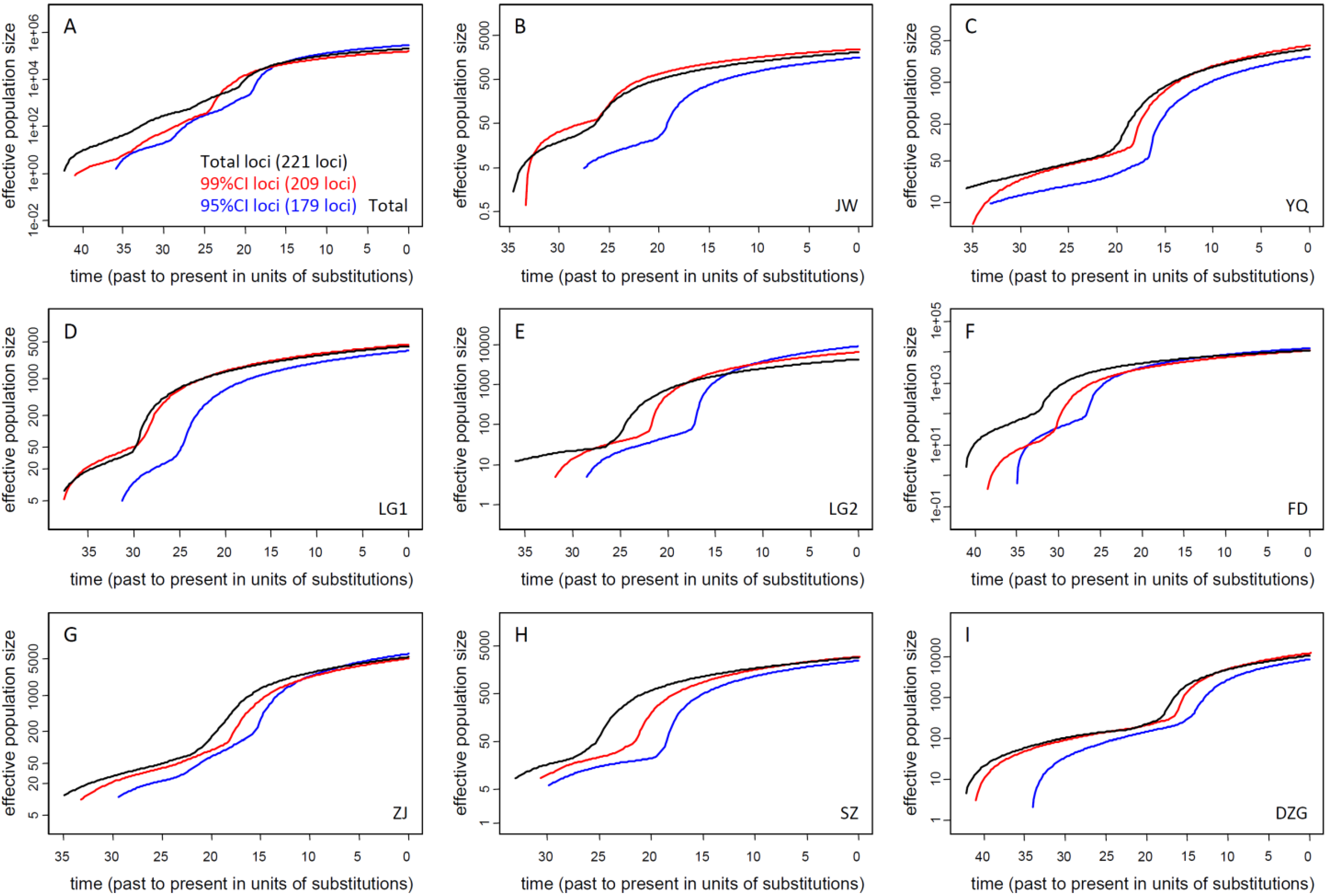
3.2. Current Ne of K. obovata
3.2.1. Ne of Natural Populations
| ALL | JW | FD | ZJ | SZ | DZG | YQ * | LG1 * | LG2 * | |
|---|---|---|---|---|---|---|---|---|---|
| Mean | 188289 | 1792 | 13922 | 6284 | 2581 | 9178 | 3094 | 3703 | 10053 |
| Median | 163113 | 1550 | 11847 | 5029 | 2201 | 7818 | 2624 | 3074 | 8666 |
| 95% CI | 65730–437432 | 554–4289 | 3644–37609 | 1273–18864 | 825–6432 | 2009–25295 | 826–8215 | 1105–10259 | 1926–26150 |
3.2.2. Ne of Reforested Populations and Ne Reduction of Biased Sampling for Restoration
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Supplementary Materials
References
- Ellstrand, N.C.; Elam, D.R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Lynch, M.; Lande, R. The critical effective size for a genetically secure population. Anim.Conserv. 1998, 1, 70–72. [Google Scholar] [CrossRef]
- Franklin, I.R.; Frankham, R. How large must populations be to retain evolutionary potential? Anim. Conserv. 1998, 1, 69–70. [Google Scholar] [CrossRef]
- Hare, M.P.; Nunney, L.; Schwartz, M.K.; Ruzzante, D.E.; Burford, M.; Waples, R.S.; Ruegg, K.; Palstra, F. Understanding and estimating effective population size for practical application in marine species management. Conserv. Biol. 2011, 25, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, I.G.; Allendorf, F.W. How does the 50/500 rule apply to MVPs? Trends Ecol. Evol. 2012, 27, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Schoen, D.J.; Brown, A.H.D. Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc. Natl. Acad. Sci. USA 1991, 88, 4494–4497. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009, 10, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Lande, R. Genetics and demography in biological conservation. Science 1988, 241, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y. Effects of plant density and age on the mating system of Kandelia candel Druce (Rhizophoraceae), a viviparous mangrove species. Hydrobiologia 2000, 432, 189–193. [Google Scholar] [CrossRef]
- Nagelkerken, I.; Blaber, S.J.M.; Bouillon, S.; Green, P.; Haywood, M.; Kirton, L.G.; Meynecke, J.O.; Pawlik, J.; Penrose, H.M.; Sasekumar, A.; Somerfield, P.J. The habitat function of mangroves for terrestrial and marine fauna: A review. Aquat. Bot. 2008, 89, 155–185. [Google Scholar] [CrossRef]
- Lewis, R.R.; Gilmore, R.G. Important considerations to achieve successful mangrove forest restoration with optimum fish habitat. Bull. Mar. Sci. 2007, 80, 823–837. [Google Scholar]
- Kathiresan, K.; Bingham, B.L. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 2001, 40, 81–251. [Google Scholar]
- Lee, S.Y. Tropical mangrove ecology: Physical and biotic factors influencing ecosystem structure and function. Aust. J. Ecol. 1999, 24, 355–366. [Google Scholar] [CrossRef]
- Feller, I.C.; Lovelock, C.E.; Berger, U.; McKee, K.L.; Joye, S.B.; Ball, M.C. Biocomplexity in mangrove ecosystems. Annu. Rev. Mar. Sci. 2010, 2, 395–417. [Google Scholar] [CrossRef]
- Liao, P.C.; Hwang, S.Y.; Huang, S.; Chiang, Y.C.; Wang, J.C. Contrasting demographic patterns of Ceriops tagal (Rhizophoraceae) populations in the South China Sea. Aust. J. Bot. 2011, 59, 523–532. [Google Scholar] [CrossRef]
- Liao, P.C.; Chiang, Y.C.; Huang, S.; Wang, J.C. Gene flow of Ceriops tagal (Rhizophoraceae) across the Kra Isthmus in the Thai-Malay Peninsula. Bot. Stud. 2009, 50, 193–204. [Google Scholar]
- Liao, P.C.; Havanond, S.; Huang, S. Phylogeography of Ceriops tagal (Rhizophoraceae) in Southeast Asia: The land barrier of the Malay Peninsula has caused population differentiation between the Indian Ocean and South China Sea. Conserv. Genet. 2007, 8, 89–98. [Google Scholar] [CrossRef]
- Ruan, Y.; Huang, B.H.; Lai, S.J.; Wan, Y.T.; Li, J.Q.; Huang, S.; Liao, P.C. Population genetic structure, local adaptation, and conservation genetics of Kandelia obovata. Tree Genet. Genomes 2013, 9, 913–925. [Google Scholar] [CrossRef]
- Geng, Q.F.; Lian, C.L.; Goto, S.; Tao, J.M.; Kimura, M.; Islam, M.D. S.; Hogetsu, T. Mating system, pollen and propagule dispersal, and spatial genetic structure in a high-density population of the mangrove tree Kandelia candel. Mol. Ecol. 2008, 17, 4724–4739. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L. Inbreeding coefficient and effective size for an X-linked locus in nonrandom mating populations. Heredity 1996, 76, 569–577. [Google Scholar] [CrossRef]
- Kimura, M.; Maruyama, T. Pattern of neutral polymorphism in a geographically structured population. Genet. Res. 1971, 18, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Caballero, A.; Hill, W.G. Effective size of nonrandom mating populations. Genetics 1992, 130, 909–916. [Google Scholar] [PubMed]
- Broadhurst, L.M.; Lowe, A.; Coates, D.J.; Cunningham, S.A.; McDonald, M.; Vesk, P.A.; Yates, C. Seed supply for broadscale restoration: Maximizing evolutionary potential. Evol. Appl. 2008, 1, 587–597. [Google Scholar] [PubMed]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [PubMed]
- Watterson, G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975, 7, 256–276. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.Y. W.; Shapiro, B. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol. Ecol. Resour. 2011, 11, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Light, J.E.; Hafner, J.C.; Upham, N.S.; Reddington, E. Conservation genetics of kangaroo mice, Genus Microdipodops. J. Mamm. Evol. 2013, 20, 129–146. [Google Scholar] [CrossRef]
- Bernard, A.M.; Shivji, M.S.; Prince, E.D.; Hazin, F.H. V.; Arocha, F.; Domingo, A.; Feldheim, K.A. Comparative population genetics and evolutionary history of two commonly misidentified billfishes of management and conservation concern. BMC Genet. 2014, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Garrick, R.C.; Kajdacsi, B.; Russello, M.A.; Benavides, E.; Hyseni, C.; Gibbs, J.P.; Tapia, W.; Caccone, A. Naturally rare versus newly rare: Demographic inferences on two timescales inform conservation of Galapagos giant tortoises. Ecol. Evol. 2015, 5, 676–694. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.Y.; Chiang, Y.C.; Chen, Y.J.; Chou, C.H.; Havanond, S.; Hong, T.N.; Huang, S. Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Mol. Ecol. 2001, 10, 2697–2710. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.B.; Ding, W.Y.; Qiu, J.B.; Wang, G.Y.; Zhou, Z.M.; Chen, J.F.; Ai, W.M.; Wang, C.Y.; Xie, Q.L. The genetic diversity of the mangrove Kandelia obovata in China revealed by ISSR analysis. Pak. J. Bot. 2010, 42, 3755–3764. [Google Scholar]
- Islam, M.S.; Lian, C.L.; Kameyama, N.; Hogetsu, T. Analyses of genetic population structure of two ecologically important mangrove tree species, Bruguiera gymnorrhiza and Kandelia obovata from different river basins of Iriomote Island of the Ryukyu Archipelago, Japan. Tree Genet. Genomes 2012, 8, 1247–1260. [Google Scholar] [CrossRef]
- Forestry Bureau, COA. Taiwan, Nature Conservation. Available online: http://conservation.forest.gov.tw/ (accessed on 26 March 2015).
- Varshney, R.K.; Chabane, K.; Hendre, P.S.; Aggarwal, R.K.; Graner, A. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci. 2007, 173, 638–649. [Google Scholar] [CrossRef]
- Ho, S.Y.; Shapiro, B. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol. Ecol. Resour. 2011, 11, 423–434. [Google Scholar] [CrossRef] [PubMed]
- AFLP loci, Dryad Digital Repository. Available online: http://dx.doi.org/10.5061/dryad.pb3tr (accessed on 23 April 2015).
- Excoffier, L.; Hofer, T.; Foll, M. Detecting loci under selection in a hierarchically structured population. Heredity 2009, 103, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A.; Balding, D.J. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 2004, 13, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Opgen-Rhein, R.; Fahrmeir, L.; Strimmer, K. Inference of demographic history from genealogical trees using reversible jump Markov chain Monte Carlo. BMC Evol. Biol. 2005, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA. 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zheng, P.P.; Hu, Y.B.; Wei, F.W. Genome-scale analysis of demographic history and adaptive selection. Protein Cell 2014, 5, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Storz, J.F.; Wheat, C.W. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution 2010, 64, 2489–2509. [Google Scholar] [CrossRef] [PubMed]
- Campbell, D.; Bernatchez, L. Generic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Mol. Biol. Evol. 2004, 21, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Tero, N.; Aspi, J.; Siikamaki, P.; Jakalaniemi, A.; Tuomi, J. Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica. Mol. Ecol. 2003, 12, 2073–2085. [Google Scholar] [CrossRef] [PubMed]
- Kuchma, O.; Vornam, B.; Finkeldey, R. Mutation rates in Scots pine (Pinus sylvestris L.) from the Chernobyl exclusion zone evaluated with amplified fragment-length polymorphisms (AFLPs) and microsatellite markers. Mutat. Res. Genet. Toxicol. Environ. 2011, 725, 29–35. [Google Scholar] [CrossRef]
- Dansgaard, W. Greenland Ice Core Studies. Palaeogeogr. Palaeocl. 1985, 50, 185–187. [Google Scholar] [CrossRef]
- Mann, M.E.; Zhang, Z.H.; Rutherford, S.; Bradley, R.S.; Hughes, M.K.; Shindell, D.; Ammann, C.; Faluvegi, G.; Ni, F.B. Global signatures and dynamical origins of the Little Ice Age and Medieval Climate Anomaly. Science 2009, 326, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Loehle, C. A 2000-year global temperature reconstruction based on non-tree ring proxies. Energy Environ. 2007, 18, 1049–1058. [Google Scholar] [CrossRef]
- Loehle, C.; McCulloch, J.H. Correction to: A 2000-year global temperature reconstruction based on non-tree ring proxies. Energy Environ. 2008, 19, 93–100. [Google Scholar] [CrossRef]
- Yamada, K.; Kamite, M.; Saito-Kato, M.; Okuno, M.; Shinozuka, Y.; Yasuda, Y. Late Holocene monsoonal-climate change inferred from Lakes Ni-no-Megata and San-no-Megata, northeastern Japan. Quat. Int. 2010, 220, 122–132. [Google Scholar] [CrossRef]
- Ryman, N.; Allendorf, F.W.; Jorde, P.E.; Laikre, L.; Hossjer, O. Samples from subdivided populations yield biased estimates of effective size that overestimate the rate of loss of genetic variation. Mol. Ecol. Resour. 2014, 14, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Jan, S.; Wang, J.; Chern, C.S.; Chao, S.Y. Seasonal variation of the circulation in the Taiwan Strait. J. Mar. Syst. 2002, 35, 249–268. [Google Scholar] [CrossRef]
- Sun, M.; Wong, K.C.; Lee, J.S.Y. Reproductive biology and population genetic structure of Kandelia candel (Rhizophoraceae), a viviparous mangrove species. Am. J. Bot. 1998, 85, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-T. Changes of mangrove (Kandelia candel (L.) Druce) distribution in Tanshui estuary. Hwa Kang Geogr. J. 2003, 16, 59–69. [Google Scholar]
- Liao, P.C.; Huang, B.H.; Huang, S. Microbial community composition of the Danshui river estuary of northern Taiwan and the practicality of the phylogenetic method in microbial barcoding. Microb. Ecol. 2007, 54, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Strasburg, J.L.; Kane, N.C.; Raduski, A.R.; Bonin, A.; Michelmore, R.; Rieseberg, L.H. Effective population size is positively correlated with levels of adaptive divergence among annual sunflowers. Mol. Biol. Evol. 2011, 28, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Nazareno, A.G.; dos Reis, M.S. At risk of population decline? An ecological and genetic approach to the threatened palm species Butia eriospatha (Arecaceae) of Southern Brazil. J. Hered. 2014, 105, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, D.J.; Gibson, D.J.; Nickrent, D.L. Genetic diversity and competitive abilities of Dalea purpurea (Fabaceae) from remnant and restored grasslands. Int. J. Plant Sci. 2002, 163, 979–990. [Google Scholar] [CrossRef]
- Lande, R. Anthropogenic, ecological and genetic factors in extinction and conservation. Res. Popul. Ecol. 1998, 40, 259–269. [Google Scholar] [CrossRef]
- Keller, M.; Kollmann, J.; Edwards, P.J. Genetic introgression from distant provenances reduces fitness in local weed populations. J. Appl. Ecol. 2000, 37, 647–659. [Google Scholar] [CrossRef]
- Liao, P.C.; Tsai, C.C.; Chou, C.H.; Chiang, Y.C. Introgression between cultivars and wild populations of Momordica charantia L. (Cucurbitaceae) in Taiwan. Int. J. Mol. Sci. 2012, 13, 6469–6491. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, B.-H.; Ruan, Y.; Li, J.-Q.; Liao, P.-C. Applying Effective Population Size Estimates of Kandelia obovata Sheue, Liu and Yong to Conservation and Restoration Management. Forests 2015, 6, 1439-1453. https://doi.org/10.3390/f6051439
Huang B-H, Ruan Y, Li J-Q, Liao P-C. Applying Effective Population Size Estimates of Kandelia obovata Sheue, Liu and Yong to Conservation and Restoration Management. Forests. 2015; 6(5):1439-1453. https://doi.org/10.3390/f6051439
Chicago/Turabian StyleHuang, Bing-Hong, Yu Ruan, Jun-Qing Li, and Pei-Chun Liao. 2015. "Applying Effective Population Size Estimates of Kandelia obovata Sheue, Liu and Yong to Conservation and Restoration Management" Forests 6, no. 5: 1439-1453. https://doi.org/10.3390/f6051439
APA StyleHuang, B.-H., Ruan, Y., Li, J.-Q., & Liao, P.-C. (2015). Applying Effective Population Size Estimates of Kandelia obovata Sheue, Liu and Yong to Conservation and Restoration Management. Forests, 6(5), 1439-1453. https://doi.org/10.3390/f6051439







