SNP-Based Genetic Analysis of Dimensional Stability and Wood Density in Eucalyptus pellita F.Muell. and Hybrids
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Area
2.2. Genetic Material and Trial Establishment
2.3. Estimation of Physical Properties
2.4. Correlation Between Physical Property, Growth, and Chemical Property Traits
2.5. Assessment of Phenotypic Variation and Genetic Association
3. Results and Discussion
3.1. Phenotypic Variation in Shrinkage Property Traits
3.2. Green Density, Oven-Dry Density, and Basic Density
3.3. Phenotypic Correlation
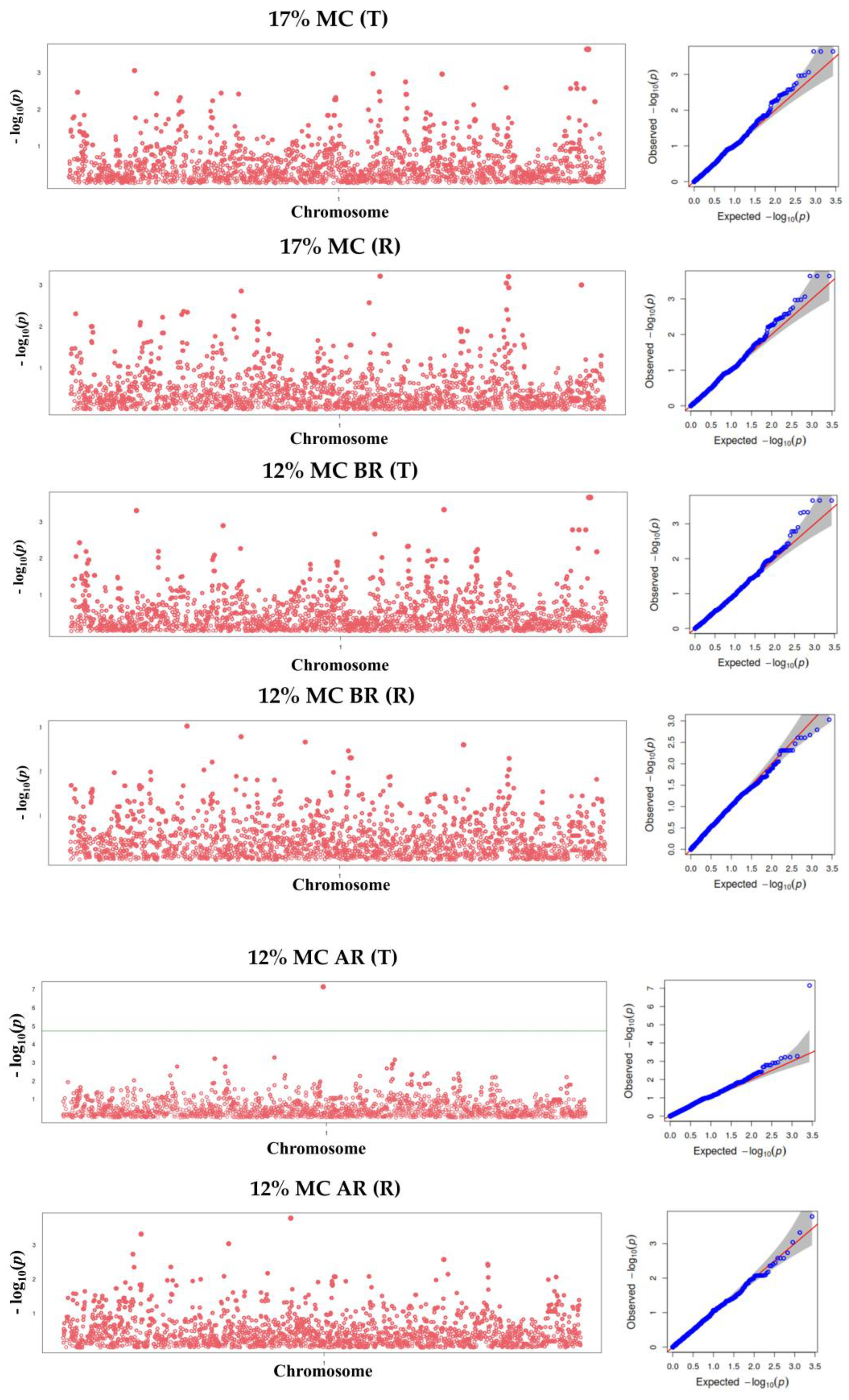
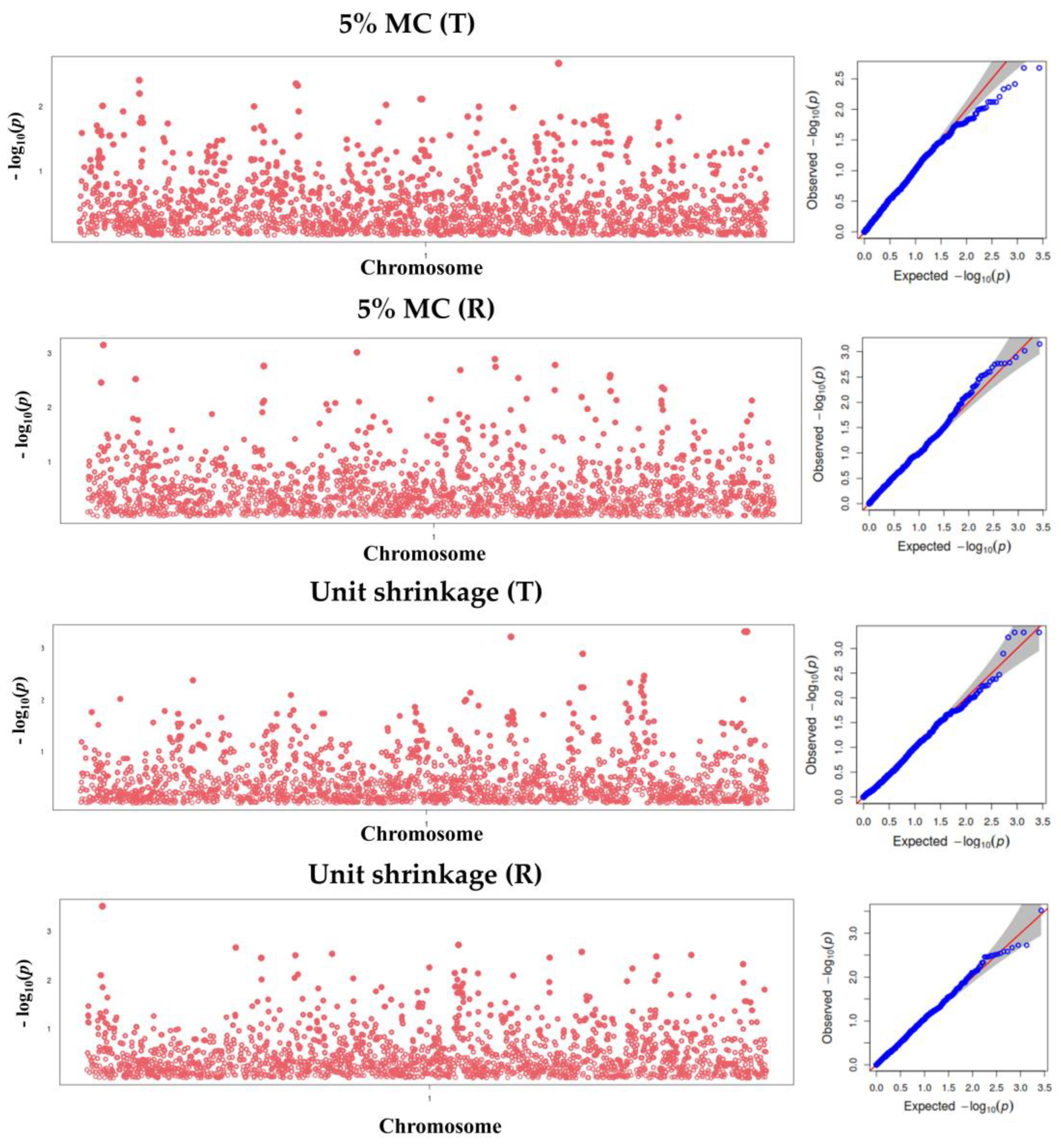
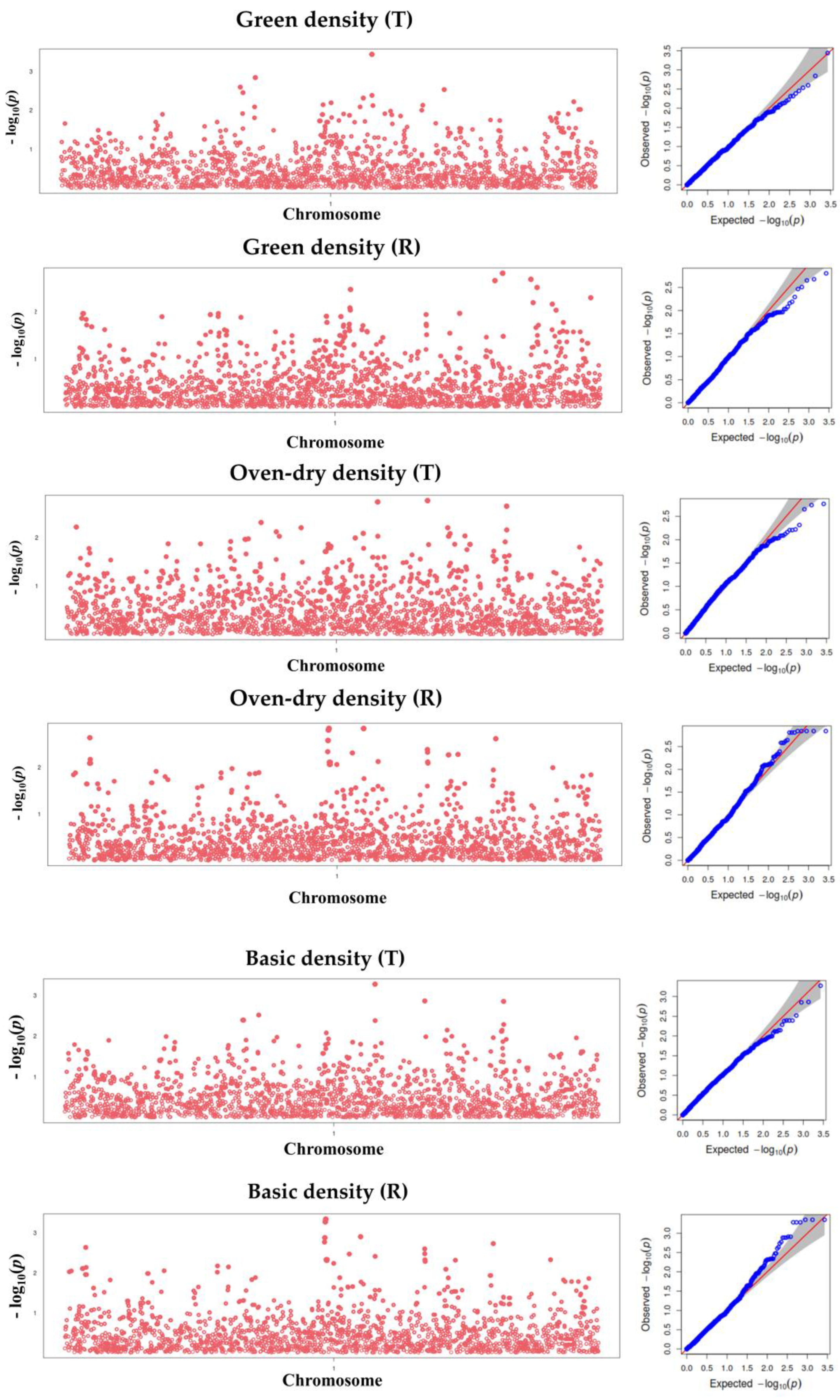
3.4. Manhattan and Q-Q Plots for Wood Shrinkage and Density Traits
3.5. Marker Identification for Wood Property Traits
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Plantation Sites/Hybrids | Green to 17% (%) | Green to 12% BR (%) | Green to 12% AR (%) | Green to 5% (%) | Unit Shrinkage (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| T | R | T | R | T | R | T | R | T | R | |
| KI64 | ||||||||||
| E. pellita | 1.15–3.48 | 0.70–1.71 | 2.82–5.18 | 1.30–3.02 | 1.87–4.13 | 0.63–2.29 | 5.23–7.05 | 3.36–5.99 | 0.16–0.24 | 0.13–0.23 |
| E. pellita × E. brassiana (BC) | 2.55–5.16 | 0.99–2.21 | 3.98–6.56 | 1.67–3.75 | 3.07–5.62 | 1.33–3.34 | 5.70–7.65 | 2.99–6.17 | 0.15–0.23 | 0.09–0.21 |
| E. pellita × E. brassiana (F1) | 2.56–5.61 | 1.06–2.45 | 4.11–7.28 | 2.11–4.50 | 2.79–5.72 | 1.45–3.75 | 6.25–8.74 | 2.96–6.61 | 0.16–0.26 | 0.06–0.20 |
| E. pellita × E. urophylla (BC) | 1.21–3.44 | 1.03–1.95 | 2.82–5.09 | 2.17–3.74 | 1.89–4.04 | 1.26–2.77 | 4.87–6.64 | 2.89–5.29 | 0.15–0.23 | 0.08–0.18 |
| E. pellita × E. urophylla (F1) | 1.96–5.00 | 0.78–2.06 | 3.58–6.52 | 1.26–3.45 | 2.56–5.53 | 0.94–3.05 | 5.54–7.77 | 1.60–4.94 | 0.11–0.22 | 0.01–0.15 |
| YP28 | ||||||||||
| E. pellita | 1.13–2.90 | 1.07–1.77 | 2.68–4.39 | 2.02–3.11 | 2.08–3.63 | 1.58–2.68 | 5.32–6.76 | 3.70–5.68 | 0.19–0.23 | 0.09–0.20 |
| E. pellita × E. brassiana (BC) | 2.12–4.46 | 0.91–1.82 | 3.81–5.96 | 1.81–3.22 | 2.18–4.04 | 1.21–2.52 | 6.02–7.84 | 2.36–4.73 | 0.19–0.25 | 0.04–0.19 |
| E. pellita × E. brassiana (F1) | 5.20–8.46 | 2.55–3.47 | 6.48–9.65 | 3.51–4.92 | 5.67–8.51 | 3.18–4.49 | 7.37–10.04 | 3.37–5.75 | 0.07–0.15 | 0.01–0.16 |
| E. pellita × E. urophylla (BC) | 1.32–3.27 | 0.92–1.67 | 2.97–4.87 | 2.09–3.26 | 2.13–3.85 | 1.68–2.87 | 5.48–7.08 | 3.51–5.67 | 0.20–0.25 | 0.10–0.22 |
| E. pellita × E. urophylla (F1) | 2.47–4.79 | 0.74–2.39 | 3.78–5.99 | 1.18–3.71 | 2.96–4.92 | 0.83–3.44 | 5.54–7.41 | 2.47–7.22 | 0.14–0.19 | 0.05–0.31 |
| Plantation sites combined | ||||||||||
| E. pellita | 1.58–2.74 | 0.73–1.93 | 3.12–4.35 | 1.75–3.01 | 2.30–3.56 | 1.21–2.51 | 5.35–6.70 | 3.90–5.30 | 0.17–0.24 | 0.13–0.19 |
| E. pellita × E. brassiana (BC) | 3.04–4.25 | 0.85–2.10 | 4.68–5.96 | 1.94–3.26 | 3.26–4.57 | 1.48–2.83 | 6.33–7.73 | 3.32–4.78 | 0.17–0.23 | 0.10–0.16 |
| E. pellita × E. brassiana (F1) | 4.39–5.92 | 1.72–2.93 | 5.73–7.38 | 3.08–4.35 | 4.51–6.17 | 2.49–3.80 | 7.11–8.90 | 3.94–5.34 | 0.15–9.23 | 0.08–0.15 |
| E. pellita × E. urophylla (BC) | 1.74–2.94 | 0.82–2.02 | 3.33–4.59 | 2.22–3.49 | 2.37–3.67 | 1.52–2.82 | 5.33–6.73 | 3.70–5.10 | 0.17–0.24 | 0.11–0.17 |
| E. pellita × E. urophylla (F1) | 2.88–4.15 | 0.30–2.00 | 4.17–5.52 | 1.22–3.05 | 3.38–4.76 | 0.89–2.73 | 5.75–7.23 | 2.41–4.40 | 0.13–0.20 | 0.06–0.15 |
| Plantation Sites/Hybrids | Green Density (kg/m3) | Oven-Dry Density (kg/m3) | Basic Density (kg/m3) | |||
|---|---|---|---|---|---|---|
| T | R | T | R | T | R | |
| KI64 | ||||||
| E. pellita | 1034.00–1135.00 | 1010.00–1079.00 | 631.00–771.00 | 529.00–649.00 | 537.00–648.00 | 480.00–583.00 |
| E. pellita × E. brassiana (BC) | 1103.00–1210.00 | 1044.00–1127.00 | 694.00–838.00 | 579.00–716.00 | 585.00–701.00 | 514.00–631.00 |
| E. pellita × E. brassiana (F1) | 1067.00–1206.00 | 1043.00–1138.00 | 683.00–879.00 | 549.00–703.00 | 550.00–705.00 | 487.00–618.00 |
| E. pellita × E. urophylla (BC) | 1038.00–1137.00 | 1026.00–1089.00 | 625.00–766.00 | 532.00–641.00 | 533.00–644.00 | 487.00–581.00 |
| E. pellita × E. urophylla (F1) | 1032.00–1154.00 | 1025.00–1113.00 | 610.00–776.00 | 527.00–683.00 | 505.00–638.00 | 478.00–612.00 |
| YP28 | ||||||
| E. pellita | 1056.00–1150.00 | 1000.00–1083.00 | 621.00–743.00 | 531.00–633.00 | 533.00–625.00 | 484.00–568.00 |
| E. pellita × E. brassiana (BC) | 1105.00–1206.00 | 1064.00–1164.00 | 694.00–835.00 | 610.00–736.00 | 583.00–694.00 | 547.00–651.00 |
| E. pellita × E. brassiana (F1) | 1031.00–1185.00 | 1015.00–1115.00 | 634.00–851.00 | 569.00–694.00 | 512.00–681.00 | 499.00–603.00 |
| E. pellita × E. urophylla (BC) | 1071.00–1170.00 | 1011.00–1102.00 | 683.00–816.00 | 564.00–675.00 | 569.00–671.00 | 506.00–597.00 |
| E. pellita × E. urophylla (F1) | 1063.00–1170.00 | 1005.00–1204.00 | 641.00–790.00 | 542.00–784.00 | 547.00–664.00 | 492.00–691.00 |
| Plantation sites combined | ||||||
| E. pellita | 1062.00–1119.00 | 1008.00–1068.00 | 645.00–725.00 | 542.00–624.00 | 551.00–614.00 | 495.00–559.00 |
| E. pellita × E. brassiana (BC) | 1132.00–1192.00 | 1072.00–1134.00 | 746.00–830.00 | 624.00–709.00 | 618.00–684.00 | 558.00–625.00 |
| E. pellita × E. brassiana (F1) | 1097.00–1172.00 | 1054.00–1114.00 | 716.00–812.00 | 586.00–670.00 | 576.00–653.00 | 519.00–584.00 |
| E. pellita × E. urophylla (BC) | 1080.00–1139.00 | 1027.00–1087.00 | 679.00–772.00 | 558.00–650.00 | 571.00–643.00 | 507.00–580.00 |
| E. pellita × E. urophylla (F1) | 1092.00–1156.00 | 1041.00–1123.00 | 692.00–780.00 | 575.00–679.00 | 577.00–646.00 | 522.00–606.00 |
References
- Rhodes, D.; Stephens, M. Planted Forest Development in Australia and New Zealand: Comparative Trends and Future Opportunities. N. Z. J. For. Sci. 2014, 44, S10. [Google Scholar] [CrossRef]
- Lindenmayer, D.; Taylor, C. Diversifying Forest Landscape Management—A Case Study of a Shift from Native Forest Logging to Plantations in Australian Wet Forests. Land 2022, 11, 407. [Google Scholar] [CrossRef]
- Downham, R.; Gavran, M. Australian Plantation Statistics 2020 Update; Australian Bureau of Agricultural and Resource Economics and Sciences (ABARES): Canberra, Australia, 2020; p. 21.
- McGavin, R.L.; Leggate, W. Comparison of Processing Methods for Small-Diameter Logs: Sawing Versus Rotary Peeling. BioResources 2019, 14, 1545–1563. [Google Scholar] [CrossRef]
- Pliura, A.; Yu, Q.; Zhang, S.Y.; MacKay, J.; Périnet, P.; Bousquet, J. Variation in Wood Density and Shrinkage and Their Relationship to Growth of Selected Young Poplar Hybrid Crosses. For. Sci. 2005, 51, 472–482. [Google Scholar] [CrossRef]
- Sargent, R. Evaluating Dimensional Stability in Solid Wood: A Review of Current Practice. J. Wood Sci. 2019, 65, 36. [Google Scholar] [CrossRef]
- Japarudin, Y.; Meder, R.; Lapammu, M.; Waburton, P.; Paul-Macdonell, P.; Brown, M.; Brawner, J. Developing Eucalyptus pellita Breeding Populations for the Solid Wood Industry of Eastern Malaysia. J. Trop. For. Sci. 2022, 34, 347–358. [Google Scholar] [CrossRef]
- Japarudin, Y.; Lapammu, M.; Alwi, A.; Warburton, P.; Macdonell, P.; Boden, D.; Brawner, J.; Brown, M.; Meder, R. Growth Performance of Selected Taxa as Candidate Species for Productive Tree Plantations in Borneo. Aust. For. 2020, 83, 29–38. [Google Scholar] [CrossRef]
- de Oliveira Castro, C.A.; dos Santos, G.A.; Takahashi, E.K.; Pires Nunes, A.C.; Souza, G.A.; de Resende, M.D.V. Accelerating Eucalyptus Breeding Strategies through Top Grafting Applied to Young Seedlings. Ind. Crops Prod. 2021, 171, 113906. [Google Scholar] [CrossRef]
- Muneera Parveen, A.B.; Muthupandi, M.; Kumar, N.; Chauhan, S.S.; Vellaichamy, P.; Senthamilselvam, S.; Rajasugunasekar, D.; Nagarajan, B.; Mayavel, A.; Bachpai, V.K.W.; et al. Quantitative Genetic Analysis of Wood Property Traits in Biparental Population of Eucalyptus camaldulensis × E. tereticornis. J. Genet. 2021, 100, 46. [Google Scholar] [CrossRef] [PubMed]
- Simiqueli, G.F.; Resende, R.T.; Takahashi, E.K.; de Sousa, J.E.; Grattapaglia, D. Realized Genomic Selection across Generations in a Reciprocal Recurrent Selection Breeding Program of Eucalyptus Hybrids. Front. Plant Sci. 2023, 14, 1252504. [Google Scholar] [CrossRef]
- Prasetyo, A.; Aiso-Sanada, H.; Ishiguri, F.; Wahyudi, I.; Wijaya, I.P.G.; Ohshima, J.; Yokota, S. Growth Characteristics and Wood Properties of Two Interspecific Eucalyptus Hybrids Developed in Indonesia. For. Prod. J. 2018, 68, 436–444. [Google Scholar] [CrossRef]
- Van Duong, D.; Hasegawa, M.; Yamauchi, S.; Do, C.H. Within-Tree Variation Regarding Ultrasonic Velocity, Wood Density and Compressive Strength of the Eucalypt Hybrid (Eucalyptus urophylla × E. pellita). Wood Mater. Sci. Eng. 2024, 20, 420–426. [Google Scholar] [CrossRef]
- Ramage, M.H.; Burridge, H.; Busse-Wicher, M.; Fereday, G.; Reynolds, T.; Shah, D.U.; Wu, G.; Yu, L.; Fleming, P.; Densley-Tingley, D.; et al. The Wood from the Trees: The Use of Timber in Construction. Renew. Sustain. Energy Rev. 2017, 68, 333–359. [Google Scholar] [CrossRef]
- Hassan Vand, M.; Tippner, J.; Brabec, M. Effects of Species and Moisture Content on the Behaviour of Solid Wood under Impact. Eur. J. Wood Wood Prod. 2024, 82, 23–34. [Google Scholar] [CrossRef]
- Teodorescu, I.; Erbasu, R.; Branco, J.M.; Tăpuşi, D. Study in the Changes of the Moisture Content in Wood. IOP Conf. Ser. Earth Environ. Sci. 2021, 664, 012017. [Google Scholar] [CrossRef]
- Tan, B.; Ingvarsson, P.K. Integrating Genome-Wide Association Mapping of Additive and Dominance Genetic Effects to Improve Genomic Prediction Accuracy in Eucalyptus. Plant Genome 2022, 15, e20208. [Google Scholar] [CrossRef] [PubMed]
- Gion, J.-M.; Carouché, A.; Deweer, S.; Bedon, F.; Pichavant, F.; Charpentier, J.-P.; Baillères, H.; Rozenberg, P.; Carocha, V.; Ognouabi, N.; et al. Comprehensive Genetic Dissection of Wood Properties in a Widely-Grown Tropical Tree: Eucalyptus. BMC Genom. 2011, 12, 301. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-L.; Lindström, H.; Nakada, R.; Ralston, J. Cell Wall Structure and Wood Properties Determined by Acoustics—A Selective Review. Holz Als Roh-Und Werkst. 2003, 61, 321–335. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, W.; Zhang, Y.; Shi, J.; Shan, S.; Cai, L. Exploring Wood Micromechanical Structure: Impact of Microfibril Angle and Crystallinity on Cell Wall Strength. J. Build. Eng. 2024, 90, 109452. [Google Scholar] [CrossRef]
- Hamilton, M.G.; Potts, B.M.; Greaves, B.L.; Dutkowski, G.W. Genetic Correlations between Pulpwood and Solid-Wood Selection and Objective Traits in Eucalyptus globulus. Ann. For. Sci. 2010, 67, 511. [Google Scholar] [CrossRef][Green Version]
- Nickolas, H.; Williams, D.; Downes, G.; Tilyard, P.; Harrison, P.A.; Vaillancourt, R.E.; Potts, B. Genetic Correlations among Pulpwood and Solid-Wood Selection Traits in Eucalyptus globulus. New For. 2020, 51, 137–158. [Google Scholar] [CrossRef]
- Ho, T.X.; Schimleck, L.R.; Sinha, A. Utilization of Genetic Algorithms to Optimize Eucalyptus globulus Pulp Yield Models Based on Nir Spectra. Wood Sci. Technol. 2021, 55, 757–776. [Google Scholar] [CrossRef]
- Kombi Kaviriri, D.; Liu, H.; Zhao, X. Estimation of Genetic Parameters and Wood Yield Selection Index in a Clonal Trial of Korean Pine (Pinus koraiensis) in Northeastern China. Sustainability 2021, 13, 4167. [Google Scholar] [CrossRef]
- Nayanathara, R.M.O.; Leng, W.; Street, J.; Zhang, X. Wood Dimensional Stability Enhancement by Multivalent Metal-Cation-Induced Lignocellulosic Microfibrils Crosslinking. Int. J. Biol. Macromol. 2024, 269, 131877. [Google Scholar] [CrossRef] [PubMed]
- Climent, J.; Alía, R.; Karkkainen, K.; Bastien, C.; Benito-Garzon, M.; Bouffier, L.; De Dato, G.; Delzon, S.; Dowkiw, A.; Elvira-Recuenco, M.; et al. Trade-Offs and Trait Integration in Tree Phenotypes: Consequences for the Sustainable Use of Genetic Resources. Curr. For. Rep. 2024, 10, 196–222. [Google Scholar] [CrossRef]
- Jurcic, E.J.; Villalba, P.V.; Dutour, J.; Centurión, C.; Munilla, S.; Cappa, E.P. Breeding Value Predictive Accuracy for Scarcely Recorded Traits in a Eucalyptus grandis Breeding Population Using Genomic Selection and Data on Predictor Traits. Tree Genet. Genomes 2023, 19, 35. [Google Scholar] [CrossRef]
- Marco de Lima, B.; Cappa, E.P.; Silva-Junior, O.B.; Garcia, C.; Mansfield, S.D.; Grattapaglia, D. Quantitative Genetic Parameters for Growth and Wood Properties in Eucalyptus “Urograndis” Hybrid Using near-Infrared Phenotyping and Genome-Wide Snp-Based Relationships. PLoS ONE 2019, 14, e0218747. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Jiang, L.; Xu, G.; Li, J.; Wang, B.; Li, Y.; Zhao, X. Evaluation and Selection Analyses of 60 Larix Kaempferi Clones in Four Provenances Based on Growth Traits and Wood Properties. Tree Genet. Genomes 2020, 16, 1–11. [Google Scholar] [CrossRef]
- Sargent, R. Evaluating Dimensional Stability in Modified Wood: An Experimental Comparison of Test Methods. Forests 2022, 13, 613. [Google Scholar] [CrossRef]
- Shen, X.; Yang, S.; Li, G.; Liu, S.; Chu, F. The Contribution Mechanism of Furfuryl Alcohol Treatment on the Dimensional Stability of Plantation Wood. Ind. Crops Prod. 2022, 186, 115143. [Google Scholar] [CrossRef]
- Owoyemi, J.M.; Falade, O.E.; Iyiola, E.A.; Oladapo, O.D. An Analysis of Impact of Furfurylation Treatments on the Physical and Mechanical Properties of Pterygota macrocarpa Wood. J. Mater. Sci. Res. Rev. 2023, 6, 6–23. [Google Scholar]
- Cao, Y.; Li, X.; Liu, L.; Xie, G.; Lai, M.; Gao, J. Increased Dimensional Stability of Eucalyptus grandis × Eucalyptus urophylla ‘Glgu9’ Wood through Palm Oil Thermal Treatment. BioResources 2023, 18, 3471–3478. [Google Scholar] [CrossRef]
- He, L.; Zhang, T.; Zhao, Y.; Gao, J.; Zhang, Y.; Yang, Y.; He, Z.; Yi, S. Effect of Natural Tung Oil on Wood Shrinkage During the Thermal Modification Process. J. Clean. Prod. 2022, 379, 134450. [Google Scholar] [CrossRef]
- Tibbits, J.F.G.; McManus, L.J.; Spokevicius, A.V.; Bossinger, G. A Rapid Method for Tissue Collection and High-Throughput Isolation of Genomic DNA from Mature Trees. Plant Mol. Biol. Rep. 2006, 24, 81–91. [Google Scholar] [CrossRef]
- Kingston, R.S.T.; Risdon, C.J.E. Shrinkage and Density of Australian and Other South-West Pacific Woods; Commonwealth Scientific and Industrial Research Organization: Melbourne, VIC, Australia, 1961; pp. 1–23. [Google Scholar]
- ASTM D2395-17; Standard Test Methods for Density and Specific Gravity (Relative Density) of Wood and Wood-Based Materials. ASTM: West Conshohocken, PA, USA, 2017.
- Poke, F.S.; Raymond, C.A. Predicting Extractives, Lignin, and Cellulose Contents Using near Infrared Spectroscopy on Solid Wood in Eucalyptus globulus. J. Wood Chem. Technol. 2006, 26, 187–199. [Google Scholar] [CrossRef]
- Downes, G.M.; Meder, R.; Bond, H.; Ebdon, N.; Hicks, C.; Harwood, C. Measurement of Cellulose Content, Kraft Pulp Yield and Basic Density in Eucalypt Woodmeal Using Multisite and Multispecies near Infra-Red Spectroscopic Calibrations. South. For. 2011, 73, 181–186. [Google Scholar] [CrossRef]
- Gondwana Genomics. Maximise Your Growth, Density, Pulp Yield. Available online: https://gondwanagenomics.com.au/ (accessed on 24 May 2025).
- Tang, Y.; Liu, X.; Wang, J.; Li, M.; Wang, Q.; Tian, F.; Su, Z.; Pan, Y.; Liu, D.; Lipka, A.E.; et al. Gapit Version 2: An Enhanced Integrated Tool for Genomic Association and Prediction. Plant Genome 2016, 9, plantgenome2015.2011.0120. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. Blink: A Package for the Next Level of Genome-Wide Association Studies with Both Individuals and Markers in the Millions. GigaScience 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gao, Y.; Fu, Z.; Zhou, Y.; Gao, X.; Zhou, F.; Cao, H. Moisture-Related Shrinkage Behavior of Wood at Macroscale and Cellular Level. Polymers 2022, 14, 5045. [Google Scholar] [CrossRef] [PubMed]
- Gezici-Koç, Ö.; Erich, S.J.F.; Huinink, H.P.; van der Ven, L.G.J.; Adan, O.C.G. Bound and Free Water Distribution in Wood During Water Uptake and Drying as Measured by 1d Magnetic Resonance Imaging. Cellulose 2017, 24, 535–553. [Google Scholar] [CrossRef]
- Baillères, H.; Hopewell, G.; McGavin, R.L. Evaluation of Wood Characteristics of Tropical Post-Mid Rotation Plantation Eucalyptus cloeziana and E. pellita: Part (C) Wood Quality and Structural Properties; PN07.3022; Forest & Wood Product Australia: Melbourne, VIC, Australia, 2008. [Google Scholar]
- WoodSolutions. Mahogany, Red—Overview. Available online: https://www.woodsolutions.com.au/wood-species/hardwood/mahogany-red (accessed on 10 November 2023).
- Sattar, M.A.; Bhattacharjee, D.K.; Kabir, M.F. Physical and Mechanical Properties and Uses of Timbers of Bangladesh; Bangladesh Forest Research Institute: Chittagong, Bangladesh; Government of the People’s Republic of Bangladesh: Dhaka, Bangladesh, 1999; pp. 1–57.
- Kabir, M.F.; Sattar, M.A. Strength Properties and Drying Characteristics of Eucalyptus Wood Grown in Bandladesh. Thai J. For. 1995, 14, 103–109. [Google Scholar]
- Júnior, L.S.; Garcia, J.N. Physical and Mechanical Properties of Eucalyptus urophylla. Braz. J. Agric. 2023, 98, 12–22. [Google Scholar] [CrossRef]
- Wu, Y.-Q.; Hayashi, K.; Liu, Y.; Cai, Y.; Sugimori, M. Relationships of Anatomical Characteristics Versus Shrinkage and Collapse Properties in Plantation-Grown Eucalypt Wood from China. J. Wood Sci. 2006, 52, 187–194. [Google Scholar] [CrossRef]
- Thomson, A.B. Shrinkage, Collapse and Dimensional Recovery of Regrowth Jarrah; Wood Utilisation Reserach Centre: Kensington, WA, Australia, 1989; pp. 1–11. [Google Scholar]
- Yang, L.; Liu, H. A Review of Eucalyptus Wood Collapse and Its Control During Drying. BioResources 2018, 13, 2171–2181. [Google Scholar] [CrossRef]
- Gonya, N.A.S.; Naghizadeh, Z.; Wessels, C.B. An Investigation into Collapse and Shrinkage Behaviour of Eucalyptus grandis and Eucalyptus grandis-urophylla Wood. Eur. J. Wood Wood Prod. 2022, 80, 139–157. [Google Scholar] [CrossRef]
- Blakemore, P.; Langrish, T.A.G. Effect of Mean Moisture Content on the Steam Reconditioning of Collapsed Eucalyptus regnans. Wood Sci. Technol. 2007, 41, 87–98. [Google Scholar] [CrossRef]
- Ibarra, L.; Hodge, G.; Acosta, J.J. Quantitative Genetics of a Hybrid Population of Eucalyptus nitens × Eucalyptus globulus: Estimation of Genetic Parameters and Implications for Breeding Strategies. Forests 2023, 14, 381. [Google Scholar] [CrossRef]
- Parveen, A.B.M.; Jayabharathi, K.; Muthupandi, M.; Kumar, N.; Chauhan, S.S.; Rajasugunasekar, D.; Dasgupta, M.G. Identification of Superior Hybrid Clones for Fibre Biometry in Eucalyptus camaldulensis × E. tereticornis Using Multi Trait Stability Index. Silvae Genet. 2024, 73, 126–141. [Google Scholar] [CrossRef]
- Japarudin, Y.; Meder, R.; Lapammu, M.; Alwi, A.; Ghaffariyan, M.; Brown, M. Compression and Flexural Properties of Plantation-Grown Eucalyptus pellita in Borneo, Malaysia. Potential for Structural Timber End Use. Aust. For. 2021, 84, 139–151. [Google Scholar] [CrossRef]
- Hii, S.Y.; Ha, K.S.; Ngui, M.L.; Ak Penguang, S.; Duju, A.; Teng, X.Y.; Meder, R. Assessment of Plantation-Grown Eucalyptus pellitain Borneo, Malaysia for Solid Wood Utilisation. Aust. For. 2017, 80, 26–33. [Google Scholar] [CrossRef]
- Bergman, R. Drying and Control of Moisture Content and Dimensional Changes. In Wood Handbook; U.S. Department of Agriculture: Washington, DC, USA, 2021; pp. 1–21. [Google Scholar]
- Li, F.; Qian, H.; Sardans, J.; Amishev, D.Y.; Wang, Z.; Zhang, C.; Wu, T.; Xu, X.; Tao, X.; Huang, X. Evolutionary History Shapes Variation of Wood Density of Tree Species across the World. Plant Divers. 2024, 46, 283–293. [Google Scholar] [CrossRef]
- Downes, G.M.; Lausberg, M.; Potts, B.M.; Pilbeam, D.L.; Bird, M.; Bradshaw, B. Application of the Iml Resistograph to the Infield Assessment of Basic Density in Plantation Eucalypts. Aust. For. 2018, 81, 177–185. [Google Scholar] [CrossRef]
- Drew, D.M.; Downes, G.M.; Seifert, T.; Eckes-Shepard, A.; Achim, A. A Review of Progress and Applications in Wood Quality Modelling. Curr. For. Rep. 2022, 8, 317–332. [Google Scholar] [CrossRef]
- Elissetche, J.P.; Alzamora, R.M.; Espinoza, Y.; Emhart, V.; Pincheira, M.; Medina, A.; Rubilar, R. Wood Basic Density Assessment of Eucalyptus Genotypes Growing under Contrasting Water Availability Conditions. Forests 2024, 15, 185. [Google Scholar] [CrossRef]
- Schulgasser, K.; Witztum, A. How the Relationship between Density and Shrinkage of Wood Depends on Its Microstructure. Wood Sci. Technol. 2015, 49, 389–401. [Google Scholar] [CrossRef]
- Pulgar, J.A.; Riesco, G. Implementing Linear Mixed Effects Models to Enhance Estimation of the Dimensional Stability of Wood of Laurus nobilis L. For. Syst. 2024, 33, e05. [Google Scholar] [CrossRef]
- Listyanto, T.; Nichols, J.D.; Glencross, K.; Schoer, L. Variation in Density and Shrinkage of Six Eucalyptus Species and Interspecific Hybrid Combinations at Three Sites in Northern New South Wales. IOP Conf. Ser. Mater. Sci. Eng. 2020, 935, 012035. [Google Scholar] [CrossRef]
- Lachenbruch, B.; Moore, J.; Evans, R. Radial Variation in Wood Structure and Function in Woody Plants, and Hypotheses for Its Occurrence. In Size- and Age-Related Changes in Tree Structure and Function; Springer: Dordrecht, The Netherlands, 2011; Volume 4, pp. 121–164. [Google Scholar]
- Xiang, W.; Leitch, M.; Auty, D.; Duchateau, E.; Achim, A. Radial Trends in Black Spruce Wood Density Can Show an Age- and Growth-Related Decline. Ann. For. Sci. 2014, 71, 603–615. [Google Scholar] [CrossRef]
- Dhaka, R.K.; Gunaga, R.P.; Sinha, S.K.; Thakur, N.S.; Dobriyal, M.J. Influence of Tree Height and Diameter on Wood Basic Density, Cellulose and Fibre Characteristics in Melia Dubia Cav. Families. J. Indian Acad. Wood Sci. 2020, 17, 138–144. [Google Scholar] [CrossRef]
- Rocha, M.F.V.; Veiga, T.R.L.A.; Soares, B.C.D.; Araújo, A.C.C.d.; Carvalho, A.M.M.; Hein, P.R.G. Do the Growing Conditions of Trees Influence the Wood Properties? Floresta E Ambient. 2019, 26, e20180353. [Google Scholar] [CrossRef]
- Kumar, A.; Jyske, T.; Petrič, M. Delignified Wood from Understanding the Hierarchically Aligned Cellulosic Structures to Creating Novel Functional Materials: A Review. Adv. Sustain. Syst. 2021, 5, 2000251. [Google Scholar] [CrossRef]
- Azmul Huda, A.S.M.; Koubaa, A.; Cloutier, A.; Hernandez, R.E.; Perinet, P.; Fortin, Y. Phenotypic and Genotypic Correlations for Wood Properties of Hybrid Poplar Clones of Southern Quebec. Forests 2018, 9, 140. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Ren, H.; Jiang, Z. Wood Density and Wood Shrinkage in Relation to Initial Spacing and Tree Growth in Black Spruce (Picea mariana). J. Wood Sci. 2021, 67, 30. [Google Scholar] [CrossRef]
- Gierlinger, N. New Insights into Plant Cell Walls by Vibrational Microspectroscopy. Appl. Spectrosc. Rev. 2018, 53, 517–551. [Google Scholar] [CrossRef]
- Kocaefe, D.; Huang, X.; Kocaefe, Y. Dimensional Stabilization of Wood. Curr. For. Rep. 2015, 1, 151–161. [Google Scholar] [CrossRef]
- Ballesta, P.; Bush, D.; Silva, F.F.; Mora, F. Genomic Predictions Using Low-Density Snp Markers, Pedigree and Gwas Information: A Case Study with the Non-Model Species Eucalyptus cladocalyx. Plants 2020, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lan, J.; Wang, J.; He, W.; Lu, W.; Lin, Y.; Luo, J. Population Structure and Genetic Diversity in Eucalyptus pellita Based on Snp Markers. Front. Plant Sci. 2023, 14, 1278427. [Google Scholar] [CrossRef] [PubMed]
- Lehnebach, R.; Bossu, J.; Va, S.; Morel, H.; Amusant, N.; Nicolini, E.; Beauchêne, J. Wood Density Variations of Legume Trees in French Guiana Along the Shade Tolerance Continuum: Heartwood Effects on Radial Patterns and Gradients. Forests 2019, 10, 80. [Google Scholar] [CrossRef]
- Raymond, C.A. Genetics of Eucalyptus Wood Properties. Ann. For. Sci. 2002, 59, 525–531. [Google Scholar] [CrossRef]
- Bano, N.; Mohammad, N.; Ansari, M.I.; Ansari, S.A. Discovery of Snps in Lignin Biosynthesis Genes (Cad1, Myb1 and Myb2) and Their Association with Wood Density in Teak. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Ma, L.; Yang, M. Identification and Characterization of Long Non-Coding Rna (Lncrna) in the Developing Seeds of Jatropha curcas. Sci. Rep. 2020, 10, 10395. [Google Scholar] [CrossRef]
- Terrett, O.M.; Dupree, P. Covalent Interactions between Lignin and Hemicelluloses in Plant Secondary Cell Walls. Curr. Opin. Biotechnol. 2019, 56, 97–104. [Google Scholar] [CrossRef]
- Kovryga, A.; Stapel, P.; van de Kuilen, J.W.G. Mechanical Properties and Their Interrelationships for Medium-Density European Hardwoods, Focusing on Ash and Beech. Wood Mater. Sci. Eng. 2020, 15, 289–302. [Google Scholar] [CrossRef]
- Nenning, T.; Tockner, A.; Konnerth, J.; Gindl-Altmutter, W.; Grabner, M.; Hansmann, C.; Lux, S.; Pramreiter, M. Variability of Mechanical Properties of Hardwood Branches According to Their Position and Inclination in the Tree. Constr. Build. Mater. 2024, 419, 135448. [Google Scholar] [CrossRef]
- Du, Q.; Lu, W.; Quan, M.; Xiao, L.; Song, F.; Li, P.; Zhou, D.; Xie, J.; Wang, L.; Zhang, D. Genome-Wide Association Studies to Improve Wood Properties: Challenges and Prospects. Front. Plant Sci. 2018, 9, 1912. [Google Scholar] [CrossRef]
- Estopa, R.A.; Paludeto, J.G.Z.; Müller, B.S.F.; de Oliveira, R.A.; Azevedo, C.F.; de Resende, M.D.V.; Tambarussi, E.V.; Grattapaglia, D. Genomic Prediction of Growth and Wood Quality Traits in Eucalyptus benthamii Using Different Genomic Models and Variable Snp Genotyping Density. New For. 2023, 54, 343–362. [Google Scholar] [CrossRef]
- Duarte, D.; Jurcic, E.J.; Dutour, J.; Villalba, P.V.; Centurión, C.; Grattapaglia, D.; Cappa, E.P. Genomic Selection in Forest Trees Comes to Life: Unraveling Its Potential in an Advanced Four-Generation Eucalyptus grandis Population. Front. Plant Sci. 2024, 15, 1462285. [Google Scholar] [CrossRef]
- Giorello, F.M.; Farias, J.; Basile, P.; Balmelli, G.; Da Silva, C.C. Evaluating the Potential of Xp-Gwas in Eucalyptus: Leaf Heteroblasty as a Case Study. Plant Gene 2023, 36, 100430. [Google Scholar] [CrossRef]
- Rocha, L.F.; Benatti, T.R.; de Siqueira, L.; de Souza, I.C.G.; Bianchin, I.; de Souza, A.J.; Fernandes, A.C.M.; Oda, S.; Stape, J.L.; Yassue, R.M.; et al. Quantitative Trait Loci Related to Growth and Wood Quality Traits in Eucalyptus grandis W. Hill Identified through Single- and Multi-Trait Genome-Wide Association Studies. Tree Genet. Genomes 2022, 18, 38. [Google Scholar] [CrossRef]
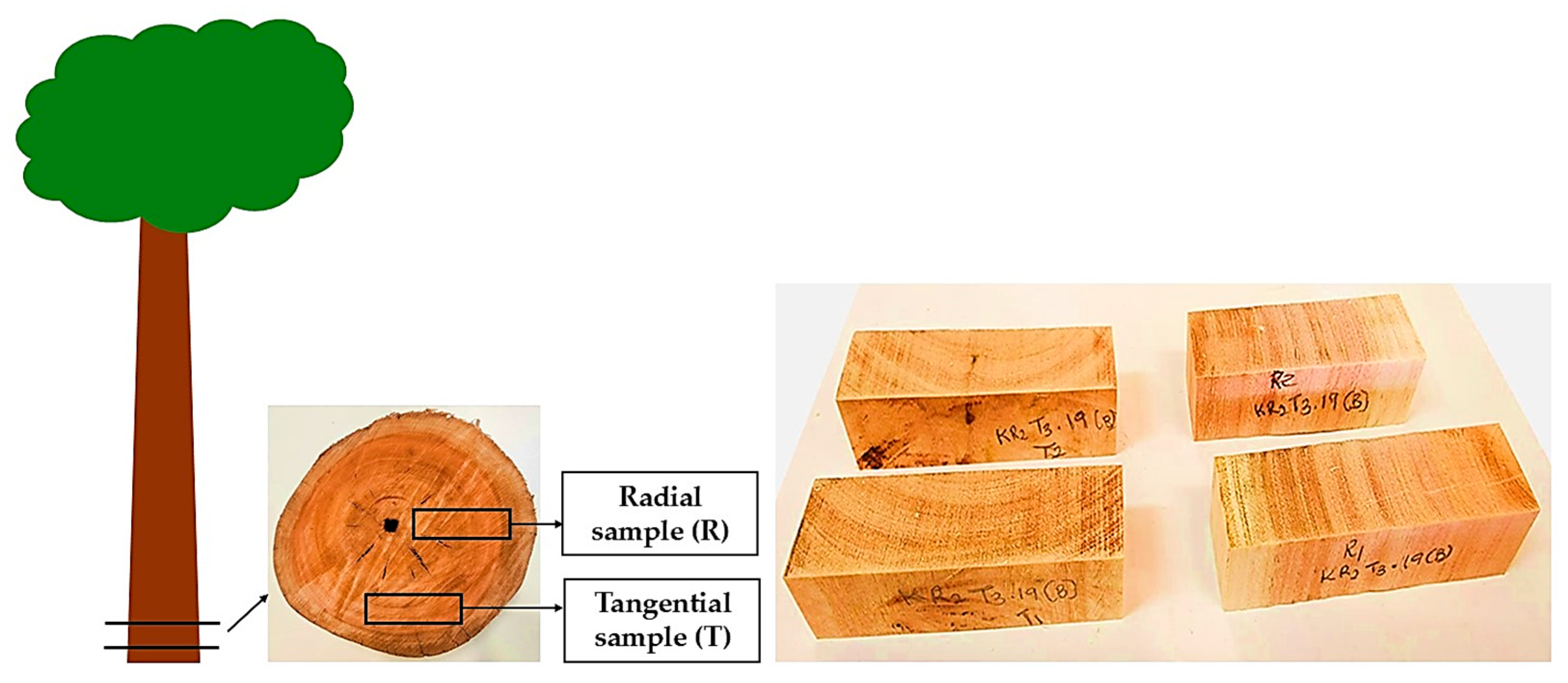
| Kilu Impini 64 | Yapilika 28 | |
| Coordinates | 11°25′6.00″ S | 11°33′45.00″ S |
| 130°30′0.00″ E | 130°34′39.00″ E | |
| Rainfall | 1200–1400 mm | 1200–1400 mm |
| Number of seedlots | 10 | 10 |
| Year planted | 2012 | 2012 |
| Number of trees planted | 1000 | 1000 |
| Number of living trees (Year 2022) | 763 | 666 |
| Soil | Red sandy soils and gray to yellow sandy soil > 2 m | Moonkinu Member sandstone > 3 m |
| Tree/plot | 25 | 25 |
| Replications | 4 | 4 |
| Spacing | 3 × 3 m | 3 × 3 m |
| Hybrid/Genotype Group | Total Genotyped | Kilu Impini 64 | Yapilika 28 | Selected Trees: Kilu Impini 64 | Selected Trees: Yapilika 28 |
|---|---|---|---|---|---|
| E. pellita | 858 | 456 | 402 | 6 | 6 |
| E. pellita × E. brassiana (BC) | 97 | 59 | 38 | 6 | 6 |
| E. pellita × E. brassiana (F1) | 25 | 17 | 8 | 6 | 5 |
| E. pellita × E. urophylla (BC) | 313 | 160 | 153 | 6 | 6 |
| E. pellita × E. urophylla (F1) | 136 | 71 | 65 | 6 | 5 |
| Total | 1429 | 763 | 666 | 30 | 28 |
| Abbreviations | Full Meaning |
|---|---|
| Physical properties traits | |
| Sh17_T | Shrinkage from green to 17% MC in tangential direction |
| Sh17_R | Shrinkage from green to 17% MC in radial direction |
| Sh12BR_T | Shrinkage from green to 12% MC before reconditioning in tangential direction |
| Sh12BR_R | Shrinkage from green to 12% MC before reconditioning in radial direction |
| Sh12AR_T | Shrinkage from green to 12% MC after reconditioning in tangential direction |
| Sh12AR_R | Shrinkage from green to 12% MC after reconditioning in radial direction |
| Sh5_T | Shrinkage from green to 5% MC in tangential direction |
| Sh5_R | Shrinkage from green to 5% MC in radial direction |
| US_T | Unit shrinkage in tangential direction |
| US_R | Unit shrinkage in radial direction |
| GD_T | Green density at tangential direction |
| GD_R | Green density in radial direction |
| OD_T | Oven-dry density in tangential direction |
| OD_R | Oven-dry density in radial direction |
| BD_T | Basic density in tangential direction |
| BD_R | Basic density in radial direction |
| Growth traits | |
| Ht | Height |
| Dbh_t | Diameter at breast height measured with tape |
| Dbh_r | Diameter at breast height measured with Resistograph |
| Chemical component traits | |
| KPY | Kraft pulp yield |
| TL | Total lignin |
| KL | Klason lignin |
| Ext | Extractives |
| Cel | Cellulose |
| Plantation Sites/Hybrids | Green to 17% (%) | Green to 12% BR (%) | Green to 12% AR (%) | Green to 5% (%) | Unit Shrinkage (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| T | R | T | R | T | R | T | R | T | R | |
| KI64 | ||||||||||
| E. pellita | 2.31 a (0.56) | 1.20 a (0.24) | 4.00 a (0.57) | 2.16 a (0.41) | 3.00 a (0.54) | 1.45 a (0.40) | 6.14 a (0.44) | 4.67 a (0.63) | 0.20 a (0.02) | 0.18 a (0.02) |
| E. pellita × E. brassiana (BC) | 3.86 a (0.62) | 1.60 a (0.29) | 5.27 a (0.62) | 2.71 a (0.49) | 4.34 a (0.61) | 2.34 a (0.47) | 6.67 a (0.47) | 4.58 a (0.74) | 0.19 a (0.02) | 0.15 a (0.03) |
| E. pellita × E. brassiana (F1) | 4.09 a (0.71) | 1.75 a (0.26) | 5.70 a (0.75) | 3.30 a (0.44) | 4.26 a (0.69) | 2.60 a (0.43) | 7.49 a (0.59) | 4.79 a (0.68) | 0.21 a (0.02) | 0.13 a (0.03) |
| E. pellita × E. urophylla (BC) | 2.33 a (0.54) | 1.49 a (0.22) | 3.95 a (0.55) | 2.95 a (0.38) | 2.96 a (0.52) | 2.02 a (0.36) | 5.76 a (0.43) | 4.09 a (0.58) | 0.19 a (0.02) | 0.13 a (0.02) |
| E. pellita × E. urophylla (F1) | 3.48 a (0.65) | 1.42 a (0.30) | 5.05 a (0.68) | 2.36 a (0.51) | 4.05 a (0.63) | 1.99 a (0.49) | 6.65 a (0.53) | 3.27 a (0.77) | 0.16 a (0.02) | 0.08 a (0.03) |
| LSD (p = 0.05) | 2.14 | 0.85 | 2.18 | 1.45 | 2.08 | 1.40 | 1.69 | 2.22 | 0.07 | 0.10 |
| LSD (p = 0.05)—Tukey | 3.15 | 1.22 | 3.16 | 2.08 | 3.06 | 2.01 | 2.42 | 3.18 | 0.10 | 0.14 |
| p-value | 0.15 | 0.62 | 0.23 | 0.41 | 0.27 | 0.41 | 0.20 | 0.60 | 0.68 | 0.24 |
| YP28 | ||||||||||
| E. pellita | 2.01 a (0.42) | 1.42 a (0.17) | 3.54 a (0.41) | 2.56 a (0.26) | 2.86 a (0.37) | 2.13 a (0.26) | 6.04 a (0.34) | 4.69 a (0.47) | 0.21 a (0.01) | 0.15 a (0.03) |
| E. pellita × E. brassiana (BC) | 3.29 a (0.55) | 1.36 a (0.21) | 4.88 a (0.51) | 2.51 a (0.33) | 3.11 a (0.44) | 1.87 a (0.31) | 6.93 a (0.43) | 3.54 a (0.56) | 0.22 a (0.01) | 0.11 a (0.03) |
| E. pellita × E. brassiana (F1) | 6.83 b (0.78) | 3.01 b (0.21) | 8.06 b (0.75) | 4.21 a (0.33) | 7.09 b (0.67) | 3.84 a (0.31) | 8.70 b (0.64) | 4.56 a (0.56) | 0.11 b (0.02) | 0.09 a (0.03) |
| E. pellita × E. urophylla (BC) | 2.30 a (0.46) | 1.29 a (0.18) | 3.92 a (0.45) | 2.68 a (0.27) | 2.99 a (0.41) | 2.28 a (0.28) | 6.28 a (0.38) | 4.59 a (0.51) | 0.22 a (0.01) | 0.16 a (0.03) |
| E. pellita × E. urophylla (F1) | 3.63 a (0.55) | 1.57 a (0.39) | 4.88 a (0.53) | 2.44 a (0.60) | 3.94 a (0.47) | 2.14 a (0.62) | 6.47 a (0.45) | 4.85 a (1.21) | 0.16 a (0.01) | 0.18 a (0.06) |
| LSD (p = 0.05) | 2.00 | 0.94 | 1.93 | 1.44 | 1.72 | 1.46 | 1.63 | 2.65 | 0.05 | 0.15 |
| LSD (p = 0.05)—Tukey | 2.88 | 1.35 | 2.78 | 2.08 | 2.49 | 2.11 | 2.35 | 3.84 | 0.07 | 0.22 |
| p-value | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.02 | 0.55 | 0.00 | 0.45 |
| Plantation combined | ||||||||||
| E. pellita | 2.16 a (0.28) | 1.33 a (0.29) | 3.74 a (0.30) | 2.38 a (0.31) | 2.93 a (0.30) | 1.86 a (0.32) | 6.03 a (0.33) | 4.60 a (0.34) | 0.20 a (0.01) | 0.16 a (0.02) |
| E. pellita × E. brassiana (BC) | 3.64 b (0.29) | 1.48 a (0.31) | 5.32 b (0.31) | 2.60 a (0.33) | 3.91 b (0.32) | 2.16 a (0.33) | 7.03 a (0.34) | 4.05 a (0.36) | 0.20 a (0.02) | 0.13 a (0.02) |
| E. pellita × E. brassiana (F1) | 5.15 c (0.38) | 2.33 a (0.29) | 6.55 c (0.41) | 3.72 b (0.31) | 5.34 c (0.41) | 3.15 a (0.32) | 8.00 b (0.45) | 4.64 a (0.34) | 0.19 a (0.02) | 0.11 a (0.02) |
| E. pellita × E. urophylla (BC) | 2.34 a (0.29) | 1.42 a (0.29) | 3.96 a (0.31) | 2.85 a (0.31) | 3.02 a (0.31) | 2.17 a (0.31) | 6.03 a (0.34) | 4.40 a (0.34) | 0.21 a (0.02) | 0.14 a (0.02) |
| E. pellita × E. urophylla (F1) | 3.52 b (0.29) | 1.15 a (0.42) | 4.84 b (0.33) | 2.14 a (0.46) | 4.07 b (0.34) | 1.81 a (0.46) | 6.49 a (0.36) | 3.41 a (0.50) | 0.16 a (0.02) | 0.11 a (0.02) |
| LSD (p = 0.05) | 0.97 | 1.03 | 1.04 | 1.11 | 1.04 | 1.11 | 1.13 | 1.21 | 0.05 | 0.05 |
| LSD (p = 0.05)—Tukey | 1.36 | 1.45 | 1.46 | 1.56 | 1.45 | 1.55 | 1.59 | 1.69 | 0.07 | 0.07 |
| Site | 0.15 | 0.31 | 0.14 | 0.36 | 0.96 | |||||
| Hybrid | 0.00 | 0.00 | 0.00 | 0.08 | 0.28 | |||||
| Position | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| Site × Hybrid | 0.03 | 0.18 | 0.04 | 0.70 | 0.25 | |||||
| Site × Position | 0.79 | 0.92 | 0.75 | 0.61 | 0.71 | |||||
| Hybrid × Position | 0.00 | 0.00 | 0.04 | 0.00 | 0.33 | |||||
| Site × Hybrid × Position | 0.50 | 0.49 | 0.45 | 0.38 | 0.17 | |||||
| Plantation Sites/Hybrids | Green Density (kg/m3) | Oven-Dry Density (kg/m3) | Basic Density (kg/m3) | |||
|---|---|---|---|---|---|---|
| T | R | T | R | T | R | |
| KI64 | ||||||
| E. pellita | 1085.00 a (24.40) | 1044.00 a (16.60) | 701.00 a (33.80) | 589.00 a (28.80) | 593.00 a (26.80) | 531.00 a (24.60) |
| E. pellita × E. brassiana (BC) | 1156.00 a (25.90) | 1085.00 a (19.50) | 766.00 a (34.80) | 647.00 a (32.80) | 643.00 a (27.90) | 573.00 a (27.80) |
| E. pellita × E. brassiana (F1) | 1136.00 a (33.40) | 1090.00 a (17.90) | 781.00 a (47.20) | 626.00 a (33.30) | 627.00 a (37.20) | 553.00 a (28.60) |
| E. pellita × E. urophylla (BC) | 1087.00 a (23.80) | 1058.00 a (15.10) | 695.00 a (33.80) | 587.00 a (26.30) | 588.00 a (26.50) | 534.00 a (22.40) |
| E. pellita × E. urophylla (F1) | 1093.00 a (29.40) | 1069.00 a (20.30) | 693.00 a (40.20) | 605.00 a (36.60) | 571.00 a (32.10) | 545.00 a (31.30) |
| LSD (p = 0.05) | 93.64 | 58.12 | 129.40 | 110.15 | 102.65 | 93.85 |
| LSD (p = 0.05)—Tukey | 134.15 | 83.30 | 184.90 | 160.85 | 146.75 | 136.75 |
| p-value | 0.23 | 0.35 | 0.36 | 0.60 | 0.41 | 0.81 |
| YP28 | ||||||
| E. pellita | 1103.00 a (22.20) | 1041.00 a (19.80) | 682.00 a (28.80) | 582.00 a (24.30) | 579.00 a (22.00) | 526.00 a (20.00) |
| E. pellita × E. brassiana (BC) | 1155.00 a (23.80) | 1114.00 a (23.60) | 765.00 a (33.40) | 673.00 a (29.40) | 638.00 a (26.40) | 599.00 a (24.40) |
| E. pellita × E. brassiana (F1) | 1108.00 a (35.90) | 1065.00 a (23.70) | 742.00 a (51.20) | 631.00 a (29.50) | 596.00 a (40.10) | 551.00 a (24.50) |
| E. pellita × E. urophylla (BC) | 1120.00 a (23.60) | 1056.00 a (21.30) | 750.00 a (31.60) | 619.00 a (26.00) | 620.00 a (24.20) | 551.00 a (21.3) |
| E. pellita × E. urophylla (F1) | 1117.00 a (25.40) | 1104.00 a (46.90) | 716.00 a (35.50) | 663.00 a (57.20) | 606.00 a (27.80) | 591.00 a (47.00) |
| LSD (p = 0.05) | 93.50 | 111.21 | 131.62 | 135.95 | 102.75 | 111.85 |
| LSD (p = 0.05)—Tukey | 135.40 | 160.55 | 189.90 | 196.10 | 148.05 | 181.30 |
| p-value | 0.50 | 0.21 | 0.37 | 0.22 | 0.52 | 0.24 |
| Plantation sites combined | ||||||
| E. pellita | 1090.00 a (13.90) | 1038.00 a (14.40) | 685.00 a (19.30) | 583.00 a (19.70) | 583.00 a (15.20) | 527.00 a (15.50) |
| E. pellita × E. brassiana (BC) | 1162.00 b (14.60) | 1103.00 a (15.10) | 788.00 b (20.20) | 666.00 a (20.60) | 651.00 a (15.90) | 591.00 a (16.30) |
| E. pellita × E. brassiana (F1) | 1135.00 b (18.50) | 1084.00 a (14.60) | 764.00 a (23.60) | 628.00 a (20.20) | 615.00 a (19.10) | 552.00 a (15.90) |
| E. pellita × E. urophylla (BC) | 1109.00 a (14.40) | 1057.00 a (14.40) | 726.00 a (21.90) | 604.00 a (21.90) | 607.00 a (17.10) | 543.00 a (17.10) |
| E. pellita × E. urophylla (F1) | 1124.00 a (15.50) | 1082.00 a (20.50) | 736.00 a (21.40) | 627.00 a (25.80) | 612.00 a (16.80) | 564.00 a (20.90) |
| LSD (p = 0.05) | 47.55 | 50.35 | 64.38 | 67.52 | 51.16 | 53.86 |
| LSD (p = 0.05)—Tukey | 66.67 | 70.57 | 90.45 | 94.80 | 71.83 | 75.55 |
| Site | 0.36 | 0.63 | 0.69 | |||
| Hybrid | 0.01 | 0.03 | 0.06 | |||
| Position | 0.00 | 0.00 | 0.00 | |||
| Site × Hybrid | 0.86 | 0.83 | 0.91 | |||
| Site × Position | 0.95 | 0.34 | 0.35 | |||
| Hybrid × Position | 0.95 | 0.69 | 0.96 | |||
| Site × Hybrid × Position | 0.262 | 0.416 | 0.540 | |||
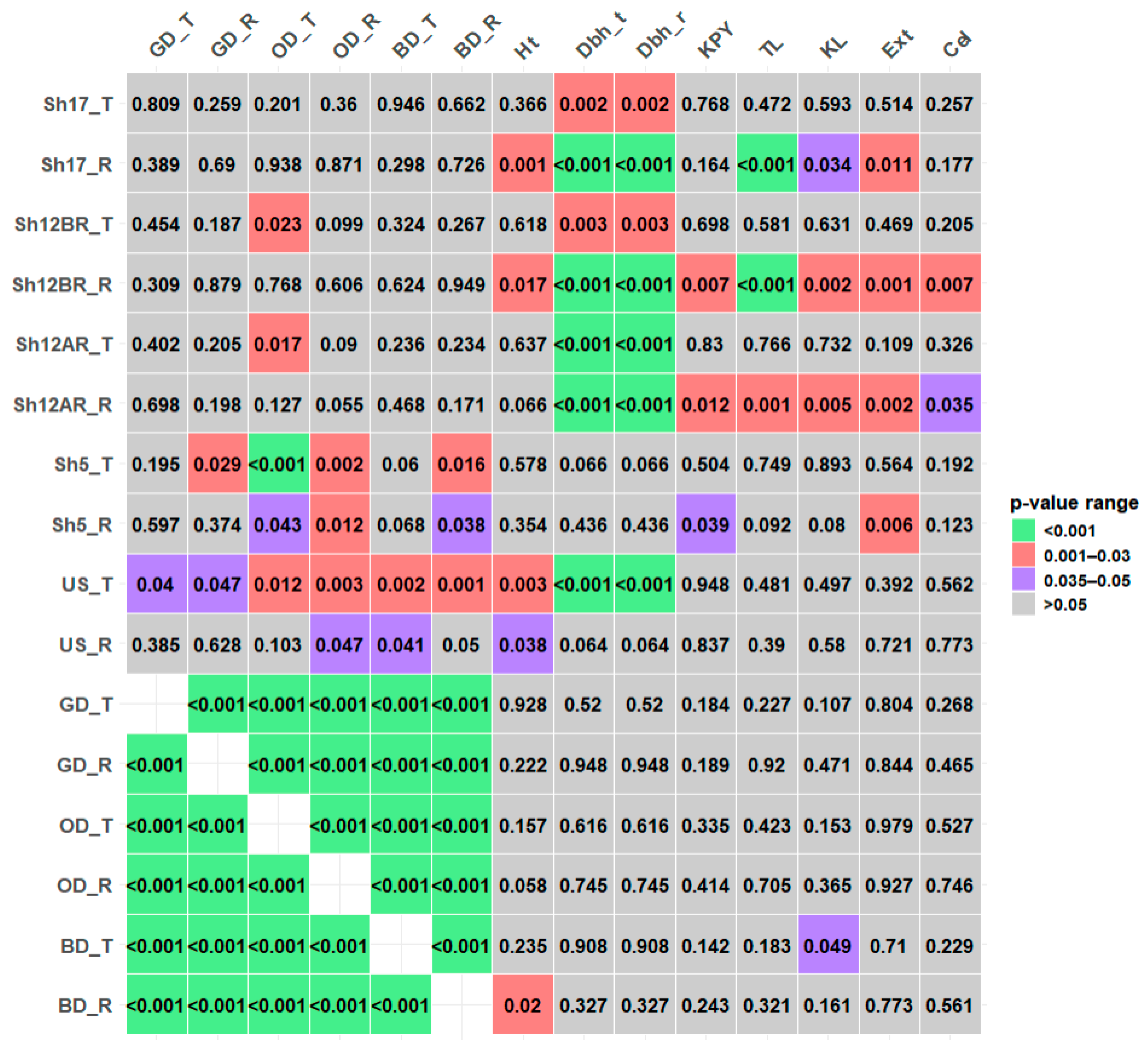
| GD_T | GD_R | OD_T | OD_R | BD_T | BD_R | Ht | Dbh_t | Dbh_r | KPY | TL | KL | Ext | Cel | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sh17_T | 0.03 | 0.17 | 0.18 | 0.14 | 0.01 | 0.07 | −0.13 | −0.43 * | −0.43 * | 0.04 | −0.10 | −0.08 | −0.09 | 0.16 |
| Sh17_R | −0.13 | 0.06 | −0.01 | 0.02 | −0.16 | −0.05 | −0.45 * | −0.71 * | −0.71 * | 0.20 | −0.49 * | −0.31 * | −0.37 * | 0.20 |
| Sh12BR_T | 0.11 | 0.20 | 0.32 * | 0.25 | 0.14 | 0.17 | −0.07 | −0.40 * | −0.40 * | 0.06 | −0.08 | −0.07 | −0.10 | 0.18 |
| Sh12BR_R | −0.16 | 0.02 | 0.05 | 0.08 | −0.08 | 0.01 | −0.34 * | −0.61 * | −0.61 * | 0.38 * | −0.54 * | −0.43 * | −0.45 * | 0.39 * |
| Sh12AR_T | 0.12 | 0.19 | 0.33 * | 0.26 | 0.17 | 0.18 | −0.07 | −0.45 * | −0.45 * | 0.03 | −0.04 | −0.05 | −0.23 | 0.14 |
| Sh12AR_R | 0.06 | 0.19 | 0.23 | 0.28 | 0.11 | 0.20 | −0.27 | −0.56 * | −0.56 * | 0.36 * | −0.45 * | −0.40 * | −0.44 * | 0.30 * |
| Sh5_T | 0.18 | 0.33 * | 0.45 * | 0.45 * | 0.27 | 0.36 * | 0.08 | −0.26 | −0.26 | 0.10 | −0.05 | −0.02 | −0.08 | 0.19 |
| Sh5_R | 0.08 | 0.13 | 0.31 * | 0.36 * | 0.28 | 0.30 * | 0.14 | −0.11 | −0.11 | 0.30 * | −0.25 | −0.26 | −0.39 * | 0.23 |
| US_T | 0.29 * | 0.30 * | 0.35 * | 0.44 * | 0.42 * | 0.47 * | 0.41 * | 0.48 * | 0.48 * | 0.01 | 0.10 | 0.10 | 0.12 | −0.08 |
| US_R | 0.13 | 0.07 | 0.25 | 0.28 * | 0.31 * | 0.28 | 0.30 * | 0.27 | 0.27 | 0.03 | 0.13 | 0.08 | −0.05 | −0.04 |
| GD_T | 0.67 * | 0.81 * | 0.64 * | 0.86 * | 0.62 * | 0.01 | −0.09 | −0.09 | −0.19 | 0.17 | 0.23 | 0.04 | −0.16 | |
| GD_R | 0.63 * | 0.79 * | 0.62 * | 0.81 * | 0.18 | 0.01 | 0.01 | −0.19 | 0.01 | 0.11 | 0.03 | −0.11 | ||
| OD_T | 0.84 * | 0.96 * | 0.80 * | 0.20 | −0.07 | −0.07 | −0.14 | 0.12 | 0.20 | 0.00 | −0.09 | |||
| OD_R | 0.80 * | 0.98 * | 0.27 | 0.05 | 0.05 | −0.12 | 0.06 | 0.13 | −0.01 | −0.05 | ||||
| BD_T | 0.78 * | 0.17 | −0.02 | −0.02 | −0.21 | 0.19 | 0.28 * | 0.05 | −0.17 | |||||
| BD_R | 0.33 * | 0.14 | 0.14 | −0.17 | 0.15 | 0.21 | 0.04 | −0.09 |
| Marker Name | Shrinkage | Alleles | Pos | p-Value | MAF | Allelic Effect | Nearest Gene |
|---|---|---|---|---|---|---|---|
| SNP2554 | Green to 12% AR (T) | T/C | 2554 | 0.000 ** | 0.167 | −2.303 | Eucgr.A00211 |
| SNP4979 | Green to 17% MC (T) | A/C | 4911 | 0.000 ** | 0.118 | −1.996 | |
| Green to 12% MC BR (T) | A/C | 4979 | 0.000 ** | 0.118 | −1.888 | ||
| Unit shrinkage (T) | A/C | 4979 | 0.000 ** | 0.118 | 0.080 | ||
| SNP4963 | Green to 17% MC (T) | T/A | 4908 | 0.000 ** | 0.118 | 1.996 | |
| Green to 12% MC BR (T) | T/A | 4963 | 0.000 ** | 0.118 | 1.888 | ||
| Unit shrinkage (T) | T/A | 4963 | 0.000 ** | 0.118 | −0.080 | ||
| SNP4976 | Green to 17% MC (T) | G/A | 2975 | 0.000 ** | 0.118 | 1.996 | |
| Green to 12% MC BR (T) | G/A | 4976 | 0.000 ** | 0.118 | 1.888 | ||
| Unit shrinkage (T) | G/A | 4976 | 0.000 ** | 0.118 | −0.080 | ||
| SNP3575 | Green to 12% MC BR (T) | C/T | 3575 | 0.000 ** | 0.167 | 1.651 | |
| Green to 5% MC (T) | C/T | 3575 | 0.002 * | 0.167 | 1.053 | ||
| SNP3577 | Green to 12% MC BR (T) | G/T | 3577 | 0.000 ** | 0.167 | 1.651 | |
| Green to 5% MC (T) | G/T | 3577 | 0.002 * | 0.167 | 1.053 | ||
| SNP1123 | Green to 12% MC BR (R) | C/T | 1123 | 0.001 ** | 0.204 | −1.112 | |
| Unit shrinkage (R) | C/T | 1123 | 0.002 * | 0.204 | 0.072 | ||
| SNP1641 | Green to 12% MC BR (R) | C/G | 1641 | 0.002 * | 0.153 | 0.998 | |
| Green to 12% MC AR (R) | C/G | 1641 | 0.001 ** | 0.153 | 1.010 | ||
| SNP2256 | Green to 12% MC BR (R) | G/A | 2256 | 0.002 * | 0.122 | 0.922 | |
| Green to 12% MC AR (R) | G/A | 2256 | 0.000 ** | 0.122 | 1.065 | ||
| SNP3769 | Green to 12% MC BR (R) | A/G | 3769 | 0.002 * | 0.357 | −0.702 | |
| Green to 12% MC AR (R) | A/G | 3769 | 0.003 * | 0.357 | −0.675 |
| Marker Name | Density | Alleles | Pos | p-Value | MAF | Allelic Effect | Nearest Gene |
|---|---|---|---|---|---|---|---|
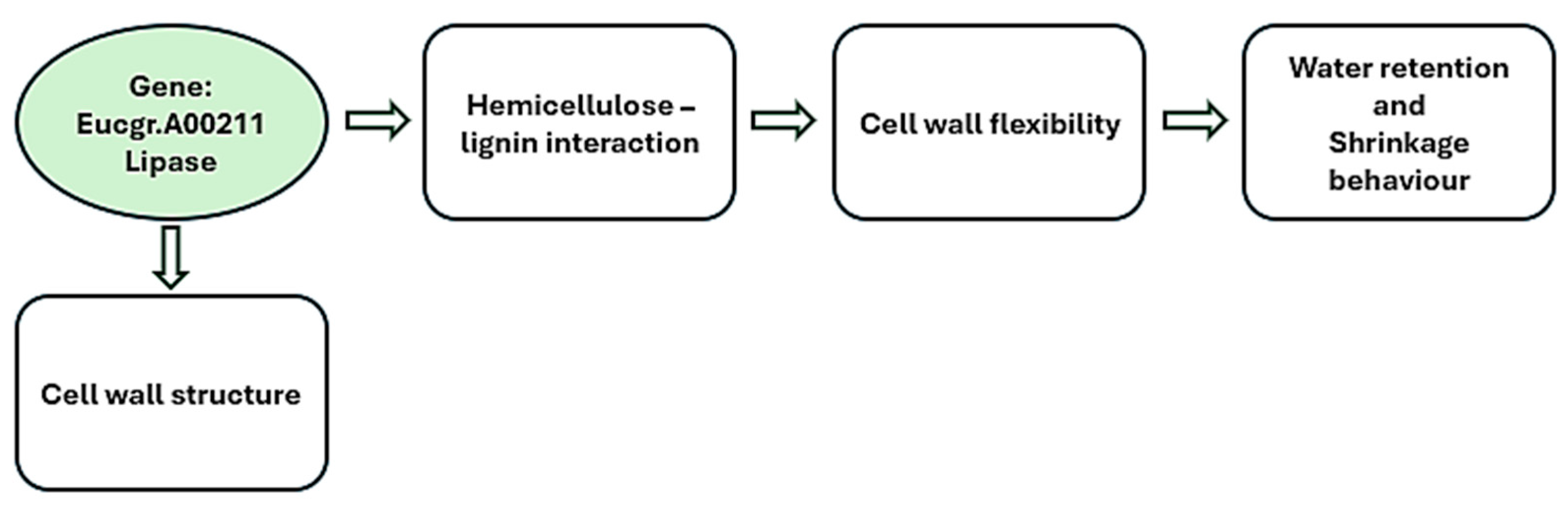
| SNP2981 | Green density (T) | G/T | 2981 | 0.000 ** | 0.098 | 69.245 | Eucgr.A00211 |
| Basic density (T) | G/T | 2981 | 0.001 ** | 0.098 | 70.362 | ||
| Oven-dry density (T) | G/T | 2981 | 0.002 * | 0.098 | 78.225 | ||
| SNP1864 | Green density (T) | G/A | 1864 | 0.001 ** | 0.461 | 35.893 | |
| Oven-dry density (T) | G/A | 1864 | 0.005 * | 0.461 | 40.701 | ||
| Basic density (T) | G/A | 1864 | 0.003 * | 0.461 | 34.975 | ||
| SNP3459 | Oven-dry density (T) | G/A | 3459 | 0.002 * | 0.333 | −46.823 | |
| Basic density (T) | G/A | 3459 | 0.001 ** | 0.333 | −39.114 | ||
| SNP4216 | Oven-dry density (T) | A/T | 4216 | 0.002 * | 0.363 | 42.607 | |
| Basic density (T) | A/T | 4216 | 0.001 ** | 0.363 | 36.333 | ||
| SNP2507 | Oven-dry density (R) | T/C | 2507 | 0.001 ** | 0.194 | −62.993 | |
| Basic density (R) | T/C | 2507 | 0.000 ** | 0.194 | −56.888 | ||
| SNP2508 | Oven-dry density (R) | T/C | 2508 | 0.001 ** | 0.194 | −62.993 | |
| Basic density (R) | T/C | 2508 | 0.000 ** | 0.194 | −56.888 | ||
| SNP2509 | Oven-dry density (R) | T/C | 2509 | 0.001 ** | 0.194 | −62.993 | |
| Basic density (R) | T/C | 2509 | 0.000 ** | 0.194 | −56.888 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falade, O.E.; Belleville, B.; Spokevicius, A.; Ozarska, B.; Bossinger, G.; Dewi, L.M.; Ibrahim, U.; Thumma, B. SNP-Based Genetic Analysis of Dimensional Stability and Wood Density in Eucalyptus pellita F.Muell. and Hybrids. Forests 2025, 16, 1301. https://doi.org/10.3390/f16081301
Falade OE, Belleville B, Spokevicius A, Ozarska B, Bossinger G, Dewi LM, Ibrahim U, Thumma B. SNP-Based Genetic Analysis of Dimensional Stability and Wood Density in Eucalyptus pellita F.Muell. and Hybrids. Forests. 2025; 16(8):1301. https://doi.org/10.3390/f16081301
Chicago/Turabian StyleFalade, Oluwatosin Esther, Benoit Belleville, Antanas Spokevicius, Barbara Ozarska, Gerd Bossinger, Listya Mustika Dewi, Umar Ibrahim, and Bala Thumma. 2025. "SNP-Based Genetic Analysis of Dimensional Stability and Wood Density in Eucalyptus pellita F.Muell. and Hybrids" Forests 16, no. 8: 1301. https://doi.org/10.3390/f16081301
APA StyleFalade, O. E., Belleville, B., Spokevicius, A., Ozarska, B., Bossinger, G., Dewi, L. M., Ibrahim, U., & Thumma, B. (2025). SNP-Based Genetic Analysis of Dimensional Stability and Wood Density in Eucalyptus pellita F.Muell. and Hybrids. Forests, 16(8), 1301. https://doi.org/10.3390/f16081301






