Genome-Wide Identification, Evolution and Expression Analysis of the G-Protein Gene Family in Poplar (Populus alba × Populus glandulosa)
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of G-Protein Gene Families in Poplar
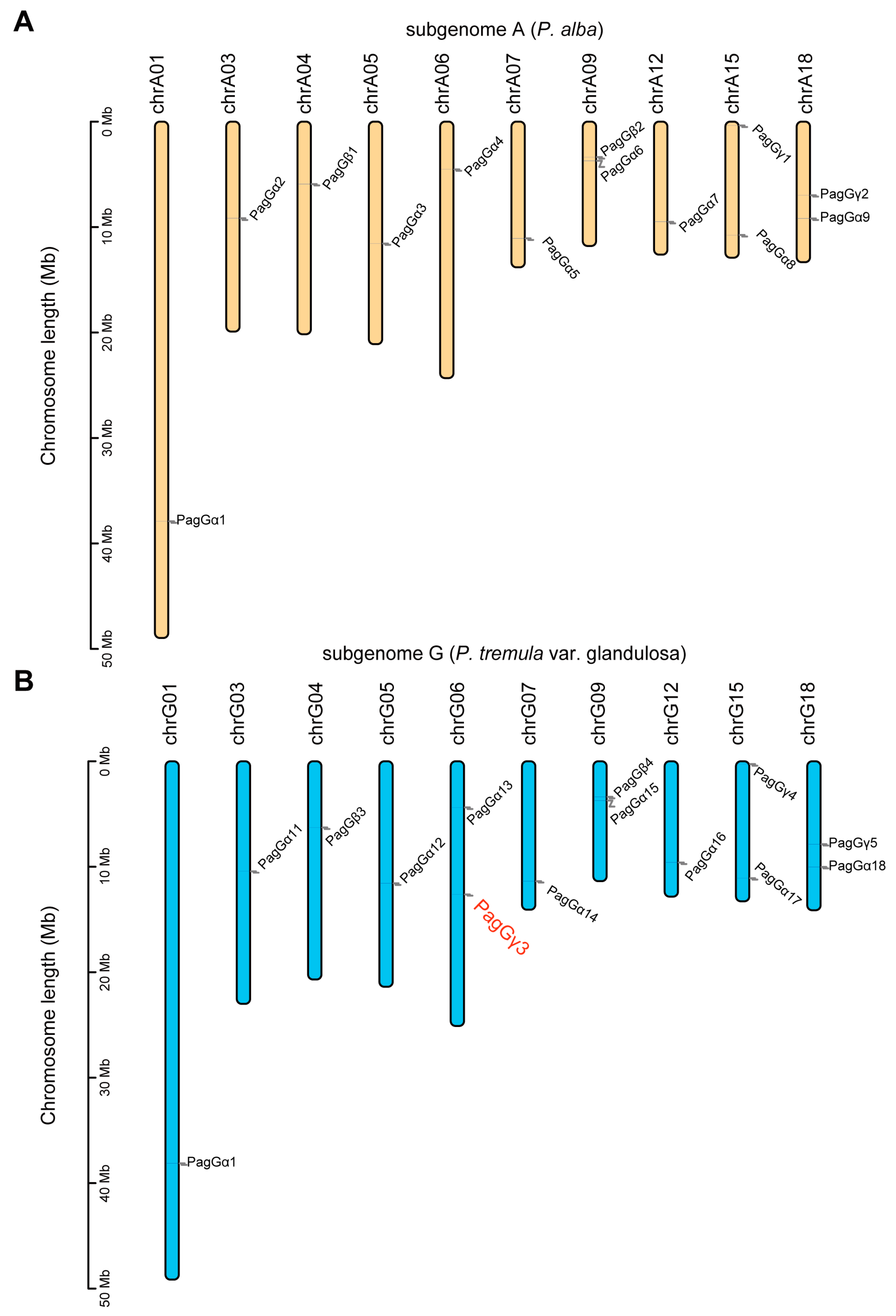
2.2. Chromosomal Localization Analysis of G-Protein Genes in the Poplar Genome
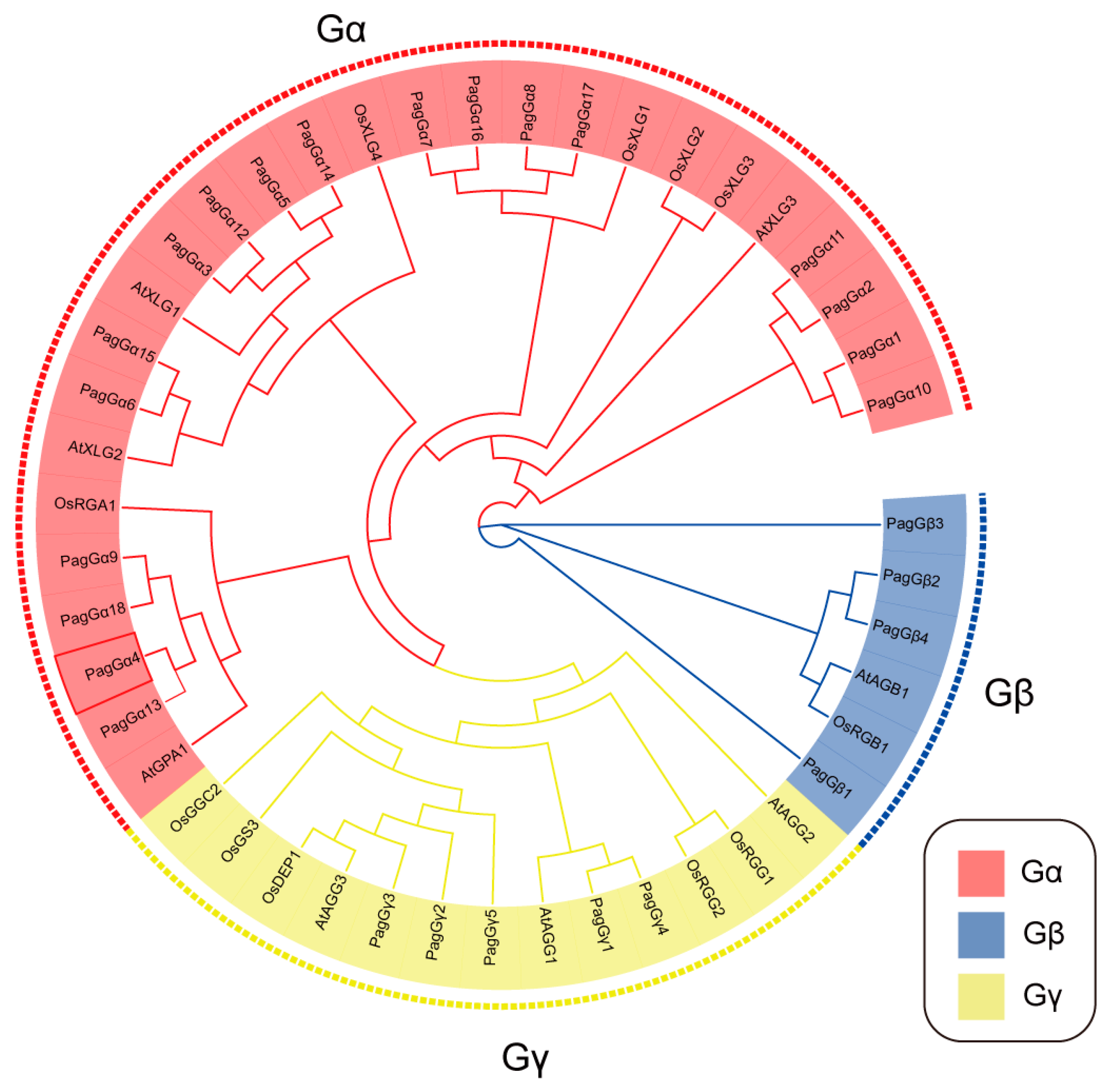
2.3. Phylogenetic Analysis of G-Protein in Poplar
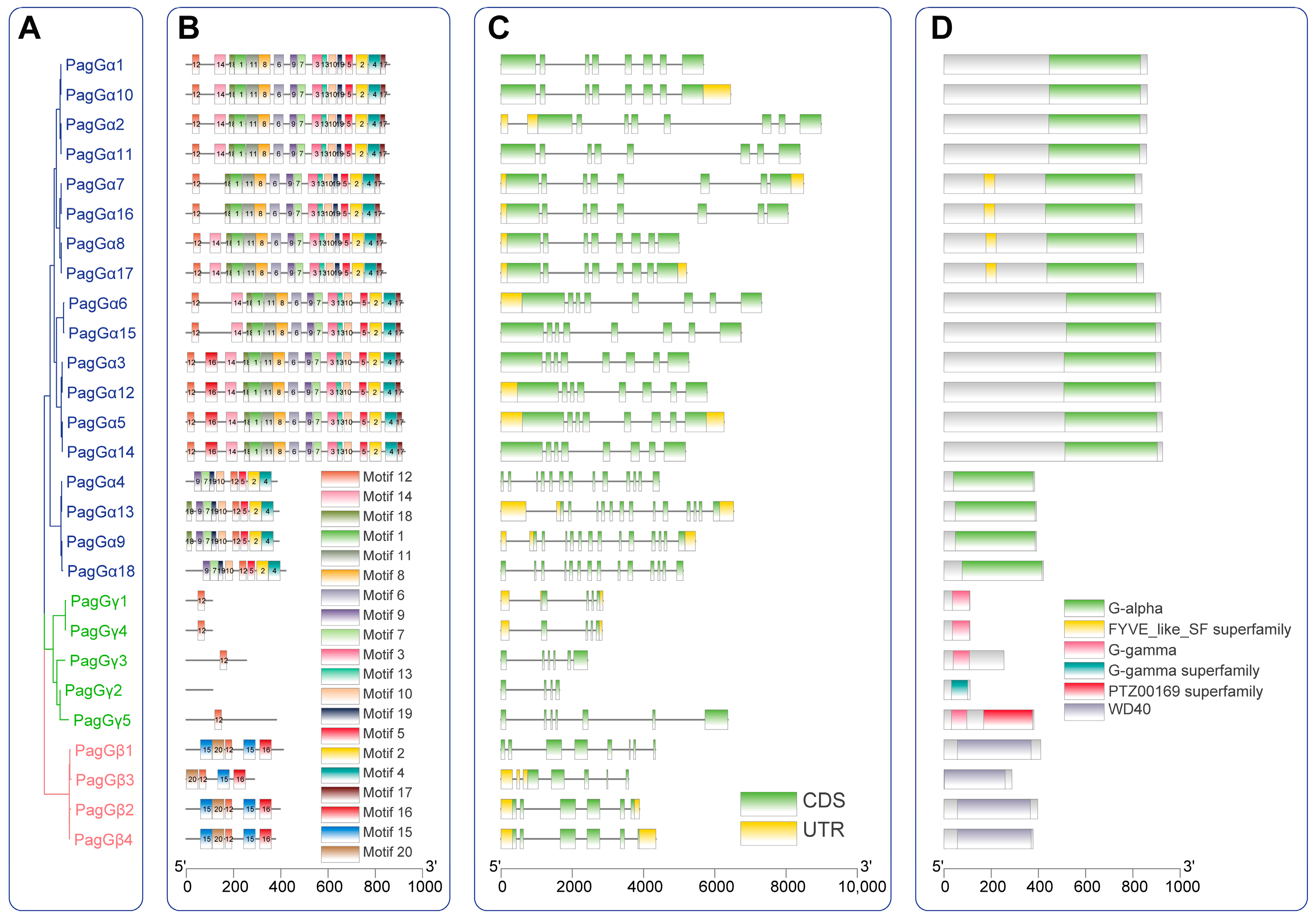
2.4. Gene Structure, Conserved Domain and Motif Analysis of G-Proteins
2.5. Cis-Acting Elements Analysis in the Promoter of G-Proteins in Poplar
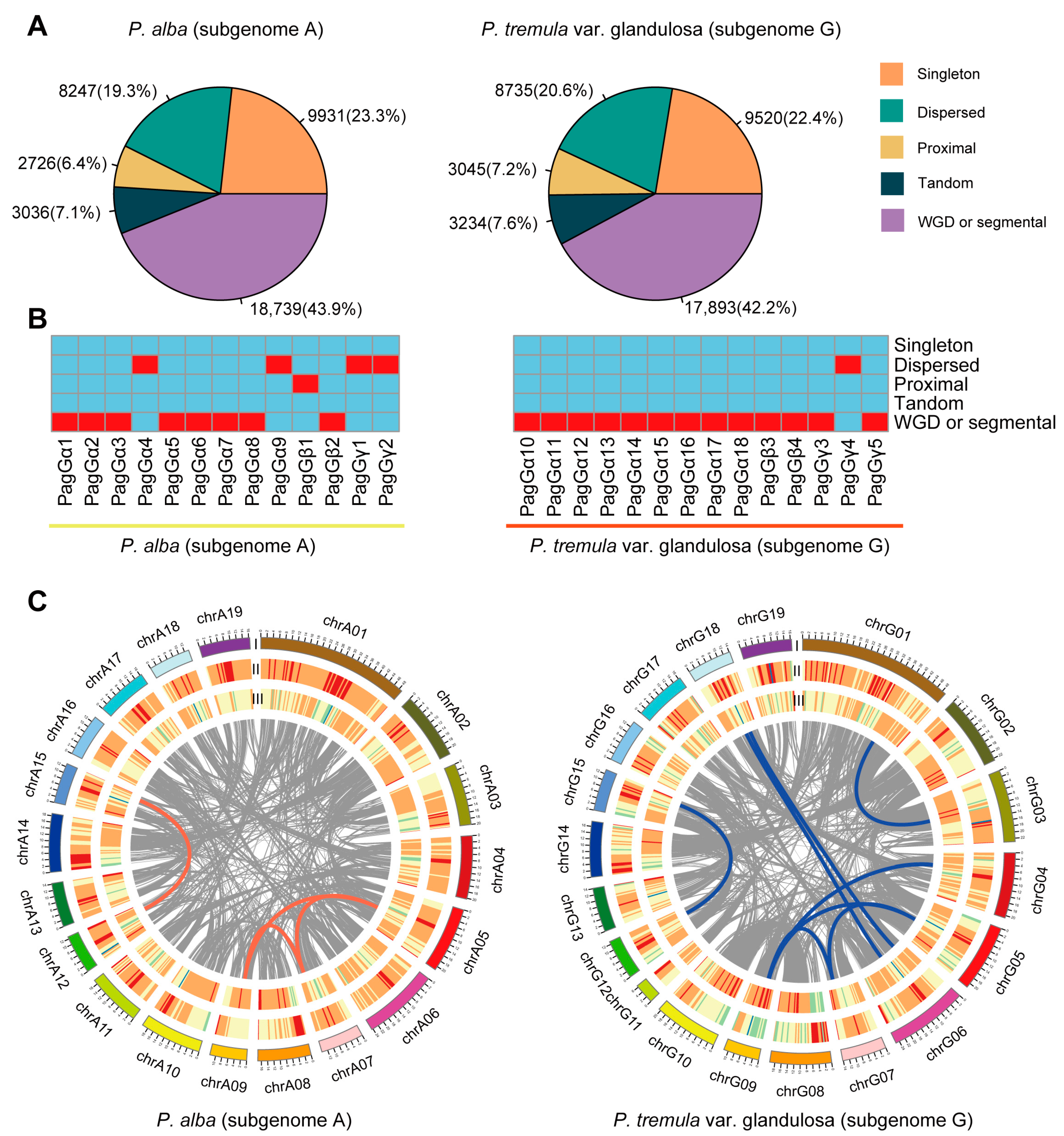
2.6. Collinearity Analysis of G-Protein Gene Family in Poplar
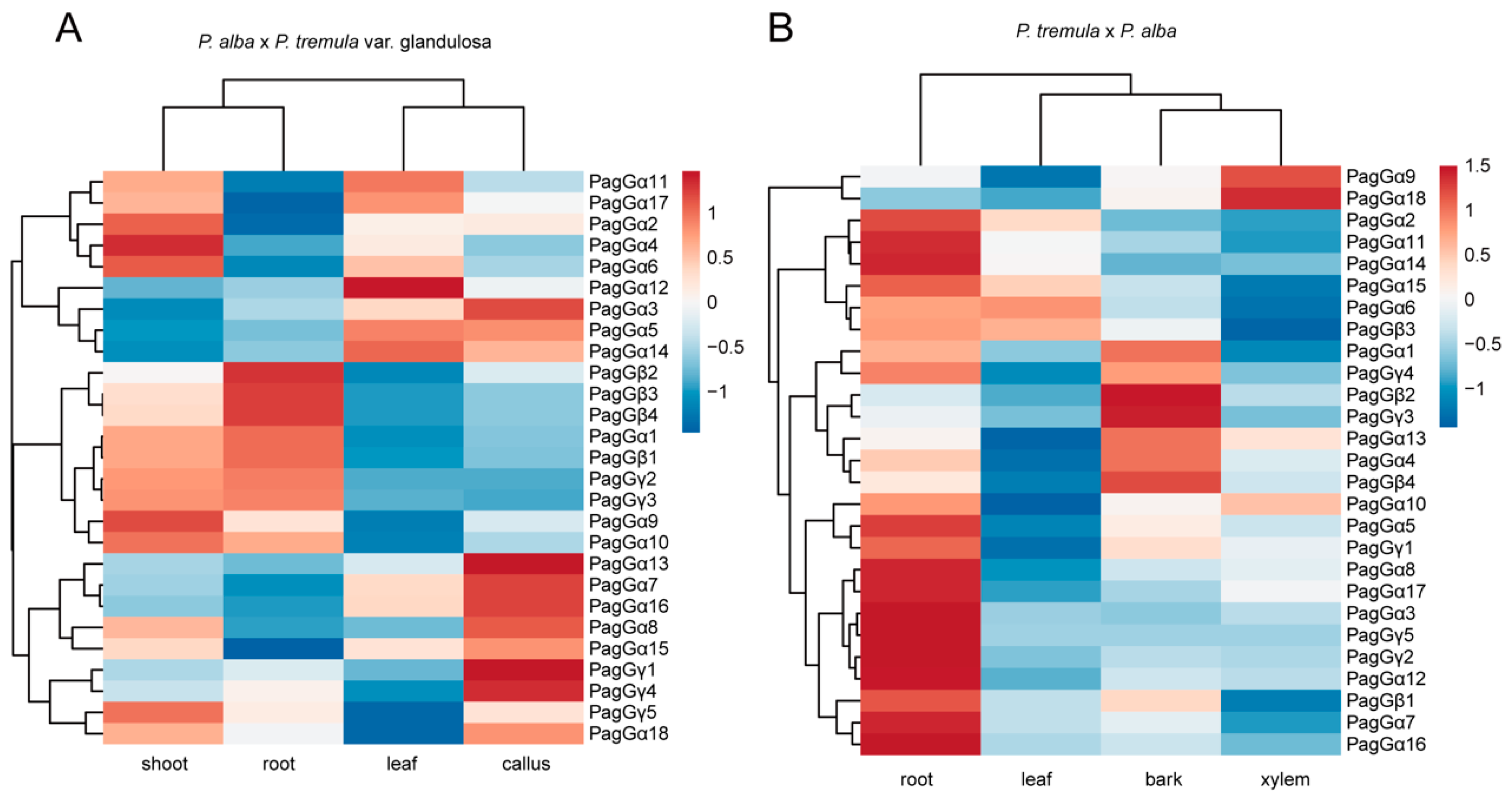
2.7. Expression Patterns of G-Protein Genes in Different Tissues of Poplar
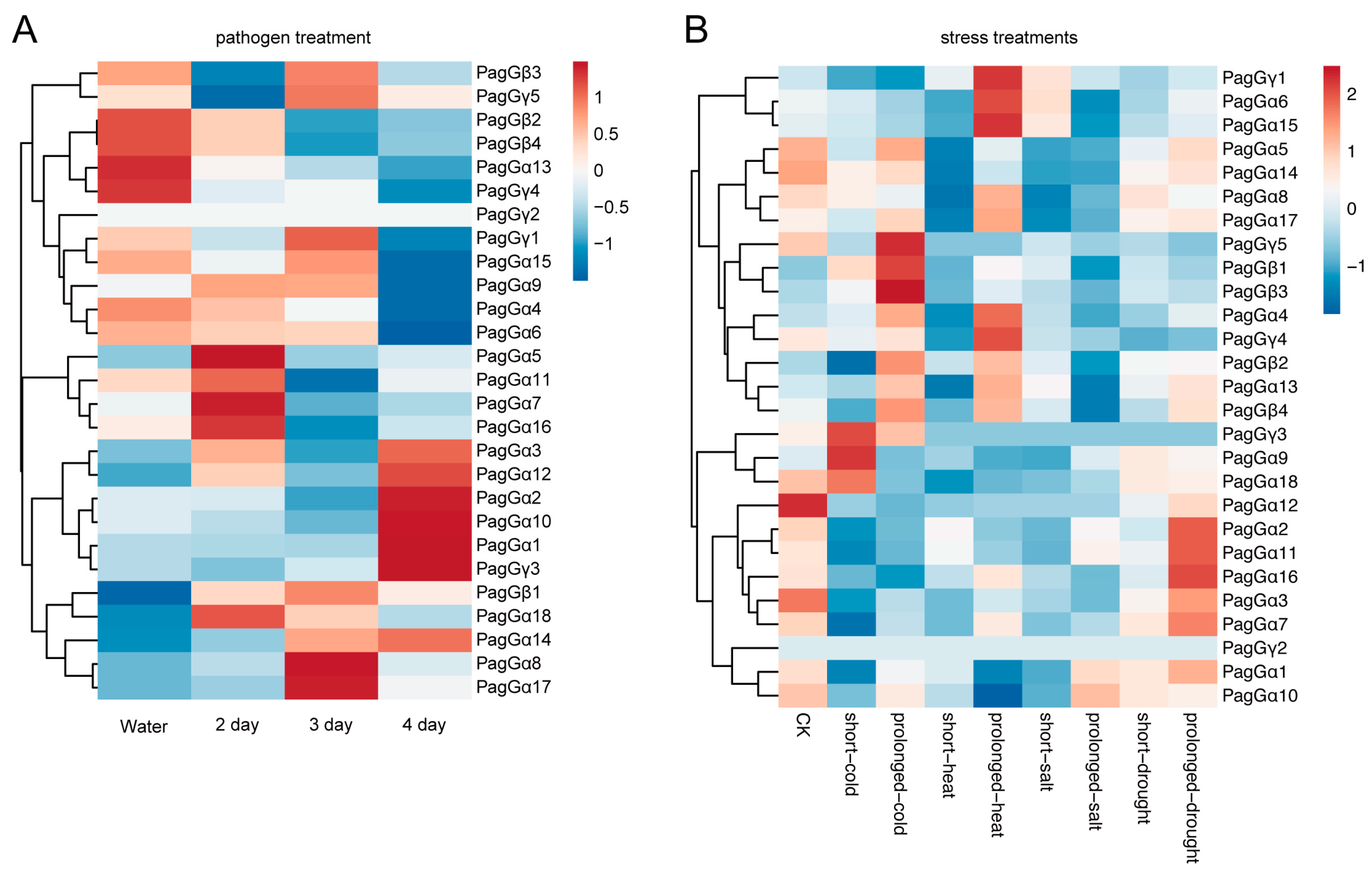
2.8. Expression Patterns of G-Protein Genes Under Biotic and Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Whole Genome Identification of G-Protein Gene Family Member in Poplar
4.2. The Physicochemical Properties Analysis of G-Proteins
4.3. Phylogenetic Analysis
4.4. Gene Structure, Conserved Domain and Motif Analysis
4.5. Cis-Elements Analysis
4.6. Collinearity Analysis of G-Protein Genes
4.7. Transcriptome Sequencing Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| pI | Theoretical isoelectric points |
| WGD | Whole genome duplication |
| HMM | Hidden Markov model |
| TE | Transposable element |
| CDS | Coding sequence |
| ABA | Abscisic acid |
| FPKMs | Fragments Per Kilobase of exon per Million mapped reads |
References
- Maruta, N.; Trusov, Y.; Jones, A.M.; Botella, J.R. Heterotrimeric G Proteins in Plants: Canonical and Atypical Gα Subunits. Int. J. Mol. Sci. 2021, 22, 11841. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Abhiman, S.; de Souza, R.F.; Aravind, L. Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene 2011, 475, 63–78. [Google Scholar] [CrossRef]
- Fan, M.; Li, J.; Zhang, T.; Huo, H.; Lü, S.; He, Z.; Wang, X.; Zhang, J. Genome-wide identification of heterotrimeric G protein genes in castor (Ricinus communis L.) and expression patterns under salt stress. BMC Genom. 2024, 25, 1115. [Google Scholar] [CrossRef]
- McCudden, C.; Hains, M.; Kimple, R.; Siderovski, D.; Willard, F. G-protein signaling: Back to the future. Cell. Mol. Life Sci. 2005, 62, 551–577. [Google Scholar] [CrossRef] [PubMed]
- Hurowitz, E.H.; Melnyk, J.M.; Chen, Y.-J.; Kouros-Mehr, H.; Simon, M.I.; Shizuya, H. Genomic characterization of the human heterotrimeric G protein α, β, and γ subunit genes. DNA Res. 2000, 7, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Birnbaumer, L. Expansion of signal transduction by G proteins: The second 15 years or so: From 3 to 16 α subunits plus βγ dimers. Biochim. Biochim. Biophys. Acta (BBA) Biomembr. 2007, 1768, 772–793. [Google Scholar] [CrossRef]
- Mason, M.G.; Botella, J.R. Completing the heterotrimer: Isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc. Natl. Acad. Sci. USA 2000, 97, 14784–14788. [Google Scholar] [CrossRef]
- Weiss, C.A.; Garnaat, C.W.; Mukai, K.; Hu, Y.; Ma, H. Isolation of cDNAs encoding guanine nucleotide-binding protein beta-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc. Natl. Acad. Sci. USA 1994, 91, 9554–9558. [Google Scholar] [CrossRef]
- Ma, H.; Yanofsky, M.F.; Meyerowitz, E.M. Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1990, 87, 3821–3825. [Google Scholar] [CrossRef]
- Cantos, C.F.; dePamphilis, C.W.; Assmann, S.M. Extra-large G proteins have extra-large effects on agronomic traits and stress tolerance in maize and rice. Trends Plant Sci. 2023, 28, 1033–1044. [Google Scholar] [CrossRef]
- Lee, Y.-R.J.; Assmann, S.M. Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): A new class of G-protein. Plant Mol. Biol. 1999, 40, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Kankanamge, D.; Tennakoon, M.; Karunarathne, A.; Gautam, N. G protein gamma subunit, a hidden master regulator of GPCR signaling. J. Biol. Chem. 2022, 298, 102618. [Google Scholar] [CrossRef]
- Gilman, A.G. G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem. 1987, 56, 615–649. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Dong, D.; Guo, L.; Dong, X.; Leng, J.; Zhao, B.; Guo, Y.-D.; Zhang, N. Research Advances in Heterotrimeric G-Protein α Subunits and Uncanonical G-Protein Coupled Receptors in Plants. Int. J. Mol. Sci. 2021, 22, 8678. [Google Scholar] [CrossRef]
- Jiang, K.; Frick-Cheng, A.; Trusov, Y.; Delgado-Cerezo, M.; Rosenthal, D.M.; Lorek, J.; Panstruga, R.; Booker, F.L.; Botella, J.R.; Molina, A.; et al. Dissecting Arabidopsis Gβ Signal Transduction on the Protein Surface. Plant Physiol. 2012, 159, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Chakravorty, D.; Botella, J.R. Diversity of heterotrimeric G-protein γ subunits in plants. BMC Res. Notes 2012, 5, 608. [Google Scholar] [CrossRef]
- Subramaniam, G.; Trusov, Y.; Lopez-Encina, C.; Hayashi, S.; Batley, J.; Botella, J.R. Type B Heterotrimeric G Protein γ-Subunit Regulates Auxin and ABA Signaling in Tomato. Plant Physiol. 2015, 170, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, G.-J.; Ding, L.; Cui, X.; Berg, H.; Assmann, S.M.; Xia, Y. Arabidopsis Extra Large G-Protein 2 (XLG2) Interacts with the Gβ; Subunit of Heterotrimeric G Protein and Functions in Disease Resistance. Mol. Plant 2009, 2, 513–525. [Google Scholar] [CrossRef]
- Jin, Y.-N.; Cui, Z.-h.; Ma, K.; Yao, J.-L.; Ruan, Y.-Y.; Guo, Z.-F. Characterization of ZmCOLD1, novel GPCR-Type G Protein genes involved in cold stress from Zea mays L. and the evolution analysis with those from other species. Physiol. Mol. Biol. Plants 2021, 27, 619–632. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 Confers Chilling Tolerance in Rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Xiong, X.-X.; Liu, Y.; Zhang, L.-L.; Li, X.-J.; Zhao, Y.; Zheng, Y.; Yang, Q.-H.; Yang, Y.; Min, D.-H.; Zhang, X.-H. G-Protein β-Subunit Gene TaGB1-B Enhances Drought and Salt Resistance in Wheat. Int. J. Mol. Sci. 2023, 24, 7337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhu, W.; Diao, S.; Wu, X.; Lu, J.; Ding, C.; Su, X. The poplar pangenome provides insights into the evolutionary history of the genus. Commun. Biol. 2019, 2, 215. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Bai, S.; Ma, J.; Zhang, L.; Shao, F.; Zhang, K.; Yang, Y.; Sun, T.; Huang, J.; Zhou, Y.; et al. The genome of Populus alba x Populus tremula var. glandulosa clone 84K. DNA Res. 2019, 26, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, S.; Marlin, M.A.; Pandey, S. The Role of Gβ Protein in Controlling Cell Expansion via Potential Interaction with Lipid Metabolic Pathways. Plant Physiol. 2019, 179, 1159–1175. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Y.; Wu, J.; Zhou, X.; Gao, H. Heterotrimeric G-proteins: Multi-dimensional regulation in plant growth, development and abiotic stress responses. Stress Biol. 2025, 5, 3. [Google Scholar] [CrossRef]
- Han, A.; Xu, Z.; Cai, Z.; Zheng, Y.; Chen, M.; Wu, L.; Shen, Q. Genome-Wide Identification and Expression Analysis of the G-Protein Gene Family in Barley Under Abiotic Stresses. Plants 2024, 13, 3521. [Google Scholar] [CrossRef]
- Gawande, N.D.; Hamiditabar, Z.; Brunetti, S.C.; Gulick, P.J. Characterization of the heterotrimeric G protein gene families in Triticum aestivum and related species. 3 Biotech 2022, 12, 99. [Google Scholar] [CrossRef]
- Bisht, N.C.; Jez, J.M.; Pandey, S. An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol. 2011, 190, 35–48. [Google Scholar] [CrossRef]
- Harding, S.A.; Hu, H.; Nyamdari, B.; Xue, L.-J.; Naran, R.; Tsai, C.-J. Tubulins, rhythms and cell walls in poplar leaves: It’s all in the timing. Tree Physiol. 2018, 38, 397–408. [Google Scholar] [CrossRef]
- Xue, L.-J.; Frost, C.J.; Tsai, C.-J.; Harding, S.A. Drought response transcriptomes are altered in poplar with reduced tonoplast sucrose transporter expression. Sci. Rep. 2016, 6, 33655. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, J.; Yang, Z.; Matsui, A.; Seki, M.; Li, S.; Yan, X.; Kohnen, M.V.; Gu, L.; Prasad, K. PtWOX11 acts as master regulator conducting the expression of key transcription factors to induce de novo shoot organogenesis in poplar. Plant Mol. Biol. 2018, 98, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Zhou, H.; Jiang, C.; Zhao, S.; Wang, L.; Li, Q.; Yang, Z.; Groover, A.; Lu, M.Z. The auxin receptor TIR 1 homolog (Pag FBL 1) regulates adventitious rooting through interactions with Aux/IAA 28 in Populus. Plant Biotechnol. J. 2019, 17, 338–349. [Google Scholar] [CrossRef]
- Ullah, H.; Chen, J.-G.; Temple, B.; Boyes, D.C.; Alonso, J.M.; Davis, K.R.; Ecker, J.R.; Jones, A.M. The β-Subunit of the Arabidopsis G Protein Negatively Regulates Auxin-Induced Cell Division and Affects Multiple Developmental Processes. Plant Cell 2003, 15, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wang, S.; Long, L.; Yu, X.; Cai, H.; Chen, P.; Gu, L.; Yang, M. Genome-wide identification and expression analysis of PtJAZ gene family in poplar (Populus trichocarpa). BMC Genom. Data 2023, 24, 55. [Google Scholar] [CrossRef] [PubMed]
- Schultheiss, H.; Dechert, C.; Kogel, K.-H.; Hückelhoven, R. Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J. 2003, 36, 589–601. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Li, S.; Liu, K.; Li, G.; Zhang, D.; Lv, B.; Gu, J.; Zhang, H.; Yang, J.; et al. OsRGA1 optimizes photosynthate allocation for roots to reduce methane emissions and improve yield in paddy ecosystems. Soil Biol. Biochem. 2021, 160, 108344. [Google Scholar] [CrossRef]
- Biswas, S.; Islam, M.N.; Sarker, S.; Tuteja, N.; Seraj, Z.I. Overexpression of heterotrimeric G protein beta subunit gene (OsRGB1) confers both heat and salinity stress tolerance in rice. Plant Physiol. Biochem. 2019, 144, 334–344. [Google Scholar] [CrossRef]
- Shi, T.; Zhang, X.; Hou, Y.; Jia, C.; Dan, X.; Zhang, Y.; Jiang, Y.; Lai, Q.; Feng, J.; Feng, J.; et al. The super-pangenome of Populus unveils genomic facets for its adaptation and diversification in widespread forest trees. Mol. Plant 2024, 17, 725–746. [Google Scholar] [CrossRef]
- Kostiou, V.D.; Theodoropoulou, M.C.; Hamodrakas, S.J. GprotPRED: Annotation of Gα, Gβ and Gγ subunits of G-proteins using profile Hidden Markov Models (pHMMs) and application to proteomes. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2016, 1864, 435–440. [Google Scholar] [CrossRef]
- Pandey, S. Plant receptor-like kinase signaling through heterotrimeric G-proteins. J. Exp. Bot. 2020, 71, 1742–1751. [Google Scholar] [CrossRef]
- Zhang, W. Roles of heterotrimeric G proteins in guard cell ion channel regulation. Plant Signal. Behav. 2011, 6, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Wang, W.; Fei, Y.; Cheng, H.-Y.; Song, B.; Zhou, Z.; Zhao, Y.; Zhang, X.; Li, L.; Chen, S.; et al. A surface-receptor-coupled G protein regulates plant immunity through nuclear protein kinases. Cell Host Microbe 2022, 30, 1602–1614.e5. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2012, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R Package 2015, 1, 790. [Google Scholar]

| Gene Name | Gene ID | Chromosome | Start | End | Strand | G Protein Type (Gα, Gβ, Gγ) | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Prediction of Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PagGα1 | Pop_A01G004180.T1 | chrA01 | 37,893,885 | 37,899,565 | + | Gα | 861 | 98,230 | 5.68 | 46.29 | 74.24 | −0.557 | nuclear |
| PagGα2 | Pop_A03G050281.T1 | chrA03 | 9,155,039 | 9,164,024 | − | Gα | 859 | 97,880 | 5.68 | 48.02 | 75.32 | −0.572 | nuclear |
| PagGα3 | Pop_A05G072994.T1 | chrA05 | 11,557,524 | 11,562,795 | − | Gα | 919 | 102,261 | 5.09 | 43.79 | 78.24 | −0.426 | nuclear |
| PagGα4 | Pop_A06G053386.T1 | chrA06 | 4,522,632 | 4,527,073 | − | Gα | 384 | 44,593 | 6.39 | 43.94 | 83.8 | −0.54 | cytoplasmic |
| PagGα5 | Pop_A07G022806.T1 | chrA07 | 11,068,517 | 11,074,776 | − | Gα | 924 | 103,003 | 5.11 | 42.29 | 77.91 | −0.434 | nuclear |
| PagGα6 | Pop_A09G026564.T1 | chrA09 | 3,705,311 | 3,712,625 | + | Gα | 918 | 103,154 | 6.07 | 46.04 | 79.81 | −0.436 | nuclear |
| PagGα7 | Pop_A12G073756.T1 | chrA12 | 9,498,343 | 9,506,833 | − | Gα | 838 | 95,958 | 5.24 | 52.44 | 70.87 | −0.67 | nuclear |
| PagGα8 | Pop_A15G064380.T1 | chrA15 | 10,744,972 | 10,749,965 | + | Gα | 845 | 96,389 | 5.23 | 53.67 | 70.3 | −0.651 | nuclear |
| PagGα9 | Pop_A18G012744.T1 | chrA18 | 9,177,768 | 9,183,224 | + | Gα | 392 | 45,599 | 5.79 | 38.43 | 87.27 | −0.493 | chloroplast |
| PagGα10 | Pop_G01G089285.T1 | chrG01 | 38,117,475 | 38,123,916 | + | Gα | 861 | 98,249 | 5.68 | 46.73 | 73.9 | −0.565 | nuclear |
| PagGα11 | Pop_G03G010649.T1 | chrG03 | 10,432,293 | 10,440,689 | − | Gα | 858 | 97,664 | 5.73 | 47.62 | 74.85 | −0.561 | nuclear |
| PagGα12 | Pop_G05G017901.T1 | chrG05 | 11,563,252 | 11,569,032 | − | Gα | 918 | 102,192 | 5.14 | 43.45 | 78.42 | −0.423 | nuclear |
| PagGα13 | Pop_G06G003473.T1 | chrG06 | 4,369,436 | 4,375,962 | − | Gα | 392 | 45,494 | 6.39 | 43.05 | 85.08 | −0.513 | mitochondrial |
| PagGα14 | Pop_G07G062643.T1 | chrG07 | 11,367,119 | 11,372,298 | − | Gα | 926 | 103,078 | 5.13 | 42.65 | 78.59 | −0.423 | nuclear |
| PagGα15 | Pop_G09G077213.T1 | chrG09 | 3,715,609 | 3,722,355 | + | Gα | 918 | 103,078 | 5.99 | 46.84 | 79.72 | −0.445 | nuclear |
| PagGα16 | Pop_G12G050751.T1 | chrG12 | 9,593,091 | 9,601,147 | − | Gα | 838 | 95,876 | 5.26 | 51.76 | 70.75 | −0.666 | nuclear |
| PagGα17 | Pop_G15G074300.T1 | chrG15 | 11,083,772 | 11,088,980 | + | Gα | 845 | 96,577 | 5.32 | 55.51 | 70.07 | −0.667 | nuclear |
| PagGα18 | Pop_G18G011084.T1 | chrG18 | 10,021,608 | 10,026,716 | + | Gα | 421 | 48,810 | 5.91 | 42.42 | 84.96 | −0.553 | cytoplasmic |
| PagGβ1 | Pop_A04G028317.T1 | chrA04 | 5,919,146 | 5,923,477 | + | Gβ | 410 | 44,959 | 6.45 | 36.19 | 78.66 | −0.25 | nuclear |
| PagGβ2 | Pop_A09G026627.T1 | chrA09 | 3,355,613 | 3,359,495 | + | Gβ | 397 | 42,965 | 6.26 | 31.33 | 86.85 | −0.094 | nuclear |
| PagGβ3 | Pop_G04G028074.T1 | chrG04 | 6,276,102 | 6,279,679 | + | Gβ | 288 | 30,900 | 5.82 | 33.37 | 73.09 | −0.221 | nuclear |
| PagGβ4 | Pop_G09G077148.T1 | chrG09 | 3,370,727 | 3,375,071 | + | Gβ | 377 | 40,795 | 6.69 | 27.31 | 82.18 | −0.144 | nuclear |
| PagGγ1 | Pop_A15G014669.T1 | chrA15 | 367,628 | 370,487 | − | Gγ | 110 | 12,409 | 4.91 | 41.67 | 70.18 | −0.677 | chloroplast |
| PagGγ2 | Pop_A18G083136.T1 | chrA18 | 6,983,288 | 6,984,925 | − | Gγ | 111 | 12,815 | 9.37 | 80.27 | 72.88 | −0.452 | nuclear |
| PagGγ3 | Pop_G06G051470.T1 | chrG06 | 12,624,041 | 12,626,469 | − | Gγ | 254 | 28,043 | 9.39 | 58.91 | 91.81 | 0.07 | nuclear |
| PagGγ4 | Pop_G15G034021.T1 | chrG15 | 265,063 | 267,901 | − | Gγ | 110 | 12,409 | 4.91 | 41.59 | 70.18 | −0.701 | chloroplast |
| PagGγ5 | Pop_G18G085849.T1 | chrG18 | 7,853,204 | 7,859,568 | + | Gγ | 381 | 42,000 | 9.75 | 54.15 | 88.64 | −0.076 | cytoplasmic |
| Subgenome | Chromosome | Start | End | Gene1 | Chromosome | Start | End | Gene2 |
|---|---|---|---|---|---|---|---|---|
| P. alba (subgenome A) | chrA05 | 11,557,524.00 | 11,562,795.00 | PagGα3 | chrA07 | 11,068,517.00 | 11,074,776.00 | PagGα5 |
| chrA05 | 11,557,524.00 | 11,562,795.00 | PagGα3 | chrA09 | 3,705,311.00 | 3,712,625.00 | PagGα6 | |
| chrA07 | 11,068,517.00 | 11,074,776.00 | PagGα5 | chrA09 | 3,705,311.00 | 3,712,625.00 | PagGα6 | |
| chrA12 | 9,498,343.00 | 9,506,833.00 | PagGα7 | chrA15 | 10,744,972.00 | 10,749,965.00 | PagGα8 | |
| P. tremula var. glandulosa (subgenome G) | chrG01 | 38,117,475.00 | 38,123,916.00 | PagGα10 | chrG03 | 10,432,293.00 | 10,440,689.00 | PagGα11 |
| chrG04 | 6,276,102.00 | 6,279,679.00 | PagGβ3 | chrG09 | 3,370,727.00 | 3,375,071.00 | PagGβ4 | |
| chrG05 | 11,563,252.00 | 11,569,032.00 | PagGα12 | chrG07 | 11,367,119.00 | 11,372,298.00 | PagGα14 | |
| chrG05 | 11,563,252.00 | 11,569,032.00 | PagGα12 | chrG09 | 3,715,609.00 | 3,722,355.00 | PagGα15 | |
| chrG06 | 4,369,436.00 | 4,375,962.00 | PagGα13 | chrG18 | 10,021,608.00 | 10,026,716.00 | PagGα18 | |
| chrG06 | 12,624,041.00 | 12,626,469.00 | PagGγ3 | chrG18 | 7,853,204.00 | 7,859,568.00 | PagGγ5 | |
| chrG07 | 11,367,119.00 | 11,372,298.00 | PagGα14 | chrG09 | 3,715,609.00 | 3,722,355.00 | PagGα15 | |
| chrG12 | 9,593,091.00 | 9,601,147.00 | PagGα16 | chrG15 | 11,083,772.00 | 11,088,980.00 | PagGα17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, B.; Liu, Q.; Zeng, Z.; Gu, Y.; Ye, W.; Fu, F.; Ming, M. Genome-Wide Identification, Evolution and Expression Analysis of the G-Protein Gene Family in Poplar (Populus alba × Populus glandulosa). Forests 2025, 16, 805. https://doi.org/10.3390/f16050805
Song B, Liu Q, Zeng Z, Gu Y, Ye W, Fu F, Ming M. Genome-Wide Identification, Evolution and Expression Analysis of the G-Protein Gene Family in Poplar (Populus alba × Populus glandulosa). Forests. 2025; 16(5):805. https://doi.org/10.3390/f16050805
Chicago/Turabian StyleSong, Bobo, Qian Liu, Zitong Zeng, Yiyang Gu, Wenxin Ye, Fangfang Fu, and Meiling Ming. 2025. "Genome-Wide Identification, Evolution and Expression Analysis of the G-Protein Gene Family in Poplar (Populus alba × Populus glandulosa)" Forests 16, no. 5: 805. https://doi.org/10.3390/f16050805
APA StyleSong, B., Liu, Q., Zeng, Z., Gu, Y., Ye, W., Fu, F., & Ming, M. (2025). Genome-Wide Identification, Evolution and Expression Analysis of the G-Protein Gene Family in Poplar (Populus alba × Populus glandulosa). Forests, 16(5), 805. https://doi.org/10.3390/f16050805






