The Hybrid Retrieval of Leaf Anthocyanin Content Using Four Machine Learning Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. In Situ Dataset
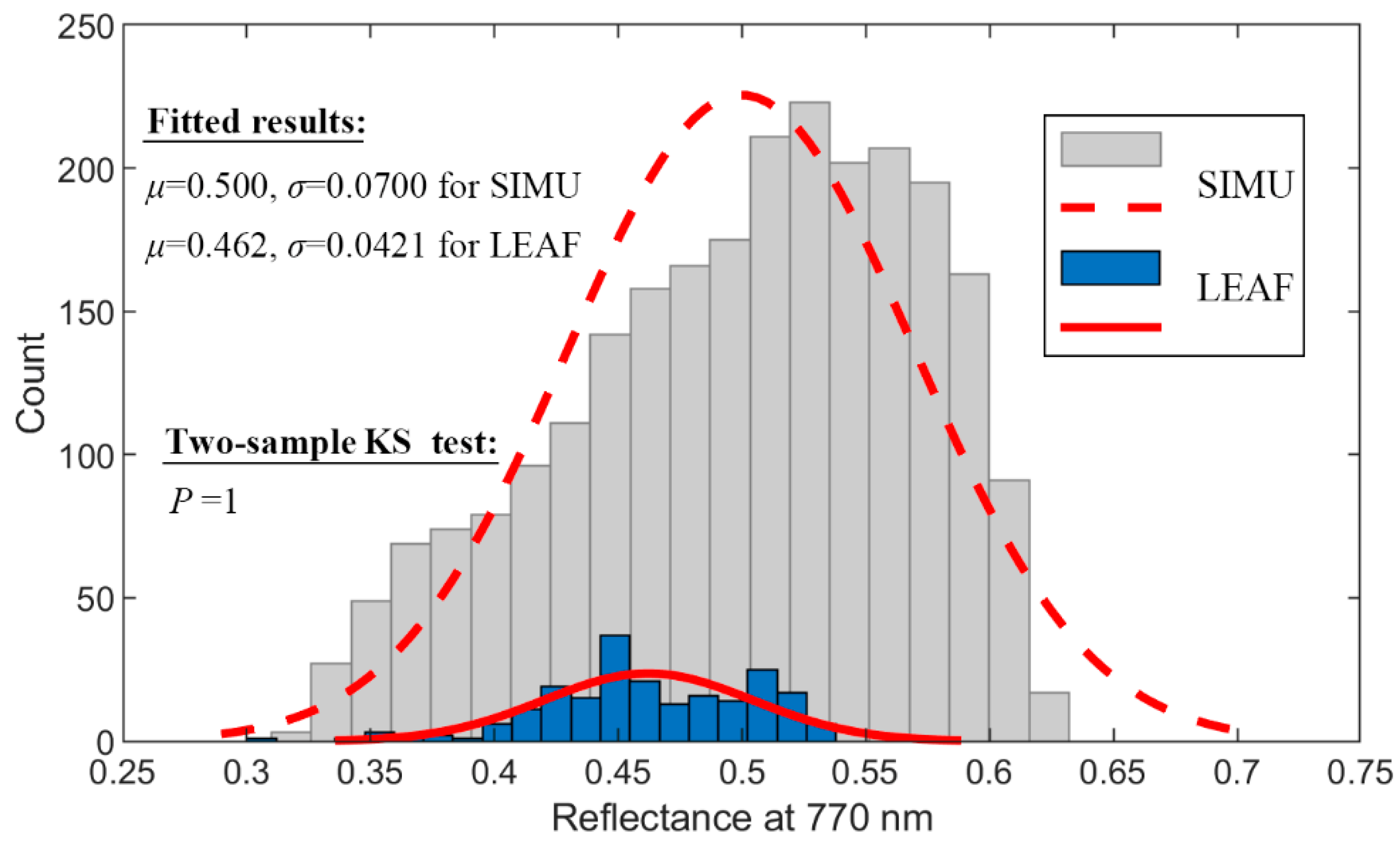
2.1.2. Simulated Dataset
2.2. The Four Machine Learning Algorithms
2.3. The Input Independent Variables
2.4. Statistical Analysis, Model Evaluation, and Accuracy Assessment
3. Results
3.1. The Statistics of Pigments in LEAF
3.2. The Selected Principal Components
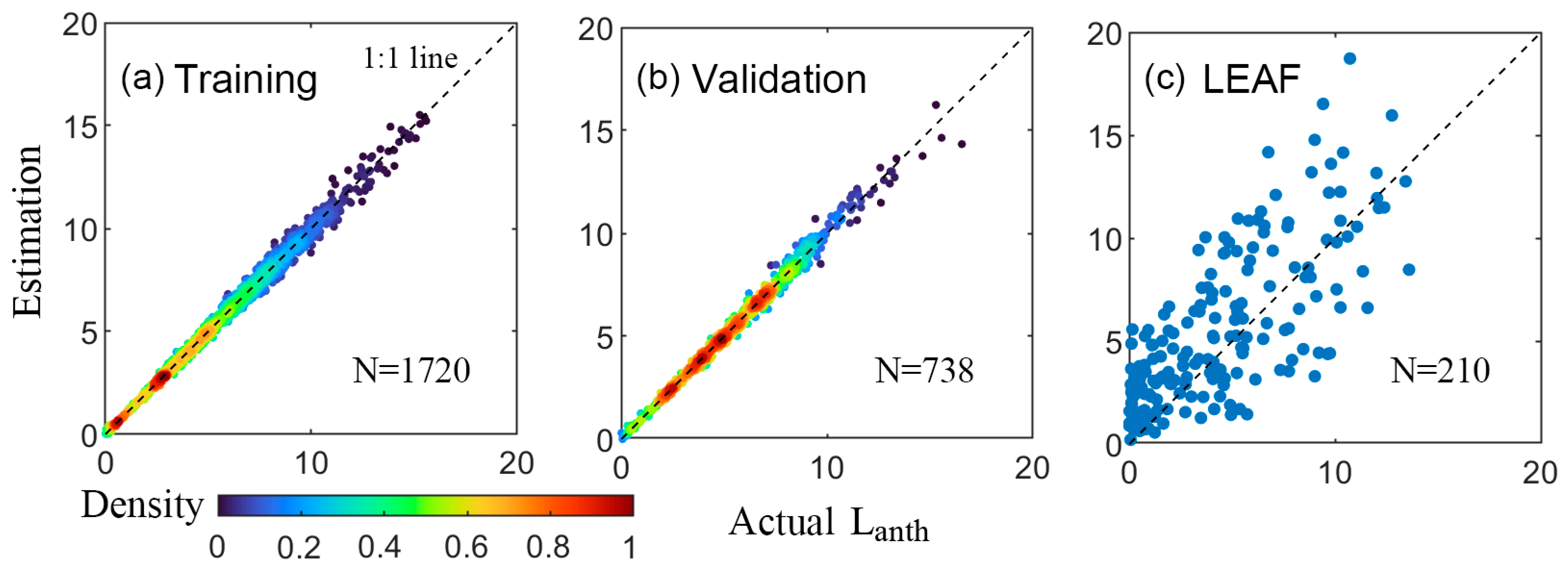
3.3. Retrieval with ANNs
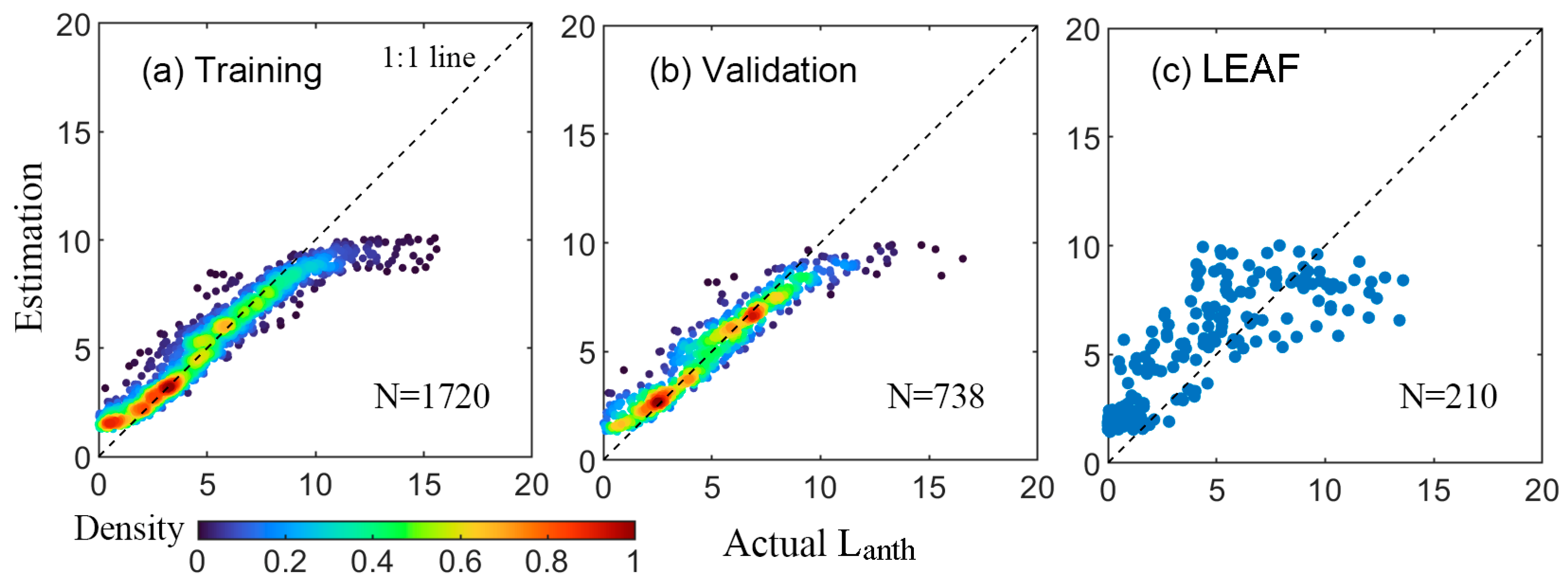
3.4. Retrieval with SVM Models
3.5. Retrieval with GPR
3.6. Retrieval with RF
4. Discussion
4.1. Comparison of the Four Machine Learning Methods
4.2. Hybrid vs. State-of-the-Art Direct Empirical Retrieval
4.3. Potential for Lanth Mapping at the Canopy Scale
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Gould, K.S. Nature’s Swiss army knife: The diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004, 2004, 314–320. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Francis, F.J. Food Colorants—Anthocyanins. Crit. Rev. Food Sci. Nutr. 1989, 28, 273–314. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Gould, K.S. Why leaves turn red—Pigments called anthocyanins probably protect leaves from light damage by direct shielding and by scavenging free radicals. Am. Sci. 2002, 90, 524–531. [Google Scholar] [CrossRef]
- Luo, L.; Chang, Q.; Gao, Y.; Jiang, D.; Li, F. Combining Different Transformations of Ground Hyperspectral Data with Unmanned Aerial Vehicle (UAV) Images for Anthocyanin Estimation in Tree Peony Leaves. Remote Sens. 2022, 14, 2271. [Google Scholar] [CrossRef]
- Verrelst, J.; Camps-Valls, G.; Munoz-Mari, J.; Pablo Rivera, J.; Veroustraete, F.; Clevers, J.G.P.W.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Brown, L.A.; Ogutu, B.O.; Dash, J. Estimating Forest Leaf Area Index and Canopy Chlorophyll Content with Sentinel-2: An Evaluation of Two Hybrid Retrieval Algorithms. Remote Sens. 2019, 11, 1752. [Google Scholar] [CrossRef]
- Estevez, J.; Salinero-Delgado, M.; Berger, K.; Pipia, L.; Rivera-Caicedo, J.P.; Wocher, M.; Reyes-Munoz, P.; Tagliabue, G.; Boschetti, M.; Verrelst, J. Gaussian processes retrieval of crop traits in Google Earth Engine based on Sentinel-2 top-of-atmosphere data. Remote Sens. Environ. 2022, 273, 112958. [Google Scholar] [CrossRef]
- Zhang, Y.; Hui, J.; Qin, Q.; Sun, Y.; Zhang, T.; Sun, H.; Li, M. Transfer-learning-based approach for leaf chlorophyll content estimation of winter wheat from hyperspectral data. Remote Sens. Environ. 2021, 267, 112724. [Google Scholar] [CrossRef]
- Guo, A.; Huang, W.; Qian, B.; Ye, H.; Jiao, Q.; Cheng, X.; Ruan, C. A hybrid model coupling PROSAIL and continuous wavelet transform based on multi-angle hyperspectral data improves maize chlorophyll retrieval. Int. J. Appl. Earth Obs. Geoinf. 2024, 132, 104076. [Google Scholar] [CrossRef]
- Ravi, J.; Nigam, R.; Bhattacharya, B.K.; Desai, D.; Patel, P. Retrieval of crop biophysical-biochemical variables from airborne AVIRIS-NG data using hybrid inversion of PROSAIL-D. Adv. Space Res. 2024, 73, 1269–1289. [Google Scholar] [CrossRef]
- Féret, J.B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef]
- Verhoef, W. Light-Scattering by Leaf Layers with Application to Canopy Reflectance Modeling—The Sail Model. Remote Sens. Environ. 1984, 16, 125–141. [Google Scholar] [CrossRef]
- Gamon, J.A.; Surfus, J.S. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys. Res. Lett. 2006, 33, 567. [Google Scholar] [CrossRef]
- Vina, A.; Gitelson, A.A. Sensitivity to Foliar Anthocyanin Content of Vegetation Indices Using Green Reflectance. IEEE Geosci. Remote Sens. Lett. 2011, 8, 464–468. [Google Scholar] [CrossRef]
- Gu, X.; Cai, W.; Fan, Y.; Ma, Y.; Zhao, X.; Zhang, C. Estimating foliar anthocyanin content of purple corn via hyperspectral model. Food Sci. Nutr. 2018, 6, 572–578. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J. Leaf Anthocyanin Content Retrieval with Partial Least Squares and Gaussian Process Regression from Spectral Reflectance Data. Sensors 2021, 21, 3078. [Google Scholar] [CrossRef] [PubMed]
- Cherif, E.; Feilhauer, H.; Berger, K.; Dao, P.D.; Ewald, M.; Hank, T.B.; He, Y.; Kovach, K.R.; Lu, B.; Townsend, P.A.; et al. From spectra to plant functional traits: Transferable multi-trait models from heterogeneous and sparse data. Remote Sens. Environ. 2023, 292, 113580. [Google Scholar] [CrossRef]
- Jiang, S.; Chang, Q.; Wang, X.; Zheng, Z.; Zhang, Y.; Wang, Q. Estimation of Anthocyanins in Whole-Fertility Maize Leaves Based on Ground-Based Hyperspectral Measurements. Remote Sens. 2023, 15, 2571. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, D.; Chang, Q.; Zheng, Z.; Fu, X.; Li, K.; Mo, H. Estimation of Anthocyanins in Leaves of Trees with Apple Mosaic Disease Based on Hyperspectral Data. Remote Sens. 2023, 15, 1732. [Google Scholar] [CrossRef]
- Miao, H.; Chen, X.; Guo, Y.; Wang, Q.; Zhang, R.; Chang, Q. Estimation of Anthocyanins in Winter Wheat Based on Band Screening Method and Genetic Algorithm Optimization Models. Remote Sens. 2024, 16, 2324. [Google Scholar] [CrossRef]
- van den Berg, A.K.; Perkins, T.D. Nondestructive estimation of anthocyanin content in autumn sugar maple leaves. HortScience 2005, 40, 685–686. [Google Scholar] [CrossRef]
- Steele, M.R.; Gitelson, A.A.; Rundquist, D.C.; Merzlyak, M.N. Nondestructive Estimation of Anthocyanin Content in Grapevine Leaves. Am. J. Enol. Vitic. 2009, 60, 87–92. [Google Scholar] [CrossRef]
- Gitelson, A.; Solovchenko, A. Non-invasive quantification of foliar pigments: Possibilities and limitations of reflectance- and absorbance-based approaches. J. Photochem. Photobiol. B-Biol. 2018, 178, 537–544. [Google Scholar] [CrossRef]
- Verrelst, J.; Munoz, J.; Alonso, L.; Delegido, J.; Pablo Rivera, J.; Camps-Valls, G.; Moreno, J. Machine learning regression algorithms for biophysical parameter retrieval: Opportunities for Sentinel-2 and -3. Remote Sens. Environ. 2012, 118, 127–139. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef] [PubMed]
- Verrelst, J.; Malenovsky, Z.; Van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.-P.; Lewis, P.; North, P.; Moreno, J. Quantifying Vegetation Biophysical Variables from Imaging Spectroscopy Data: A Review on Retrieval Methods. Surv. Geophys. 2019, 40, 589–629. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, Y.; Hao, F.; Zhang, X.; Chen, J.; de Beurs, K.; He, Y.; Fu, Y.H. Comparison of different machine learning algorithms for predicting maize grain yield using UAV-based hyperspectral images. Int. J. Appl. Earth Obs. Geoinf. 2023, 124, 103528. [Google Scholar] [CrossRef]
- Moreno-Martinez, A.; Camps-Valls, G.; Kattge, J.; Robinson, N.; Reichstein, M.; van Bodegom, P.; Kramer, K.; Cornelissen, J.H.C.; Reich, P.; Bahn, M.; et al. A methodology to derive global maps of leaf traits using remote sensing and climate data. Remote Sens. Environ. 2018, 218, 69–88. [Google Scholar] [CrossRef]
- Xu, M.; Liu, R.; Chen, J.M.; Shang, R.; Liu, Y.; Qi, L.; Croft, H.; Ju, W.; Zhang, Y.; He, Y.; et al. Retrieving global leaf chlorophyll content from MERIS data using a neural network method. ISPRS J. Photogramm. Remote Sens. 2022, 192, 66–82. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Chivkunova, O.B.; Merzlyak, M.N. Nondestructive estimation of anthocyanins and chlorophylls in anthocyanic leaves. Am. J. Bot. 2009, 96, 1861–1868. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Chivkunova, O.B.; Solovchenko, A.E.; Naqvi, K.R. Light absorption by anthocyanins in juvenile, stressed, and senescing leaves. J. Exp. Bot. 2008, 59, 3903–3911. [Google Scholar] [CrossRef]
- Quan, X.; He, B.; Li, X. A Bayesian Network-Based Method to Alleviate the Ill-Posed Inverse Problem: A Case Study on Leaf Area Index and Canopy Water Content Retrieval. IEEE Trans. Geosci. Remote 2015, 53, 6507–6517. [Google Scholar] [CrossRef]
- Feret, J.-B.; Francois, C.; Gitelson, A.; Asner, G.P.; Barry, K.M.; Panigada, C.; Richardson, A.D.; Jacquemoud, S. Optimizing spectral indices and chemometric analysis of leaf chemical properties using radiative transfer modeling. Remote Sens. Environ. 2011, 115, 2742–2750. [Google Scholar] [CrossRef]
- Shi, S.; Xu, L.; Gong, W.; Chen, B.; Chen, B.; Qu, F.; Tang, X.; Sun, J.; Yang, J. A convolution neural network for forest leaf chlorophyll and carotenoid estimation using hyperspectral reflectance. Int. J. Appl. Earth Obs. Geoinf. 2022, 108, 102719. [Google Scholar] [CrossRef]
- Li, Y.; Liang, S. Evaluation of Reflectance and Canopy Scattering Coefficient Based Vegetation Indices to Reduce the Impacts of Canopy Structure and Soil in Estimating Leaf and Canopy Chlorophyll Contents. IEEE Trans. Geosci. Remote 2023, 61, 3266500. [Google Scholar] [CrossRef]
- Beale, R.; Jackson, T. Neural Computing: An Introduction; IOP Publishing Ltd.: Bristol, UK, 1990. [Google Scholar]
- Ojha, V.K.; Abraham, A.; Snasel, V. Metaheuristic design of feedforward neural networks: A review of two decades of research. Eng. Appl. Artif. Intell. 2017, 60, 97–116. [Google Scholar] [CrossRef]
- Vapnik, V.N. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1995. [Google Scholar]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Sun, D.; Li, Y.; Wang, Q. A Unified Model for Remotely Estimating Chlorophyll a in Lake Taihu, China, Based on SVM and <i>In Situ</i> Hyperspectral Data. IEEE Trans. Geosci. Remote 2009, 47, 2957–2965. [Google Scholar] [CrossRef]
- Mehdizadeh, S.; Behmanesh, J.; Khalili, K. Using MARS, SVM, GEP and empirical equations for estimation of monthly mean reference evapotranspiration. Comput. Electron. Agric. 2017, 139, 103–114. [Google Scholar] [CrossRef]
- Rasmussen, C.E.; Williams, C.K.I. Gaussian Processes for Machine Learning; The MIT Press: Cambridge, MA, USA, 2006. [Google Scholar]
- Camps-Valls, G.; Verrelst, J.; Munoz-Mari, J.; Laparra, V.; Mateo-Jimenez, F.; Gomez-Dan, J. A Survey on Gaussian Processes for Earth-Observation Data Analysis A comprehensive investigation. IEEE Geosci. Remote Sens. Mag. 2016, 4, 58–78. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carre, G.; Garcia Marquez, J.R.; Gruber, B.; Lafourcade, B.; Leitao, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- de Sa, N.C.; Baratchi, M.; Hauser, L.T.; van Bodegom, P. Exploring the Impact of Noise on Hybrid Inversion of PROSAIL RTM on Sentinel-2 Data. Remote Sens. 2021, 13, 648. [Google Scholar] [CrossRef]
- Verrelst, J.; Pablo Rivera, J.; Veroustraete, F.; Munoz-Mari, J.; Clevers, J.G.P.W.; Camps-Valls, G.; Moreno, J. Experimental Sentinel-2 LAI estimation using parametric, non-parametric and physical retrieval methods—A comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Ashourloo, D.; Aghighi, H.; Matkan, A.A.; Mobasheri, M.R.; Rad, A.M. An Investigation Into Machine Learning Regression Techniques for the Leaf Rust Disease Detection Using Hyperspectral Measurement. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 4344–4351. [Google Scholar] [CrossRef]
- Vafaei, S.; Soosani, J.; Adeli, K.; Fadaei, H.; Naghavi, H.; Pham, T.D.; Bui, D.T. Improving Accuracy Estimation of Forest Aboveground Biomass Based on Incorporation of ALOS-2 PALSAR-2 and Sentinel-2A Imagery and Machine Learning: A Case Study of the Hyrcanian Forest Area (Iran). Remote Sens. 2018, 10, 172. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.; Gara, T.W.; Heurich, M. Machine learning methods’ performance in radiative transfer model inversion to retrieve plant traits from Sentinel-2 data of a mixed mountain forest. Int. J. Digit. Earth 2021, 14, 106–120. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. Prospect—A Model of Leaf Optical-Properties Spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Allen, W.A.; Gausman, H.W.; Richardson, A.J.; Thomas, J.R. Interaction of Isotropic Light with a Compact Plant Leaf. J. Opt. Soc. Am. 1969, 59, 1376–1379. [Google Scholar] [CrossRef]
- le Maire, G.; François, C.; Dufrêne, E. Towards universal broad leaf chlorophyll indices using PROSPECT simulated database and hyperspectral reflectance measurements. Remote Sens. Environ. 2004, 89, 1–28. [Google Scholar] [CrossRef]
- Berger, K.; Rivera Caicedo, J.P.; Martino, L.; Wocher, M.; Hank, T.; Verrelst, J. A Survey of Active Learning for Quantifying Vegetation Traits from Terrestrial Earth Observation Data. Remote Sens. 2021, 13, 287. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Yuan, Q.; Shen, H.; Li, T.; Li, Z.; Li, S.; Jiang, Y.; Xu, H.; Tan, W.; Yang, Q.; Wang, J.; et al. Deep learning in environmental remote sensing: Achievements and challenges. Remote Sens. Environ. 2020, 241, 111716. [Google Scholar] [CrossRef]
- Xiao, Z.; Liang, S.; Wang, J.; Chen, P.; Yin, X.; Zhang, L.; Song, J. Use of General Regression Neural Networks for Generating the GLASS Leaf Area Index Product From Time-Series MODIS Surface Reflectance. IEEE Trans. Geosci. Remote 2014, 52, 209–223. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for Remote Sensing with Unmanned Aerial Vehicles in Precision Agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef] [PubMed]



| Parameter | Symbol | Unit | Range | Mean, STD |
|---|---|---|---|---|
| Leaf structure | N | unitless | 1–3 | 2, 1 |
| Chlorophyll content | Cab | μg/cm2 | 0.02–55 | 11.9, 11.5 |
| Carotenoid content | Ccar | μg/cm2 | 0.04–10 | 2.8, 1.7 |
| Anthocyanin content | Lanth | μg/cm2 | 0–17 | 4.1, 3.8 |
| Brown pigment content | Cbrown | arbitrary | Fixed at 0 | |
| Equivalent water thickness | Cw | cm | 0.0002–0.038 | 0.016, 0.007 |
| Dry matter content | Cm | g/cm2 | 0.0001–0.029 | 0.0086, 0.0035 |
| N | Cab | Ccar | Lanth | Cw | Cm | |
|---|---|---|---|---|---|---|
| N | 1.00 | −0.15 | −0.02 | −0.03 | 0.30 | 0.13 |
| Cab | −0.15 | 1.00 | 0.65 | −0.12 | 0.19 | 0.50 |
| Ccar | −0.02 | 0.65 | 1.00 | 0.25 | 0.25 | 0.40 |
| Lanth | −0.03 | −0.12 | 0.25 | 1.00 | 0.20 | 0.30 |
| Cw | 0.30 | 0.19 | 0.25 | 0.20 | 1.00 | 0.70 |
| Cm | 0.13 | 0.50 | 0.40 | 0.30 | 0.70 | 1.00 |
| Name | Formula | Reference |
|---|---|---|
| Red/Green | R680/R550 | [22] |
| ARI | 1/R550 − 1/R700 | [18] |
| mARI | (1/R550 − 1/R700) × R770 | [19] |
| mACI | R770/R550 | [28] |
| Pigment | Range | Mean | Standard Deviation | Pearson’s Correlation Coefficient (r) | ||
|---|---|---|---|---|---|---|
| Chlorophylls | Carotenoids | Anthocyanins | ||||
| Chlorophylls | 0.06–48.30 | 11.72 | 11.37 | 1.00 | 0.64 1 | −0.10 2 |
| Carotenoids | 0.08–6.75 | 2.32 | 1.57 | 0.64 1 | 1.00 | 0.23 1 |
| Anthocyanins | 0–13.58 | 3.99 | 3.62 | −0.10 2 | 0.23 1 | 1.00 |
| Spectral Data | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 | PC10 |
|---|---|---|---|---|---|---|---|---|---|---|
| LEAF | 76.97 | 92.24 | 96.40 | 98.07 | 98.99 | 99.72 | 99.85 | 99.91 | 99.95 | 99.97 |
| log-LEAF | 75.65 | 95.09 | 97.67 | 98.72 | 99.48 | 99.76 | 99.85 | 99.92 | 99.95 | 99.97 |
| SIMU | 77.12 | 91.59 | 98.35 | 99.10 | 99.73 | 99.95 | 99.98 | 99.99 | 100.00 | 100.00 |
| log-SIMU | 82.23 | 96.51 | 98.63 | 99.54 | 99.86 | 99.97 | 99.99 | 100.00 | 100.00 | 100.00 |
| Independent Variable | Neuron(s) | R2 | RMSE (μg/cm2) | ||||
|---|---|---|---|---|---|---|---|
| Training | Validation | In situ | Training | Validation | In Situ | ||
| Red/Green | 1 | 0.42 | 0.42 | 0.39 | 2.36 | 2.28 | 3.60 |
| ARI | 3 | 0.83 | 0.84 | 0.72 | 1.28 | 1.21 | 4.32 |
| mARI | 4 | 0.93 | 0.93 | 0.75 | 0.81 | 0.77 | 5.32 |
| mACI | 10 | 0.77 | 0.75 | 0.42 | 1.48 | 1.52 | 5.03 |
| (R550, R710, R770) | 1 | 0.93 | 0.91 | 0.79 | 0.81 | 0.88 | 5.12 |
| log-(R550, R710, R770) | 13 | 0.99 | 0.99 | 0.48 | 0.26 | 0.33 | 5.51 |
| R-(PC1–PC6) | 6 | 0.99 | 0.99 | 0.58 | 0.25 | 0.28 | 2.96 |
| R-(PC1–PC8) | 4 | 0.99 | 0.99 | 0.54 | 0.29 | 0.36 | 3.87 |
| log-(PC1–PC6) | 3 | 0.99 | 0.99 | 0.52 | 0.27 | 0.31 | 3.46 |
| log-(PC1–PC8) | 3 | 0.99 | 0.99 | 0.48 | 0.23 | 0.29 | 3.58 |
| Independent Variable | R2 | RMSE (μg/cm2) | ||||
|---|---|---|---|---|---|---|
| Training | Validation | In Situ | Training | Validation | In Situ | |
| Red/Green | 0.44 | 0.43 | 0.36 | 2.32 | 2.28 | 3.61 |
| ARI | 0.84 | 0.82 | 0.54 | 1.26 | 1.27 | 4.14 |
| mARI | 0.93 | 0.94 | 0.37 | 0.82 | 0.76 | 4.35 |
| mACI | 0.77 | 0.75 | 0.20 | 1.49 | 1.51 | 5.27 |
| (R550, R710, R770) | 0.89 | 0.88 | 0.70 | 1.12 | 1.10 | 3.34 |
| log-(R550, R710, R770) | 0.61 | 0.59 | 0.70 | 2.70 | 2.63 | 3.22 |
| R-(PC1–PC6) | 0.81 | 0.79 | 0.62 | 1.83 | 1.79 | 2.58 |
| R-(PC1–PC8) | 0.80 | 0.79 | 0.53 | 1.56 | 1.53 | 2.60 |
| log-(PC1–PC6) | 0.99 | 0.99 | 0.58 | 0.27 | 0.33 | 3.23 |
| log-(PC1–PC8) | 0.99 | 0.98 | 0.56 | 0.33 | 0.37 | 3.11 |
| Independent Variable | R2 | RMSE (μg/cm2) | ||||
|---|---|---|---|---|---|---|
| Training | Validation | In Situ | Training | Validation | In Situ | |
| Red/Green | 0.44 | 0.43 | 0.32 | 2.31 | 2.26 | 3.80 |
| ARI | 0.83 | 0.84 | 0.68 | 1.29 | 1.19 | 4.60 |
| mARI | 0.93 | 0.94 | 0.45 | 0.81 | 0.77 | 4.92 |
| mACI | 0.77 | 0.75 | 0.52 | 1.50 | 1.52 | 5.65 |
| R550, R710, R770 | 0.99 | 0.99 | 0.86 | 0.27 | 0.32 | 7.49 |
| log-(R550, R710, R770) | 0.99 | 0.99 | 0.79 | 0.27 | 0.32 | 7.76 |
| R-(PC1–PC3) | 0.97 | 0.96 | 0.59 | 0.53 | 0.60 | 2.69 |
| log-(PC1–PC3) | 0.83 | 0.82 | 0.76 | 1.28 | 1.27 | 2.24 |
| Independent Variable | MinLeafSize (Search Range) | R2 | RMSE (μg/cm2) | ||||
|---|---|---|---|---|---|---|---|
| Training | Validation | In Situ | Training | Validation | In Situ | ||
| Red/Green | 93 (50–100) | 0.45 | 0.43 | 0.38 | 2.30 | 2.27 | 3.65 |
| ARI | 298 (250–300) | 0.78 | 0.79 | 0.61 | 1.44 | 1.37 | 3.32 |
| mARI | 148 (90–150) | 0.91 | 0.91 | 0.64 | 0.93 | 0.93 | 3.75 |
| mACI | 147 (90–150) | 0.76 | 0.73 | 0.45 | 1.53 | 1.58 | 4.42 |
| (R550, R710, R770) | 60 (50–100) | 0.88 | 0.85 | 0.59 | 1.39 | 1.42 | 3.23 |
| log-(R550, R710, R770) | 95 (50–100) | 0.83 | 0.79 | 0.58 | 1.58 | 1.59 | 3.21 |
| R-(PC1–PC6) | 57 (50–100) | 0.90 | 0.87 | 0.56 | 1.16 | 1.26 | 2.65 |
| R-(PC1–PC8) | 51 (50–100) | 0.90 | 0.86 | 0.54 | 1.09 | 1.22 | 2.70 |
| log-(PC1–PC6) | 59 (50–100) | 0.92 | 0.90 | 0.66 | 1.05 | 1.10 | 2.33 |
| log-(PC1–PC8) | 77 (50–100) | 0.91 | 0.89 | 0.65 | 1.11 | 1.15 | 2.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yi, Q.; Chen, Y. The Hybrid Retrieval of Leaf Anthocyanin Content Using Four Machine Learning Methods. Forests 2025, 16, 804. https://doi.org/10.3390/f16050804
Li Y, Yi Q, Chen Y. The Hybrid Retrieval of Leaf Anthocyanin Content Using Four Machine Learning Methods. Forests. 2025; 16(5):804. https://doi.org/10.3390/f16050804
Chicago/Turabian StyleLi, Yingying, Qiuxiang Yi, and Yaoliang Chen. 2025. "The Hybrid Retrieval of Leaf Anthocyanin Content Using Four Machine Learning Methods" Forests 16, no. 5: 804. https://doi.org/10.3390/f16050804
APA StyleLi, Y., Yi, Q., & Chen, Y. (2025). The Hybrid Retrieval of Leaf Anthocyanin Content Using Four Machine Learning Methods. Forests, 16(5), 804. https://doi.org/10.3390/f16050804






