Abstract
Plant-associated microbes play a crucial role in host growth, health, and stress resistance. Host plants significantly influence the assembly of their microbial communities. To examine the influence of plant provenance on bacterial composition and plant growth promoting (PGP) properties, we compared readily culturable bacteria isolated from the root interior and rhizosphere of Tripterygium wilfordii Hook f., a woody Chinese medicinal plant, across three provenances grown in different regions. Regardless of the host growing environment, the abundance of endophytic bacterial colonies followed the trend: Fujian provenances > Hunan provenances > Hubei provenances. A total of 227 isolates were classified into 21 genera from the root interior and 40 genera from the rhizosphere, with Pseudomonas and/or Bacillus as the dominant genera. The taxonomic composition varied across plant provenances and host growing environments. Plants from the same provenance but cultivated in different locations exhibited greater similarity in their endophytic bacterial composition. The majority of the assayed strains displayed one or more PGP traits. Local plants harbored a higher proportion of PGP strains than non-local plants. Our findings indicate that plant provenance significantly influences the composition of root-associated culturable bacteria, particularly endophytic communities. Plants in their native environments may recruit PGP bacteria to enhance their fitness.
1. Introduction
Plants are closely associated with highly diverse and abundant microbes, which play crucial roles in plant growth, nutrient absorption, disease resistance, and responses to adverse environmental conditions [1,2]. Plant microbial spectra are influenced by numerous internal and external factors, such as host plant species and genotype, environmental conditions like soil chemistry and geographical location, and plant management practices [3,4]. Soil pH value, nutrient characteristics, and other environment variables, such as temperature, water availability, and sun radiation, can affect microbial communities [5,6,7,8]. Dissimilarity in plant microbial communities between host habitats was observed commonly [9]. Plants recruit microbes from extremely diverse soil microbial communities, and subsequently shape a taxonomically restricted root-associated microbiome [10]. Different plant species and even cultivars and genotypes within a species can support distinct bacterial communities in their rhizosphere, endosphere, and phyllosphere [8,11,12,13]. For example, endophytic bacteria in cultivated Ulleung-sanmaneul were less diverse than wild plants [14]. The root microbiome is documented to be related to plant evolution. As the phylogenetic distance between plants increases, the differences in their microbiomes become more pronounced [15]. In addition, potential sources of microbiome variation include plant age and developmental stage, as these factors may influence the assembly of microbial communities through alterations in the expression of plant functional traits [16,17,18,19]. The biochemical and physiological traits that different plants express affect the assembly of bacterial communities [20]. Plant root exudates, which vary greatly in types and amounts between different plants, can selectively recruit specific bacteria in the rhizosphere [21]. Various plant defense strategies associated with microbial infection drive the selection of surrounding microbial communities to shape the microbiome [22]. The innate immune system of plants also affects the colonization of beneficial microbes in hosts [23]. Plant provenances refer to the geographic origin of seed or plant material, which contain local genetic variations. Provenance selection is frequently employed in the genetic improvement of woody plants [24]. However, genetic variation among provenances within a single species may alter their microbiome and thereby affect plant performance.
Plant growth promoting (PGP) bacteria are beneficial microbes improving plant growth. The PGP bacteria benefit their hosts through several mechanisms, including nitrogen fixation, phosphate solubilization, and the production of indole acetic acid (IAA), 1-aminocyclopropane-1-carboxylate (ACC) deaminase enzyme, and siderophores [25]. As members of host-associated microbiota, PGP bacteria are also influenced by different plant species, and even different plant varietas. Modern strains of rice were found fostering a greater abundance of nitrogen-fixing endophytes than wild strains [26]. However, little is understood about the influence of plant provenances on PGP bacteria.
Tripterygium wilfordii Hook f., a perennial woody vine of the family Celastraceae, is a Chinese medicinal plant. T. wilfordii produces more than 70 compounds with various biological activities and has been extensively applied in clinics for the treatment of rheumatoid arthritis, systemic lupus erythematosus, chronic nephritis, and other autoimmune diseases [27]. T. wilfordii is mainly distributed in southern China, such as provinces of Fujian, Hubei, Hunan, Yunnan, Guizhou, Zhejiang, and Anhui. Due to different growing environments and resulting long-term selection, medicinal plants from different regions may present some differences in morphotypes and quality. In a forest farm in Fujian province of China, T. wilfordii plants from different regions were collected for screening of the optimal genotypes according to biomass production and accumulation of bioactive compounds. Among T. wilfordii populations of different provenances, development, physiology, and production of medicinal substances are distinguishable [28,29,30]. This study focuses on root-associated bacteria as they play an important role in plant performance by interacting with their hosts. We hypothesize that the taxonomic composition of culturable root-associated bacteria in T. wilfordii is influenced by plant provenance, and this variation may affect their PGP properties. Accordingly, our objective is to explore the role of plant provenance in shaping the bacterial communities and to identify which communities are more effective in promoting plant growth for potential agricultural and forestry applications.
2. Materials and Methods
2.1. Sample Collection
Five types of plant samples from three T. wilfordii provenances were collected from their native habitats and transplanted fields for bacterial isolation. These samples included (i) Fujian provenance plants from the native habitat located in Sanming (E 117°56′28″, N 25°48′22″), Fujian, China, i.e., local Fujian T. wilfordii (LFJ); (ii) Hubei provenance plants from the native habitat located in Xianning (E 113°57′28″, N 29°11′29″), Hubei, China, i.e., local Hubei T. wilfordii (LHB); (iii) Hunan provenance plants from the native habitat located in Xiangtan (E 112°31′35″, N 27°40′59″), Hunan, China, i.e., local Hunan T. wilfordii (LHN); (iv) Hubei provenance plants from the transplanted field located in Fujian, i.e., non-local Hubei T. wilfordii (NHB); and (v) Hunan provenance plants from the transplanted field located in Fujian, i.e., non-local Hunan T. wilfordii (NHN). The non-local Hubei and Hunan T. wilfordii (NHB and NHN) plants originated from Hubei and Hunan, respectively, but were transplanted to a national forest farm in Fujian via cutting propagation in 2013. These non-local plants were grown in the same forest farm as local Fujian T. wilfordii (LFJ). Cutting propagation was performed with 12–15 cm branches, following the method of Lin et al. [31]. The environmental conditions of the three sampling locations differ (Table 1). Fujian has the highest annual average temperature, precipitation, and soil temperature, but the lowest soil soluble potassium. The soil chemistry of Hunan is similar to that of Fujian, except for a significant decrease in organic matter. Hubei exhibits higher soil nutrient levels than the other two sampling locations. To isolate root-associated bacteria, four independent plants per provenance were selected from the three geographical locations for sampling. Roots with soil from the four plants were collected at a depth of 5 to 20 cm and bulked to obtain a representative composite sample. All samples were placed into plastic bags and immediately transported to the laboratory on ice.

Table 1.
Environmental conditions of sampling locations.
2.2. Isolation and Culture of Bacterial Strains
Loose soil was shaken off the roots, while the tightly adhering soil was retained as the rhizosphere sample. Root segments (3–10 cm in length) with attached rhizosphere soil were transferred into a centrifuge tube containing 10 mL of sterile distilled water and vortexed for approximately 1 min. The soil suspension was then diluted successively up to 10−7 with sterile water. The remaining root segments were surface sterilized with 2% sodium hypochlorite solution for 20 s, and 70% ethanol for 10 min, followed by five rinses with sterilized distilled water. To verify root surface sterilization, the water from the last rinse was placed on nutrient agar (NA) plate and the plates were incubated at 30 °C for 3 d. No colony growing on the plates was used to confirm surface sterility of the roots. Surface disinfected tissues were ground using sterilized pestles and mortars. Extracts of root tissues were serially diluted from 10−1 to 10−5. Aliquots of 100 μL of each dilution were spread on the NA plates and incubated at 30 °C for 2–3 d. The total number of colonies was counted for calculating the colony number per gram fresh weight (FW). Based on bacteria’s morphological characteristics, such as size, shape, color, edge, and texture, different colonies were picked up at random and re-streaked on fresh NA plates for purification.
2.3. 16S rRNA Molecular Identification
The pure strains obtained from isolation on NA plates were grown separately in nutrient broth (NB) media for 24 h incubation at 30 °C with shaking at 180 rpm. Bacterial cells were collected via centrifugation and genomic DNA was extracted. Then, 16S rRNA gene sequences were amplified using universal primers 27F and 1492R on the GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA, USA). The 20 μL PCR mixture contained 2.0 μL 10×Ex Taq buffer, 1.6 μL 2.5 mM dNTP mixture, 0.8 μL 5 pmol F primer, 0.8 μL 5 pmol R primer, 1 μL (0.1 μg) template DNA, 0.2 μL 5 u Ex Taq, and 13.6 μL ddH2O. The PCR reaction was initialized by a denaturing step at a 95 °C for 5 min, followed by 10 cycles of 30 s at 95 °C, 30 s at 62 °C, 1 min 30 s at 72 °C, 35 cycles of 30 s at 95 °C, 30 s at 57 °C, 1 min 30 s at 72 °C, and a final extension for 10 min at 72 °C. Amplification products were examined by agarose gel electrophoresis. Sequencing was performed by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) on an ABI 3730XL sequencer (Applied Biosystems). The results were processed and edited with BioEdit software (v7.2). The 16S rRNA gene sequences were deposited to GenBank under the accession numbers: MG516108-MG516210, MK611652, MK611769, MK611774, MK611793, MK615137-MK615144, MH024379, MH024380, and MG571645-MG7516756. Homologous sequence analysis and multiple sequence alignment were conducted using the Basic Local Alignment Search Tool (BLAST) within the National Center for Biotechnology Information (NCBI) database and EzBioCloud Database.
2.4. Plant Growth Promoting Activities of Isolates
Production of IAA in isolated strains was determined according to Glickmann and Dessaux [32]. Each strain (100 μL inoculum with approximately 106 cfu/mL) was inoculated in NB media supplemented with 0.5 g/L L-tryptophan and incubated at 30 °C and 180 rpm for 48 h. After centrifugation at 10,000× g for 10 min, 2 mL culture supernatant was placed into test tubes to mix with 4 mL of Salkowski’s reagent (1 mL 0.5 M FeCl3 + 49 mL 35% HClO4). The intensity of the pink color after incubation for 30 min in the dark at room temperature was measured spectrophotometrically at 530 nm. The IAA concentration was calculated using a calibration curve of pure IAA as a standard.
Phosphate solubilizing activity of each bacterial isolate was determined by observing a clear yellowish color zone around the colonies formed by solubilization of insoluble tricalcium phosphate on inorganic phosphate medium based on tricalcium phosphate [33], which contained (NH4)2SO4 0.5 g, NaCl 0.3 g, MgSO4·7H2O 0.3 g, MnSO4·H2O 0.03 g, FeSO4·7H2O 0.03 g, KCl 0.3 g, glucose 10.0 g, Ca3(PO4)2 5.0 g, agar 18.0 g, and d.H2O 1 L with pH 7.0. The plates were incubated for 7 d at 30 °C. The ratio of the halo zone diameter to the colony diameter was measured as phosphate solubilizing level.
Capability of strains to grow in a nitrogen-free media was evaluated. Components of the media are sucrose 10.0 g, KH2PO4 0.2 g, MgSO4·7H2O 0.2 g, NaCl 0.2 g, CaCO3 5.0 g, agar 20.0 g, d.H2O 1 L, and pH 7.0–7.2. Each strain was spotted on the media and incubated at 30 °C with a transfer to a fresh nitrogen-free media every 3 d for 5 times. The growth of colonies was recorded as positive result.
Siderophore production of isolates was determined by the modified method of Schwyn and Neilands [34]. Bacterial isolates were streaked on blue agar plates containing Chrome Azurol S (CAS) dye (sucrose 2 g, casein acids hydrolysate 3 g, 1 mM CaCl2 0.1 mL, 1 mM MgSO4·7H2O 2 mL, agar 18 g, d.H2O 1 L, CAS indicator solution 50 mL, and 0.1 M phosphate-buffer solution 50 mL) and incubated at 30 °C for 5 d. Appearance of yellow halos around the inoculated colonies was recorded as a positive result.
2.5. Data Analyses
Alpha diversity was estimated using species richness and the Shannon–Wiener diversity index [35]. Species richness refers to the total number of species identified from a sample. Relative proportion was calculated as the percentage of bacterial colonies belonging to a specific species or genus relative to the total number of bacterial colonies recovered from a given sample. Shannon–Wiener diversity index was computed based on the relative proportion at the species level. Beta diversity were estimated using the Jaccard index [36] in terms of presence/absence of genera and the Bray–Curtis index [37] based on the relative proportion at the genus level. All diversity indices were computed using the vegan package in R software (v4.1.2) [38]. The Venn diagram at the genus level was plotted at https://www.bioinformatics.com.cn (accessed on 3 January 2025) and labeled with the specific genus. The similarity metric was visualized using the ggplot2 package in R software [39].
3. Results
3.1. Population Abundance
Plant provenance and host growing conditions affected the abundance of culturable bacteria of T. wilfordii, with colony counts ranging from 3.86 × 102 to 1.37 × 105 cfu/g FW in roots and from 2.17 × 105 to 2.61 × 106 cfu/g FW in rhizosphere soils (Table 2). Regardless of host growing environments, the quantity of endophytic bacteria from Fujian provenances (LFJ) consistently exhibited the highest levels among the samples of three different provenances, followed by Hunan provenances (LHN and NHN). Hubei provenances (LHB and NHB) consistently exhibited the lowest number of endophytic bacterial colonies. However, the pattern of rhizobacterial population densities among different provenances was inconsistent with that of endophytic bacteria, although the most abundant bacteria were also supported by the LFJ rhizosphere. For the samples from the same provenance but different growing environments, the LHB samples presented more endophytic bacteria and less rhizobacteria in comparison to the NHB samples. However, LHN had less endophytic bacteria but more abundant rhizobacteria than the NHN samples.

Table 2.
Number of culturable endophytic bacteria and rhizobacteria from plants of different provenances.
3.2. Taxonomic Composition and Diversity of Isolations
A total of 115 endophytic strains were obtained, including 23 from LFJ, 17 from NHN, 31 from NHB, 22 from LHN, and 22 from LHB, based on their distinct morphological appearance on NA media (Table 2). These endophytic isolates were grouped into 52 species from 21 genera of three phyla by comparing 16S rRNA sequences (Table S1). The most diverse group was Pseudomonas, followed by Bacillus. The endophytic bacteria isolated from LFJ were represented by one phylum, namely Pseudomonadota, and assigned to 14 species from seven genera (Table 3). Pseudomonas was the predominant taxon, accounting for 42.9% of the species. The endophytic bacteria isolated from NHN were represented by Pseudomonadota. All the strains isolated were affiliated with nine species from five genera. Pseudomonas was the predominant genus with 33.3% of the species. The endophytic bacteria isolated from NHB were grouped into Bacillota and Pseudomonadota, and most of the isolates belonged to Bacillota groups. All the isolates were assigned to 13 species from six genera. Bacillus, covering 69.2% of the species, was the major genus. The root endophytic bacteria isolated from LHN belonged to two phyla, Pseudomonadota and Bacillota, with Pseudomonadota being more prevalent. All the strains isolated were represented by 15 species from nine genera. Members of genera Pseudomonas and Bacilus were predominant, accounting for 26.7% and 20.0% of the species, respectively. Compared with NHN, LHN showed higher species richness. The endophytic strains isolated from LHB were grouped into two phyla, Pseudomonadota and Bacillota. Pseudomonadota were the major phylum. All isolates belonged to 12 species from eight genera in which Pseudomonas, containing 25.0% of the species, were the primary members. Overall, endophytic bacteria from different provenances exhibited similar species richness, with the exception of that from NHN. Additionally, the Shannon–Wiener diversity indices were comparable across most provenances from different locations, except for NHN, where the diversity was much lower (Figure 1).

Table 3.
Pattern of taxonomic richness for root-associated bacteria across the provenances and host growing locations.
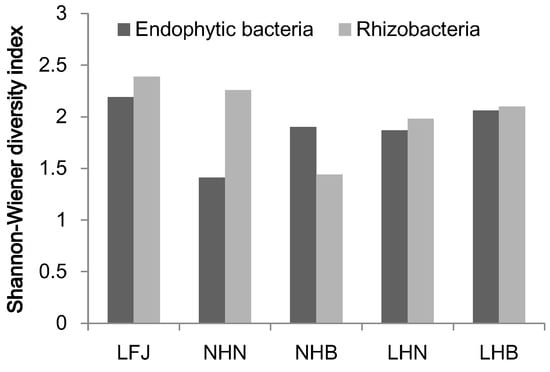
Figure 1.
Shannon–Wiener diversity index of endophytic bacteria and rhizobacteria from sample groups of different provenances based on the relative proportion at the species level.
A total of 112 strains were isolated from the rhizosphere (Table 2). The diversity of morphotypes was highest in the LHB samples. The 112 strains were classified into 56 species from 40 genera of three phyla, including Pseudomonadota, Actinomycetota, and Bacillota (Table S1). Pseudomonadota was most frequently recovered. The most diverse genus was Pseudomonas with 17 species. The rhizobacteria isolated from LFJ were affiliated to 15 species from eight genera of two phyla (Table 3). Pseudomonas, the predominant taxon, accounted for 40.0% of the species. The strains isolated from NHN were assigned to 14 species from seven genera of Pseudomonadota. Pseudomonas in NHN contained 57.1% of the species. The strains isolated from NHB rhizosphere samples were assigned to 12 species from seven genera of two phyla. Pseudomonas and Burkholderia were most frequently recovered. The strains isolated from LHN rhizosphere samples belonged to 14 species from 11 genera of two phyla, which indicated comparable species richness to that from NHN. Pseudomonas covered 21.4% of the species in LHN. The strains from LHB belonged to 19 species from nine genera of three phyla, indicating higher species richness than NHB. Pseudomonas covered 29.4% of the species. Except for the lower Shannon–Wiener diversity index observed in rhizobacteria from NHB, rhizobacteria from the other samples exhibited similar diversity (Figure 1).
3.3. Common and Specific Taxa Between Different Provenances
A distinct root-associated bacterial composition characterized three provenances from different habitats (Table S1 and Table 3). Only members of Pseudomonadota were isolated from all samples. LHB and NHB shared two endophytic and rhizosphere phyla, respectively. However, LHN and NHN, as well as LFJ, NHN, and NHB, shared only one phylum. Regardless of endophytic or rhizosphere bacteria, Pseudomonas was found in all samples (Figure 2), which could be a core population. Variovorax and Paenarthrobacter co-occurred exclusively in root and/or rhizosphere samples of LHN and NHN, indicating that they could be a population specific to Hunan T. wilfordii. Bacillus, which co-occurred exclusively in both root and rhizosphere samples of LHB and NHB, exhibited a strong association with Hubei T. wilfordii. Four root endophytic genera and five rhizobacterial genera, including Herbaspirillum and Rhizobium commonly observed in root and rhizosphere, were presented only in LHN, whereas endophytic Ralstonia and rhizosphere Leclercia were presented only in NHN. Three root endophytic genera and two rhizobacterial genera, including Rahnella commonly observed, were recovered only from LHB, whereas endophytic Brevibacterium, Lysinibacillus, and rhizosphere Flavobacteria were recovered only from NHB. These unique populations maintained a strong relationship with growing environments of their hosts. In addition, endophytic Enterobacter, which was shared exclusively among LFJ, LHB, and LHN, exhibited a strong association with local plants.
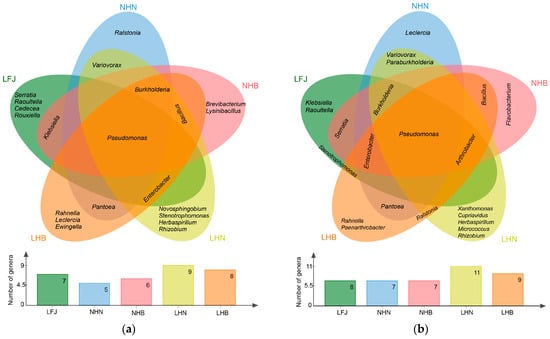
Figure 2.
Shared genus of (a) endophytic bacteria and (b) rhizobacteria among sample groups of different provenances.
Both the Jaccard index based on shared and unique genera and the Bray–Curtis index based on population abundance indicated that the similarity coefficients of endophytic bacteria between NHB and LHB, as well as between NHN and LHN, were relatively higher, exceeding those observed among LFJ, NHB, and NHN (Figure 3). This revealed greater similarities between samples from the same provenance but different growing environments, compared to those from identical growing environments but different provenances. Moreover, the similarity between NHN and NHB, based on similarity coefficients of endophytic bacteria, was lower than that between LHN and LHB, despite NHN and NHB originating from the same growing environment. These results indicated that provenance played a more significant role in shaping the endophytic communities than the host growing environment. Additionally, the similarity coefficients of endophytic bacteria between NHN and NHB, as well as between LHN and LHB, were both higher than those involving LFJ, reflecting that among the three provenances, Fujian T. wilfordii was less similar to Hunai or Hubei T. wilfordii compared to the similarity between Hunan and Hubei T. wilfordii. In contrast to endophytic bacteria, the similarity coefficients of rhizobacteria between NHB and LHB, as well as between NHN and LHN, were comparable to those among LFJ, NHB, and NHN. Furthermore, according to the similarity coefficients, NHN and NHB from the same growing environment showed higher rhizobacterial similarity than LHN and LHB from different growing environments. These findings indicated that the composition of rhizobacterial taxa was highly impacted by both the growth environment and plant provenance.
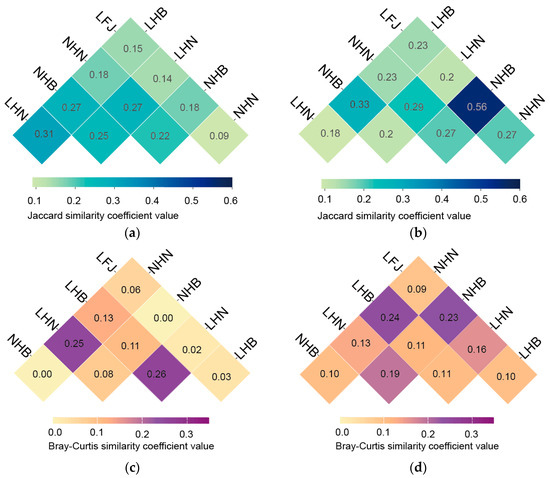
Figure 3.
Heatmap of Jaccard similarity coefficients of (a) endophytic bacteria and (b) rhizobacteria, and Bray–Curtis similarity coefficients of (c) endophytic bacteria and (d) rhizobacteria at genus level.
3.4. Plant Growth Promoting Traits
In the present study, 134 representative root endophytic and rhizobacterial strains were selected to assay their plant growth promoting traits, according to their 16S rRNA sequencing and sample distribution. Most of the assayed strains displayed one or more PGP traits (Table S2). The majority of the strains were able to produce IAA at levels up to 49.15 μg/mL. Nine endophytic strains and five rhizobacterial strains exhibited a relatively high production of IAA with levels exceeding 10 μg/mL. These strains involved five genera, i.e., Enterobacter, Rouxiella, Klebsiella, Pantoea, and Raoultella. Genus Enterobacter including seven strains, FJG6, FJG10, YHBG16, YHNG17, FJT14, HBT4, and YHBT2, was the most active producer, followed by the genera Klebsiella including strains FJG12, HBG30, and FJT2. Strains HBG30 (49.15 μg/mL) in Klebsiella, HBT4 (44.90 μg/mL), YHBG16 (31.68 μg/mL), FJT14 (23.49 μg/mL), YHBT2 (23.32 μg/mL), and YHNG17 (20.18 μg/mL) in Enterobacter, and YHBG10 (26.06 μg/mL) in Pantoea were the top-ranked IAA producers. Twelve strains produced a moderate amount (5–10 μg/mL) of IAA, of which Pseudomonas strains were the most abundant. Among the five types of samples, LFJ had the largest number of high IAA producers, with seven strains producing IAA exceeding 10 μg/mL and eight strains producing moderate levels of IAA. LHB contained four strains capable of producing high levels of IAA, and two strains displayed moderate IAA production levels. Strains HBG30 and HBT4, identified as the two highest IAA producers, and two moderate IAA producers were isolated from NHB. Only one endophytic strain from LHN produced high levels of IAA. The strains from NHN all displayed low IAA production levels. Those strains able to produce a relatively large amount of IAA were isolated more commonly from LFJ and LHB than NHB and NHN.
About 39.7% of endophytic strains and 35.2% of rhizobacterial strains exhibited phosphate solubilization activity by forming yellow halos in inorganic phosphorus media (Table S2). These phosphate solubilizers were affiliated with 14 genera, of which Pseudomonas and Burkholderia were the dominant strains. Strains YHBG18 in Rahnella and YHBT18 in Pantoea exhibited the highest phosphate solubilization, followed by strain HNT12 in Burkholderia. LHN contained the most abundant phosphate-solubilizing strains, followed by LHB and LFJ. The quantities of phosphate solubilizing strains isolated from local T. wilfordii were much more than those from non-local plants.
In the present study, 28.9% of the assayed strains were identified as potential nitrogen fixers. These nitrogen-fixing strains belonged to 15 genera, of which the predominant bacteria were Enterobacter. LHB showed the most abundant nitrogen-fixing strains, followed by LFJ. NHB exhibited a much lower quantity of nitrogen-fixing strains compared to LHB. Similarly to the abundance of phosphate-solubilizing strains, nitrogen-fixing strains isolated from the local T. wilfordii were more than those from the non-local plants.
Approximately 94.8% of the assessed strains exhibited the ability to produce siderophores. Among these, Pseudomonas strains were the most abundant, followed by Bacillus and Burkholderia strains. Different from the distribution of phosphate-solubilizing strains and nitrogen-fixing strains, the five types of samples all possessed a high proportion of siderophore-producing strains. This suggested that the ability to produce siderophores was commonly found in root-associated bacteria.
4. Discussion
4.1. Isolation of Root-Associated Bacteria
In this study, root-associated bacteria of T. wilfordii, an important medicine plant of China, were isolated. The endophytic bacterial population density from the different provenances ranged from 3.80 × 102 to 1.37 × 105 cfu/g root on NA media (Table 2). The quantity of endophytes isolated was in the range reported previously for cotton [40,41], potato [42], Salicornia europaea L. [43], etc. Rhizobacterial population density (2.17 × 105~2.61 × 106 cfu/g soil) of T. wilfordii fell within the range reported for other plants, such as avocado trees [44], S. europaea [43], and pineapple plant [45]. In present study, the population densities of rhizosphere and especially endophytic bacteria were variable among the three different provenance samples in spite of the host growing environments and also among the samples from the same host provenances but different growing environments (Table 2). Bacterial population densities are influenced by a variety of factors, including host plant species and genotype, soil conditions, and environmental factors [46,47,48,49]. It is possible that variation in culturable bacterial number among our samples is a result of plant provenances and growing region selection on microbes. Our findings indicated that regardless of the host growing environment, the number of endophytic bacterial colonies among the three provenances exhibited a consistent trend, i.e., LFJ > LHN and NHN > LHB and NHB (Table 2). In contrast, no similar pattern was observed in the rhizobacteria across the provenances. This suggested that the endophytic bacteria were influenced more by plant provenances than by the soil condition and environmental factors. Cordero et al. [47] also observed that community structure, species richness, and diversity of root endophytic bacteria were mainly impacted by the host crop, whereas the rhizobacterial communities varied among crop species and (or) sampling locations. Bacterial quantities in LHB and LHN samples were not necessarily higher than those in NHB and NHN, indicating that plants in the local environment did not necessarily promote bacterial colonization.
Endophytic and rhizosphere bacteria isolated from the different provenances represent 21 genera from four phyla and 40 genera of three phyla, respectively (Table S1). Pseudomonas and/or Bacillus were the dominant bacteria. In addition, of all the genera identified, only Pseudomonas was ubiquitous throughout the samples (Table 3 and Figure 2). Strains of Bacillus and Pseudomonas strains have been extensively reported to dominate the roots and rhizosphere of various plants, such as lavender [50], Panax ginseng [51], and Sorghum [52]. These bacteria can thrive in diverse environmental conditions and exhibit resistance to various stresses, which contributes to their dominance [53,54]. Various Pseudomonas sp. function as plant probiotic bacteria with plant growth promoting capability [55,56] and antagonistic activity against plant pathogens [57,58]. Bacillus strains have been reported possessing plant growth promoting traits [59] and bioactivity, able to produce antimicrobials, siderophores, and phytotoxins [60]. The extensive colonization of Pseudomonas and Bacillus within the rhizosphere may be crucial for plant growth promotion and disease suppression.
4.2. Influence of Host Provenance and Growing Environment on the Taxonomic Composition
The variation in the composition of root-associated bacteria from the three different provenances, including those grown in the same regions (Table 3), revealed the difference in the genetic background of the host provenances. Plant gene expression plays a significant role in root exudation [61] which is considered a key determinant of root-associated bacterial communities [62,63,64]. Differences in host genetics can cause variation in the composition of microbes associated with the roots [65]. Distinct endophytic bacterial diversity and composition were observed among three Pitayas provenances, although the difference was less pronounced than that observed between species [66]. It is possible that genetic variations among T. wilfordii provenances lead to differences in root exudation, which are sufficient to recruit specific bacteria as rhizosphere members or endophytes. In the present study, Variovorax and Paenarthrobacter and Bacillus were identified as bacterial groups potentially specific to Hunan T. wilfordii and Hubei T. wilfordii, respectively (Figure 2).
Similarity coefficients indicated host provenance, rather than growing environment, was a crucial factor associated with the taxonomic composition of endophytic bacteria (Figure 3), which was in accordance with the results with colony quantities (Table 2). Blain et al. [67] also found that similar plant species had largely similar compositions of root endophytic bacteria, despite different sampling locations. Bonito et al. [68] referred that bacterial communities in the root were more tightly structured by plant host species than by soil origin. The absence of correlation between endophytic bacterial abundance and soil factors confirmed that root endophytic bacteria of crops was influenced more by factors associated with the host plant than by soil characteristics [47]. These findings supported the idea that the host plant plays a more important role in the assembly of the endosphere microbiome than that of the rhizosphere compartment [3]. In fact, diverse root exudates and various defense mechanisms of different plants affect their capacity to recruit various bacteria as endophytes [23,69,70,71]. Additional factors involving growth habits, developmental stage, health statuses, and response to environmental stresses also influences the selection of specific endophytic bacteria [72,73]. Geographical isolation is an important driver of the variance of plants [74]. In this study, a greater genetic variance was expected between Fujian and Hunan/Hubei provenances than that between Hunan and Hubei provenances due to the increased geographic distance and the more diverse natural environmental conditions. As a result, larger differences in endophytic bacterial composition between Fujian and Hunan/Hubei provenances with lower similar coefficients were observed, compared to that between Hunan and Hubei provenances (Figure 3). Our results confirmed that the plant provenance and homogeneity serve as selective pressures on the root endophytic bacterial populations.
Owing to the influence of climate, soil, and other environmental conditions, the composition of plant-associated microbial community may have regional specificity [48,75,76]. For example, more bacterial species and genera were isolated from tomato roots grown under tropical conditions in Indonesia than those grown under temperate conditions in Germany, and more than 60% of species were recovered exclusively from one of the regions [77]. In the present study, root-associated bacteria exhibited distinct taxonomic compositions across three regions, each characterized by unique populations (Table 3 and Figure 2). For the plants of the same provenance, similarity coefficients of rhizobacteria from the same region were higher than from different regions (Figure 3). These findings revealed the potential of environmental dependence of root-associated bacteria, especially rhizobacteria. Soil conditions and environmental factors can influence root bacterial communities by altering soil microbial communities which act as a source community [75]. Resource availability and soil pH has been found to be drivers of soil bacterial communities [78,79]. Plant–microbial interactions, influenced by soil conditions and environmental factors, also play a significant role in shaping root bacterial communities [73]. The effects of soil properties on endophytic bacteria are typically attenuated due to the selectivity and mechanical barrier of plants to soil microorganisms [80]. However, soil factors can still indirectly influence endophytic bacteria by mediating the physiology or root exudation of plants [48].
4.3. Plants in Their Native Soil Support Abundant Plant Growth Promoting Bacteria
In the present study, root-associated bacteria in five types of samples exhibited distinguishing differences in PGP traits, including IAA production, phosphate solubilization, and nitrogen fixation (Table S2). Plants can enrich their rhizosphere with beneficial bacteria regardless of the environment [81]. A previous study revealed a cultivar-dependent relationship between plant growth and the composition of the recruited microbiome [69]. Modern barley cultivars harbored more abundant plant growth promoting rhizobacterial genus Pseudomonas. Due to the varied metabolites produced by different bacteria, their structural diversity leads to functional diversity [82]. Environmental conditions play an important role not only in the diversity but also the functional traits of culturable microbiomes [83]. In our study, local T. wilfordii, LFJ, LHN, and LHB, showed a greater abundance of functional bacteria with abilities of high and moderate IAA production, N fixation, and phosphate solubilization than non-local T. wilfordii (Table S2). Beneduzi et al. [84] also observed that some PGP activities of bacterial strains isolated from sugarcane of native varieties varied among the six distinct production regions of Sul State, Brazil. PGP microbes were isolated from the rhizosphere of plants treated with native soil, whereas they are absent in the rhizosphere of greenhouse soil-treated plants [85]. These findings suggest that the adaptability of plants influences their recruitment of PGP bacteria. The better adaptability of plants to their native habitat appears to form more effective symbiotic relationships with PGP bacteria. As a result, the improved growth, survival, or stress resistance of plants in their native habitat may also be supported by these recruited functional bacteria. Plants treated with native soil, which harbored more PGP bacteria, showed improved growth compared to those treated with non-native soil [85]. Microbiomes in the native soils rather than the disinfected soil stimulated an immune response in tomato plants, thereby providing protection against bacterial wilt disease [86]. Some isolates of arbuscular mycorrhizal fungi and bacteria from indigenous bahiagrass facilitate the growth of their host in native soil [87]. These evidences confirm that functional microbes recruited from native soils can be an important contributor to the fitness of their host plants within those environments.
5. Conclusions
Composition of culturable bacteria isolated from T. wilfordii indicated that plant provenances were colonized by specific bacterial consortia associated with their roots. This result suggests that genetic variations among plant provenances are sufficient to influence bacterial recruitment. Plants from the same provenance grown in different locations exhibited greater similarity in endophytic bacterial composition than those from different provenances grown in the same location. However, the similarity of rhizobacteria among the same provenance grown in different locations was comparable to that observed among different provenances grown in the same location. These findings indicate that endophytic bacteria have a closer association with host plants than rhizobacteria. Local plants harbored a higher proportion of strains with PGP potentials compared to non-local plants, suggesting that plants in their native environments recruit more beneficial bacteria to support their growth. Consequently, in provenance selection, attention should be given not only to plant phenotypes but also to their associated microbiomes. Utilizing native plants facilitates the natural recruitment of beneficial microbes, offering a potential strategy to enhance plant health and growth, thereby reducing dependence on chemical fertilizers and pesticides. As this study was limited to culturable bacteria, further research using next-generation sequencing technologies is needed to uncover broader microbial diversity and reveal additional beneficial microbes that were not captured in the current study. Future investigation should also examine how soil properties and climate interact with native plants to influence beneficial microbial recruitment, which is essential for fully harnessing the potential of plant-associated microbes in sustainable agricultural and forestry practices.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/f16040637/s1, Table S1: Classification of the root-associated bacteria based on the 16S rRNA gene sequence similarity; Table S2: Multiple plant growth promoting activities of root endophytic and rhizosphere strains from different provenances.
Author Contributions
Conceptualization, P.S. and C.W.; Methodology, P.S. and Y.W.; Software, R.D., C.Y. and D.M.; Formal analysis, P.S.; Investigation, Y.W., Z.L. and L.F.; Data curation, R.D. and C.Y.; Writing—original draft, P.S.; Writing—review and editing, P.S. and L.F.; Visualization, R.D., C.Y., and D.M.; Supervision, P.S. and L.F.; Funding acquisition, L.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31000264, 31470577), the Natural Science Foundation of Fujian Province of China (2022J01124), and the Special Project for Science and Technology Innovation of Fujian Agriculture and Forestry University (KFB24120A).
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| PGP | Plant growth promoting |
| IAA | Indole acetic acid |
| LFJ | Local Fujian T. wilfordii |
| NHB | Non-local Hubei T. wilfordii |
| NHN | Non-local Hunan T. wilfordii |
| LHB | Local Hubei T. wilfordii |
| LHN | Local Hunan T. wilfordii |
| NA | Nutrient agar |
| NB | Nutrient broth |
| FW | Fresh weight |
| CAS | Chrome Azurol S |
References
- Tian, L.; Lin, X.; Tian, J.; Ji, L.; Chen, Y.; Tran, L.-S.P.; Tian, C. Research advances of beneficial microbiota associated with crop plants. Int. J. Mol. Sci. 2020, 21, 1792. [Google Scholar] [CrossRef] [PubMed]
- Daraz, U.; Ahmad, I.; Li, Q.; Zhu, B.; Saeed, M.; Li, Y.J.; Wang, X. Plant growth promoting rhizobacteria induced metal and salt stress tolerance in Brassica juncea through ion homeostasis. Ecotoxicol. Environ. Saf. 2023, 267, 115657. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Themaat, E.V.L.v.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Chen, S.; Tang, J.; Xu, J.; Peng, L.; Wu, P.; Li, Q. The impact of abandoned iron ore on the endophytic bacterial communities and functions in the root systems of three major crops in the local area. Front. Microbiol. 2025, 16, 1536083. [Google Scholar] [CrossRef]
- Deng, S.; Ke, T.; Li, L.; Cai, S.; Zhou, Y.; Liu, Y.; Guo, L.; Chen, L.; Zhang, D. Impacts of environmental factors on the whole microbial communities in the rhizosphere of a metal-tolerant plant: Elsholtzia haichowensis Sun. Environ. Pollut. 2018, 237, 1088–1097. [Google Scholar] [CrossRef]
- Silva, I.; Alves, M.; Malheiro, C.; Silva, A.R.R.; Loureiro, S.; Henriques, I.; Gonzalez-Alcaraz, M.N. Short-term responses of soil microbial communities to changes in air temperature, soil moisture and UV radiation. Genes 2022, 13, 850. [Google Scholar] [CrossRef]
- Tabassum, N.; Ahmed, H.I.; Parween, S.; Sheikh, A.H.; Saad, M.M.; Krattinger, S.G.; Hirt, H. Host genotype, soil composition, and geo-climatic factors shape the fonio seed microbiome. Microbiome 2024, 12, 11. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Hartman, K.; Tringe, S.G. Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef]
- Matthews, A.C.; Pierce, S.; Hipperson, H.; Raymond, B. Rhizobacterial community assembly patterns vary between crop species. Front. Microbiol. 2019, 10, 581. [Google Scholar] [CrossRef] [PubMed]
- Morella, N.M.; Weng, F.C.-H.; Joubert, P.M.; Metcalf, C.J.E.; Lindow, S.; Koskella, B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. USA 2020, 117, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Maestro-Gaitán, I.; Granado-Rodríguez, S.; Redondo-Nieto, M.; Battaglia, A.; Poza-Viejo, L.; Matías, J. Unveiling changes in rhizosphere-associated bacteria linked to the genotype and water stress in quinoa. Microb. Biotechnol. 2023, 16, 2326–2344. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Kim, Y.S.; Lee, Y.H. Characteristics of rhizosphere and endogenous bacterial community of Ulleung-sanmaneul, an endemic plant in Korea: Application for alleviating salt stress. Sci. Rep. 2022, 12, 21124. [Google Scholar] [CrossRef]
- Bouffaud, M.-L.; Poirier, M.-A.; Muller, D.; Moenne-Loccoz, Y. Root microbiome relates to plant host evolution in maize and other Poaceae. Environ. Microbiol. 2014, 16, 2804–2814. [Google Scholar] [CrossRef]
- Bressan, M.; Roncato, M.-A.; Bellvert, F.; Comte, G.; Haichar, F.e.Z.; Achouak, W.; Berge, O. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 2009, 3, 1243–1257. [Google Scholar] [CrossRef]
- Karina, B.; Rodolfo, D.; David, S.; Paul, B. Ontogenetic switches from plant resistance to tolerance: Minimizing costs with age? Ecol. Lett. 2007, 10, 177–187. [Google Scholar] [CrossRef]
- Pongrac, P.; Vogel-Mikus, K.; Regvar, M.; Tolra, R.; Poschenrieder, C.; Barcelo, J. Glucosinolate profiles change during the life cycle and mycorrhizal colonization in a Cd/Zn hyperaccumulator Thlaspi praecox (Brassicaceae). J. Chem. Ecol. 2008, 34, 1038–1044. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; del Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.B.; Pulkkinen, P. Genotype-environment interaction and stability in growth of aspen hybrid clones. For. Ecol. Manag. 2003, 173, 25–35. [Google Scholar] [CrossRef]
- Fanai, A.; Bohia, B.; Lalremruati, F.; Lalhriatpuii, N.; Lalmuanpuii, R.; Singh, P.K. Plant growth promoting bacteria (PGPB)-induced plant adaptations to stresses: An updated review. PeerJ 2024, 12, e17882. [Google Scholar] [CrossRef]
- Engelhard, M.; Hurek, T.; Reinhold-Hurek, B. Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ. Microbiol. 2000, 2, 131–141. [Google Scholar] [CrossRef]
- Kumar, D.S.S.; Cheung, H.Y.; Lau, C.S.; Chen, F.; Hyde, K.D. In vitro studies of endophytic fungi from Tripterygium wilfordii with anti-proliferative activity on human peripheral blood mononuclear cells. J. Ethnopharmacol. 2004, 94, 295–300. [Google Scholar] [CrossRef]
- Liang, A.; Yu, C.; Wu, C.; Tu, Y.-h.; Lin, Z.-s.; Hong, W.; Li, J. Seasonal dynamics of nutrient and tripolide contents in leaves of Tripterygium wilfordii from different provenances. Chin. J. Appl. Environ. Biol. 2018, 24, 299–306. [Google Scholar]
- Lin, Z.; Tian, Y.; Tu, Y. Comparison of the growing diversities of Tripterygium wilfordii seedling in different clones. J. Fujian Coll. For. 2012, 32, 226–231. [Google Scholar]
- Long, F.; Chen, X.; Wu, C.-z.; Tu, Y.-h.; Lin, Z.-s.; Hong, W.; Li, J. Comparison of the photosynthetic characteristics of Tripterygium wilfordii from 24 different provenances. J. Xiamen Univ. (Nat. Sci.) 2017, 56, 525–530. [Google Scholar]
- Lin, Z.; Tian, Y.; Tu, Y.; Wu, C. Breeding methods for seedlings of Tripterygium wilfordii Hook f. Pract. For. Technol. 2013, 23–25. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Ricotta, C. Bridging the gap between ecological diversity indices and measures of biodiversity with Shannon’s entropy: Comment to Izsák and Papp. Ecol. Model. 2002, 152, 1–3. [Google Scholar] [CrossRef]
- Jaccard, P. Étude comparative de la distribution florale dans une portion des Alpes et du Jura. Bull. Soc. Vaud. Sci. Nat. 1901, 37, 547–579. [Google Scholar]
- Chao, A.; Chazdon, R.; Colwell, R.K.; Shen, T.-J. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 2005, 8, 148–159. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpsonj, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, R Package Version 2.6-10. Available online: https://CRAN.R-project.org/package=vegan (accessed on 15 February 2025).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.; Kloepper, J. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- McInroy, J.A.; Kloepper, J.W. Studies on indigenous endophytic bacteria of sweet corn and cotton. In Molecular Ecology of Rhizosphere Microorganisms: Biotechnology and the Release of GMOs; O’Gara, D.D.F., Boesten, B., Eds.; VCH Verlagsgesellschaft: Weinheim, Germany, 1994; pp. 19–28. [Google Scholar]
- Krechel, A.; Faupel, A.; Hallmann, J.; Ulrich, A.; Berg, G. Potato-associated bacteria and their antagonistic potential towards plant-pathogenic fungi and the plant-parasitic nematode Meloidogyne incognita (Kofoid & White) Chitwood. Can. J. Microbiol. 2002, 48, 772–786. [Google Scholar] [CrossRef]
- Szymanska, S.; Plociniczak, T.; Piotrowska-Seget, Z.; Hrynkiewicz, K. Endophytic and rhizosphere bacteria associated with the roots of the halophyte Salicornia europaea L.—community structure and metabolic potential. Microbiol. Res. 2016, 192, 37–51. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Shaharoona, B.; Arshad, M.; Crowley, D.E. Population density and functional diversity of plant growth promoting rhizobacteria associated with avocado trees in saline soils. Appl. Soil Ecol. 2012, 62, 147–154. [Google Scholar] [CrossRef]
- Ginting, M.; Manalu, K.; Nasution, R.A. Population and characterization of rhyzospheric bacteria of pineapple plant (Ananas comosus L. Merr) on the highland land of Lumban Sihite Village, Regency Dairi. J. Biol. Trop. 2024, 24, 535–540. [Google Scholar] [CrossRef]
- Adams, P.D.; Kloepper, J.W. Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant Soil 2002, 240, 181–189. [Google Scholar] [CrossRef]
- Cordero, J.; de Freitas, J.R.; Germida, J.J. Bacterial microbiome associated with the rhizosphere and root interior of crops in Saskatchewan, Canada. Can. J. Microbiol. 2020, 66, 71–85. [Google Scholar] [CrossRef]
- Brigham, L.M.; de Mesquita, C.P.B.; Spasojevic, M.J.; Farrer, E.C.; Porazinska, D.L.; Smith, J.G.; Schmidt, S.K.; Suding, K.N. Drivers of bacterial and fungal root endophyte communities: Understanding the relative influence of host plant, environment, and space. FEMS Microbiol. Ecol. 2023, 99, fiad034. [Google Scholar] [CrossRef]
- Mushtaq, S.; Shafiq, M.; Tariq, M.R.; Sami, A.; Nawaz-ul-Rehman, M.S.; Bhatti, M.H.T.; Haider, M.S.; Sadiq, S.; Abbas, M.T.; Hussain, M.; et al. Interaction between bacterial endophytes and host plants. Front. Plant Sci. 2023, 13, 1092105. [Google Scholar] [CrossRef]
- Pereira, S.; Monteiro, C.; Vega, A.; Castro, P.M. Endophytic culturable bacteria colonizing Lavandula dentata L. plants: Isolation, characterization and evaluation of their plant growth-promoting activities. Ecol. Eng. 2016, 87, 91–97. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, D.; Ding, W.; Ying, Y. Isolation of endophytic bacteria in roots of Panax ginseng and screening of antagonistic strains against phytopathogens prevalent in P. ginseng. China J. Chin. Mater. Medica 2012, 37, 1532–1535. [Google Scholar]
- Chiniquy, D.; Barnes, E.M.; Zhou, J.; Hartman, K.; Li, X.; Sheflin, A.; Pella, A.; Marsh, E.; Prenni, J.; Deutschbauer, A.M.; et al. Microbial community field surveys reveal abundant Pseudomonas population in sorghum rhizosphere composed of many closely related phylotypes. Front. Microbiol. 2021, 12, 598180. [Google Scholar] [CrossRef]
- Ali, M.; Walait, S.; Ul Haque, M.F.; Mukhtar, S. Antimicrobial activity of bacteria associated with the rhizosphere and phyllosphere of Avena fatua and Brachiaria reptans. Environ. Sci. Pollut. Res. 2021, 28, 68846–68861. [Google Scholar] [CrossRef]
- Schillaci, M.; Raio, A.; Sillo, F.; Zampieri, E.; Mahmood, S.; Anjum, M.; Khalid, A.; Centritto, M. Pseudomonas and Curtobacterium strains from olive rhizosphere characterized and evaluated for plant growth promoting traits. Plants 2022, 11, 2245. [Google Scholar] [CrossRef] [PubMed]
- Hol, W.; Bezemer, T.M.; Biere, A. Getting the ecology into interactions between plants and the plant growth-promoting bacterium Pseudomonas fluorescens. Front. Plant Sci. 2013, 4, 81. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wei, Z.; Weidner, S.; Friman, V.-P.; Xu, Y.-C.; Shen, Q.-R.; Jousset, A. Probiotic Pseudomonas communities enhance plant growth and nutrient assimilation via diversity-mediated ecosystem functioning. Soil Biol. Biochem. 2017, 113, 122–129. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Friman, V.-P.; Gu, S.-h.; Wang, X.-f.; Eisenhauer, N.; Yang, T.-j.; Ma, J.; Shen, Q.-r.; Xu, Y.-c. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 2016, 7, e1790-16. [Google Scholar]
- Susilowati, A.; Wahyudi, A.T.; Lestari, Y.; Suwanto, A.; Wiyono, S. Potential Pseudomonas isolated from soybean rhizosphere as biocontrol against soilborne phytopathogenic fungi. HAYATI J. Biosci. 2011, 18, 51–56. [Google Scholar] [CrossRef]
- Jasim, B.; Jimtha, C.J.; Jyothis, M.; Radhakrishnan, E. Plant growth promoting potential of endophytic bacteria isolated from Piper nigrum. Plant Growth Regul. 2013, 71, 1–11. [Google Scholar] [CrossRef]
- Miller, K.I.; Qing, C.; Sze, D.M.Y.; Neilan, B.A. Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS ONE 2012, 7, e35953. [Google Scholar] [CrossRef]
- Rengel, Z. Genetic control of root exudation. Plant Soil 2002, 245, 59–70. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martinez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef] [PubMed]
- Aleklett, K.; Leff, J.W.; Fierer, N.; Hart, M. Wild plant species growing closely connected in a subalpine meadow host distinct root-associated bacterial communities. PeerJ 2015, 3, e804. [Google Scholar] [CrossRef] [PubMed]
- Zhen, R.; Shukun, T.; Yi, J.; Mingxing, J.; Shangyong, Z.; Wenjing, L.; Zhili, Y.; Shuping, S.; Zebin, C.; Tiyuan, X.; et al. High-throughput sequencing analysis of endophytic bacteria diversity in fruits of white and red pitayas from three different origins. Pol. J. Microbiol. 2018, 67, 27–35. [Google Scholar] [CrossRef]
- Blain, N.P.; Helgason, B.L.; Germida, J.J. Endophytic root bacteria associated with the natural vegetation growing at the hydrocarbon-contaminated Bitumount Provincial Historic site. Can. J. Microbiol. 2017, 63, 502–515. [Google Scholar] [CrossRef]
- Bonito, G.; Reynolds, H.; Robeson, M.S.; Nelson, J.; Hodkinson, B.P.; Tuskan, G.; Schadt, C.W.; Vilgalys, R. Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants. Mol. Ecol. 2014, 23, 3356–3370. [Google Scholar] [CrossRef]
- Pacheco-Moreno, A.; Bollmann-Giolai, A.; Chandra, G.; Brett, P.; Davies, J.; Thornton, O.; Poole, P.; Ramachandran, V.; Brown, J.K.M.; Nicholson, P.; et al. The genotype of barley cultivars influences multiple aspects of their associated microbiota via differential root exudate secretion. PLoS Biol. 2024, 22, e3002232. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Teixeira, P.J.P.L. Root-exuded coumarin shapes the root microbiome. Proc. Natl. Acad. Sci. USA 2018, 115, 5629–5631. [Google Scholar] [CrossRef]
- Wen, T.; Zhao, M.; Yuan, J.; Kowalchuk, G.A.; Shen, Q. Root exudates mediate plant defense against foliar pathogens by recruiting beneficial microbes. Soil Ecol. Lett. 2020, 3, 42–51. [Google Scholar] [CrossRef]
- Wu, W.; Chen, W.; Liu, S.; Wu, J.; Zhu, Y.; Qin, L.; Zhu, B. Beneficial relationships between endophytic bacteria and medicinal plants. Front. Plant Sci. 2021, 12, 646146. [Google Scholar] [CrossRef]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. Microbial diversity in soil: Selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef]
- Schemske, D.W. Understanding the origin of species. Evolution 2000, 54, 1069–1073. [Google Scholar] [CrossRef]
- Adair, K.L.; Douglas, A.E. Making a microbiome: The many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 2017, 35, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; De Frenne, P.; Boon, N.; Brunet, J.; Cousins, S.A.O.; Decocq, G.; Kolb, A.; Lemke, I.; Liira, J.; Naaf, T.; et al. Plant species identity and soil characteristics determine rhizosphere soil bacteria community composition in European temperate forests. FEMS Microbiol. Ecol. 2019, 95, fiz063. [Google Scholar] [CrossRef] [PubMed]
- Munfi, A. Studies on the importance of endophytic bacteria for the biological control of the root-knot nematode Meloidogyne incognita on tomato. Bachelor’s Thesis, University of Bonn, Bonn, Germany, 2001. [Google Scholar]
- Chen, L.; Xin, X.; Zhang, J.; Marc, R.-G.; Nie, G.; Wang, Q. Soil characteristics overwhelm cultivar effects on the structure and assembly of root-associated microbiomes of modern maize. Pedosphere 2019, 29, 360–373. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; van Themaat, E.V.L.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef]
- Saikkonen, K.; Ion, D.; Gyllenberg, M. The persistence of vertically transmitted fungi in grass metapopulations. Proc. R. Soc. Lond. Ser. Biol. Sci. 2002, 269, 1397–1403. [Google Scholar] [CrossRef]
- Venkatachalam, S.; Ranjan, K.; Prasanna, R.; Ramakrishnan, B.; Thapa, S.; Kanchan, A. Diversity and functional traits of culturable microbiome members, including cyanobacteria in the rice phyllosphere. Plant Biol. 2016, 18, 627–637. [Google Scholar] [CrossRef]
- Beneduzi, A.; Moreira, F.; Costa, P.B.; Vargas, L.K.; Lisboa, B.B.; Favreto, R.; Baldani, J.I.; Passaglia, L.M.P. Diversity and plant growth promoting evaluation abilities of bacteria isolated from sugarcane cultivated in the South of Brazil. Appl. Soil Ecol. 2013, 63, 94–104. [Google Scholar] [CrossRef]
- Ganesh, J.; Singh, V.; Hewitt, K.; Kaundal, A. Exploration of the rhizosphere microbiome of native plant Ceanothus velutinus—an excellent resource of plant growth-promoting bacteria. Front. Plant Sci. 2022, 13, 979069. [Google Scholar] [CrossRef] [PubMed]
- Chialva, M.; Salvioli di Fossalunga, A.; Daghino, S.; Ghignone, S.; Bagnaresi, P.; Chiapello, M.; Novero, M.; Spadaro, D.; Perotto, S.; Bonfante, P. Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 2018, 220, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Liu, X.; Qin, Y.; Feng, G.; Zhou, Y.; Zhu, H.; Yao, Q. Cooperation of arbuscular mycorrhizal fungi and bacteria to facilitate the host plant growth dependent on soil pH. Front. Microbiol. 2023, 14, 1116943. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).