Abstract
The camphor tree (Cinnamomum camphora) is of great economic and ecological value, and the WRKY transcription factor (TF) family plays a crucial role in regulating plant growth and development as well as the responses toward environmental changes. However, the research on WRKY TFs in C. camphora remains scarce, and their roles in the leaf expansion period are unknown. In this study, we identified WRKY TFs across the C. camphora genome, followed by a phylogenetic analysis. Then, we conducted RNA sequencing and qPCR experiments on leaves collected from three distinct stages during leaf expansion (S1, S2, and S3) to determine which WRKY genes showed significant up-regulation during these stages. Here, a total of 72 CcWRKY TFs were found in the C. camphora genome, and they were phylogenetically clustered with corresponding subfamilies of Arabidopsis thaliana. These CcWRKY proteins were divided into three major groups (I, II, and III), where group II consisted of five subgroups. We found that three genes (CcWRKY24, CcWRKY42, and CcWRKY70) were upregulated from both S1 to S2 and from S1 to S3. The expression level of CcWRKY24 increased gradually from S1 to S3, while CcWRKY42 and CcWRKY70 exhibited higher expression levels in S2 and S3 than in S1. These predicted gene expression profiles were further confirmed by qPCR experiments. In summary, this study analyzed WRKY TFs in C. camphora from a genome-wide perspective and paves the way for future research on the functions of CcWRKYs.
1. Introduction
WRKY is a key transcription factor (TF) mainly found in plants. The first WRKY TF was isolated from the sweet potato (Ipomoea batatas) by Ishiguro and Nakamura [1]. Since then, as research on the WRKY gene family has continued to deepen, many WRKY TFs have been identified in various plants, including thale cress (Arabidopsis thaliana), rice (Oryza sativa), and wheat (Triticum aestivum), among many others [2,3]. The WRKY TF name was derived from the seven conserved amino acids, namely WRKYGQK [4,5]. Traditionally, WRKY TFs can be classified into three groups (groups I, II, and II) based on the number of WRKY domains and the zinc finger types. The group I WRKY proteins contain two WRKY domains, while the other two WRKY groups are composed of one WRKY domain, but the zinc finger motif is different between the two WRKY groups [4]. Meanwhile, group II can be further divided into five subgroups (IIa, IIb, IIc, IId, and IIe). Nevertheless, phylogenetic analyses showed that group II domains designated by Eulgem et al. [4] were not monophyletic but formed three distinct clades instead (IIa + IIb, IIc, and IId + IIe) [6]. Among these groups, group IIc was close to group I while group III was clustered with group IId + IIe.
A body of research indicates that many WRKY TFs are involved in plant growth, development, senescence, and response to environmental changes [2,4,5,7,8,9]. For instance, numerous studies have confirmed that WRKY TFs are involved in the regulation of seed size (A. thaliana and soybean) [10,11], pollen development (A. thaliana) [12], flowering time (soybean) [13], fruit development (pepper and black wolfberry) [14,15], leaf senescence (A. thaliana and the Chinese flowering cabbage) [16,17,18], and petal senescence (the wallflower and tulips) [19,20]. Meanwhile, the WRKY TFs are involved in the responses to high temperature and drought stress (rice, birchleaf pear, and wheat) [21,22,23] and ion stress (wheat) [24]. WRKY TFs were extensively studied in A. thaliana. For example, previous studies have demonstrated that AtWRKY44 (also known as TTG2) controls the epidermal color of A. thaliana seeds by regulating vacuolar transport steps in the proanthocyanidin biosynthesis pathway [25]. Also, the overexpression of AtWRKY57 can increase the drought tolerance of A. thaliana [26]. Expression levels of AtWRKY4/6/7 and 11 would be enhanced in senescent leaves [4]. The AtWRKY12 and AtWRKY13 TFs control the flowering time of A. thaliana through the gibberellin signaling pathway and the functions of these two TFs differ in this process. For example, the knockout of the AtWRKY12 gene can delay the flowering time, while the loss of the AtWRKY13 function would promote the flowering process [27,28]. In short, WRKY TFs are crucial to numerous plant functions.
The camphor tree (Cinnamomum camphora (L.) J. Presl), a member of the Lauraceae family, is renowned for its rich aromatic oils and versatile values in multiple aspects, including ornamental, economic, ecological, and evolutionary ones [29,30,31,32,33,34]. Its attractive foliage and fragrance make it a popular ornamental plant [30]. In China, it starts rapid leaf expansion from April to May, during which its floral organs start to develop simultaneously [35]. Economically, the camphor tree is a significant industrial tree species globally, as it provides excellent wood for furniture making and sculpture crafting [29,30]. Meanwhile, its roots, stems, leaves, and fruits are abundant in essential oils and are widely utilized in cosmetics, food additives, insect repellents, and traditional Chinese herbal medicine [29,36,37]. Moreover, the camphor tree plays a critical role in the ecological benefits of forest ecosystems due to its considerable height, with some individuals reaching up to 40 meters [38,39]. For instance, it can fix a substantial amount of CO2, effectively regulating local climate and improving soil quality, thereby playing a crucial role in both carbon fixation and mitigating global climate change [34,39]. Beyond its inherent values, the camphor tree is also a well-known fast-growing species for afforestation, significantly contributing to greening efforts worldwide [40]. Furthermore, the camphor tree has a vast distribution, with extensive distributions throughout major tropical and subtropical regions, as well as the transition zone between subtropical and temperate climates. This not only emphasizes its immense ecological significance but also showcases its remarkable adaptability to diverse environments [34,39].
In summary, C. camphora is of great economic and ecological value and provides us with an ideal material to study plant adaptation. Notwithstanding the importance of WRKY TFs and the camphor tree, previous works have mainly focused on the TPS, MYB, or NAC TF families, leaving the WRKY TFs relatively understudied [34,41,42,43] and poorly understood. Nevertheless, some preliminary results shed light on the importance of WRKY TFs in several biological processes in C. camphora. For example, Yang et al. (2021) [41] found that several WRKY genes interacted with TPS genes to participate in borneol biosynthesis. More recently, Li et al. (2023) [34] determined that some WRKY TFs were involved in triglyceride biosynthesis and circadian rhythm. Nevertheless, we still lack a comprehensive understanding of the distribution of WRKY TFs across the genome of C. camphora and their roles in response to changing environments (such as increasing temperatures) or in the regulation during individual development (such as the leaf expansion period), leading to an evident knowledge gap that calls for deeper investigations.
In the present study, we conducted a genome-wide analysis of CcWRKY TFs to determine their physicochemical properties, chromosomal localization, phylogenetic relationship, conserved motif determination, and expression patterns during the leaf expansion period of the camphor tree. Highlighting several novel findings, our work provides a thorough characterization of CcWRKY TFs across the C. camphora genome, laying a solid foundation for future research in functional genomics regarding this important tree species.
2. Results
2.1. Identification of WRKY Protein Family
In this study, a total of 72 WRKY candidate proteins were identified in C. camphora (Table 1). These candidate proteins were submitted to the NCBI CDD database (Table S1) and the SMART program (Table S2) for conserved domain validations. The results showed that all the 72 candidate proteins contained WRKY domains. These WRKY proteins were renamed, and their physicochemical properties are shown in Table 1. The lengths of CcWRKY proteins varied greatly, ranging from 154 aa (CcWRKY29) to 831 aa (CcWRKY4), with an average length of approximately 379 aa. The molecular weight ranged from 17,619.86 Da (CcWRKY29) to 91,314.11 Da (CcWRKY4). The isoelectric point (pI) ranged from 4.81 (CcWRKY12) to 9.73 (CcWRKY44), among which 40 pIs < 7 and 32 pIs > 7. The instability index (II) ranged from 30.09 (CcWRKY10) to 70.14 (CcWRKY2). Among them, CcWRKY5/8/10/32/38 had instability indices of less than 40, indicating that they were stable proteins. The aliphatic index ranged from 42.12 (CcWRKY42) to 82.71 (CcWRKY36). The grand average of hydropathicity (GRAVY) ranged from −1.05 (CcWRKY42) to −0.37 (CcWRKY6), indicating that the CcWRKY TFs were all hydrophilic proteins. Subcellular localization predictions indicated that 71 of the 72 CcWRKY proteins were localized in the nucleus, with CcWRKY29 predicted to be in the chloroplast. This suggested that CcWRKY proteins predominantly function in the nucleus (Table 1). Targeting protein predictions showed that only CcWRKY29 contained a signal peptide. Meanwhile, nuclear localization signal (NLS) predictions revealed that 13 of the 72 CcWRKY proteins possessed an NLS motif (Table 1).

Table 1.
Basic information of CcWRKY TF members.
In addition, the distribution of 70 CcWRKY genes on 12 chromosomes of C. camphora was statistically random overall (X-squared = 10.452, df = 12, p = 0.4602), while two CcWRKYs (CcWRKY71, CcWRKY72) were not anchored to chromosomes (Figure 1). Nevertheless, we observed a clustered pattern in their distribution. For instance, chromosome 3 contained the largest number of CcWRKYs, with 15 genes showing some clusterings, while chromosome 4 contained nine CcWRKYs, and chromosome 11 had eight CcWRKYs. In contrast, chromosome 9 only contained one CcWRKY gene, and chromosome 12 contained three CcWRKYs (Figure 1).
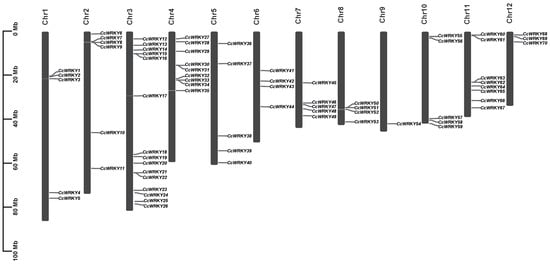
Figure 1.
The distribution of CcWRKY genes on chromosomes. The scale bar on the left corresponds to the length of the chromosomes.
2.2. Classification and Phylogenetic Analysis
The phylogenetic analysis showed that CcWRKY proteins were divided into three groups (I, II, and III), and group II contained five subgroups (IIa, II b, II c, II d, and IIe) (Figure 2). Among these groups, group III was clustered with group IId + IIe. Members of different subfamily CcWRKY TFs clustered with the corresponding AtWRKY subfamilies. There were 14 AtWRKY proteins and 14 CcWRKY proteins in group I. Also, the number of CcWRKY proteins was the same as that of AtWRKY proteins in group IIb (both of them were eight). There were 17 AtWRKY proteins and 15 CcWRKY proteins in group IIc, seven AtWRKY proteins and nine CcWRKY proteins in group IId, eight AtWRKY proteins and nine CcWRKY proteins in group IIe, and 14 AtWRKY proteins and 11 CcWRKY proteins in group III. However, in group IIa, there were three AtWRKY proteins, whereas the number of CcWRKY proteins was six, which was twice the number of AtWRKY. Though the number of WRKY TFs varied between C. camphora and A. thaliana across different groups, the chi-square test demonstrated that the difference was not statistically significant (X-squared = 1.7869, df = 6, p = 0.9382).
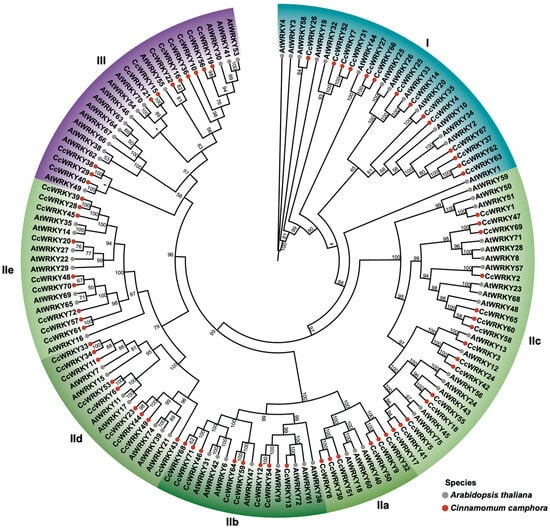
Figure 2.
Phylogenetic tree of WRKY family in C. camphora and A. thaliana. The consensus maximum likelihood (ML) phylogenetic tree was inferred using IQ-TREE with 1000 ultra-fast bootstrap (BS) replicates. The WRKY proteins were clustered into three groups (I, II, and III), and group II contained five subgroups (II a, II b, II c, II d, and II e). The different species and groups are indicated by different colors. The BS values of nodes are shown.
2.3. Motif Analysis of the WRKY in C. camphora
The visualization and comparison of 72 CcWRKY protein sequences showed that most of the TFs contained a highly conserved heptapeptide, WRKYGQK, and a zinc finger motif (C2H2 type/C2HXC type) (Figure S1), while some sequence variations occurred (Figure S2). Specifically, the corresponding heptapeptides for CcWRKY1/10/29/34/38 were WRKYGKK, WRKYAQE, WKKYGQK, GKKYGQK, and WKKYGQK, respectively. Furthermore, we did not observe a corresponding zinc finger structure following the conserved heptapeptide sequences in CcWRKY6 and WRKY10.
Subsequently, we identified ten conserved motifs among 72 CcWRKY TFs (Figure 3 and Figure S3). Motif 1 and motif 3 represented the conserved heptapeptide WRKYGQK, while motif 2 contained most of the zinc finger structure. Collectively, motif 1 and motif 2 formed a conserved WRKY domain (Figure S3). Generally, most CcWRKY proteins (70 out of 72) possessed both motif 1 and motif 2. Additionally, group III not only had motif 1 and motif 2 but also possessed motif 3, corresponding to two WRKY domains. Furthermore, we found that the motif composition and distribution were relatively conservative among members within the same clade. For example, motifs 1/2/5/7 were present in every WRKY protein in the IIa subgroup clade, with motif 1 and motif 2 located between motif 5 and motif 7, motif 5 located at the N-terminal, and motif 7 located at the C-terminal of the WRKY protein sequences. This indicates that motif patterns could be related to the function of WRKY proteins. Meanwhile, closely related clades shared motif similarities but also exhibited clade-specific attributes. For example, motif 5 and motif 7 were exclusively present in the IIa clade and the IIb clade, while motif 9 only appeared in the latter clade.
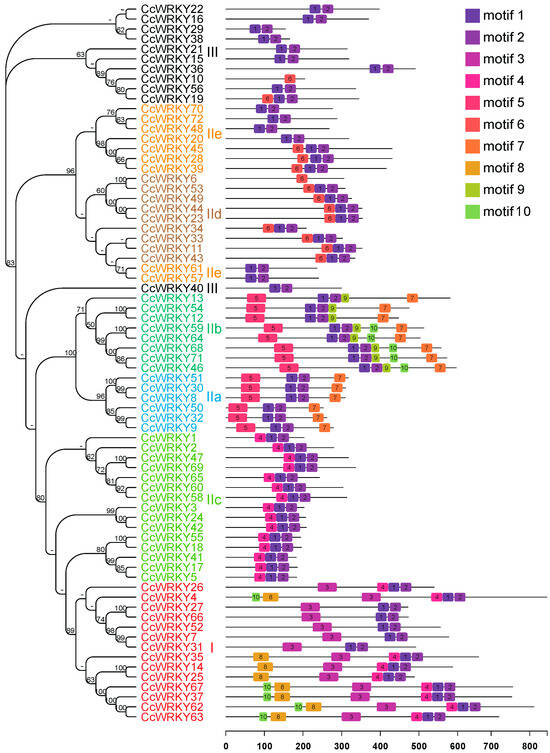
Figure 3.
Conserved motifs for CcWRKY proteins in C. camphora. Different motifs are shown with colored boxes and numbers (1–10). The lines represent the non-conserved sequences. The lengths of motifs can be estimated using the scale at the bottom.
Finally, we conducted a cis-acting regulatory elements analysis. The results indicated that, in addition to the core CAAT-box and TATA-box elements, as well as some non-functional elements, a total of 61 cis-regulatory elements were identified in the promoter regions of CcWRKY genes (Table S3, Figure S4). These elements were mostly related to hormone response, light response, plant growth and development, as well as stress response. Most CcWRKY genes contained 20 to over 30 cis-acting elements. Nevertheless, the number of cis-elements varied among CcWRKY genes. CcWRKY16 contained the fewest cis-acting elements (12), whereas CcWRKY21 contained the most (49). Among the CcWRKY genes, cis-acting elements related to abscisic acid responsiveness (ABRE), anaerobic induction (ARE), light responsiveness (Box4, G-box), and MeJA responsiveness (CGTCA-motif, TGACG-motif) were notably prevalent. These cis-acting elements collectively exceed 100 across the CcWRKY genes, with most genes containing at least one of them (Table S3).
2.4. Identification of Differential Expression Patterns of WRKY Genes
We isolated total RNA from tree leaves collected from healthy camphor trees during three distinct leaf-expansion stages (Figure 4A). Qualified RNA samples were subject to RNA sequencing, and an average of 6.06 Gb of clean data (see Section 4) were generated for each sample (Table S4).
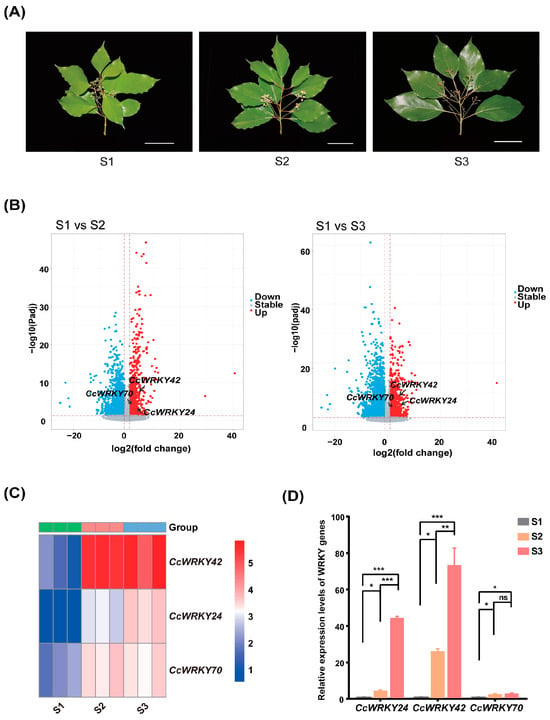
Figure 4.
(A) The morphological illustrations of C. camphora at three stages of leaf development. Scale bar: 5 cm. (B) Volcano plots displaying the DEGs between different stages. Each point on the plot represents an individual gene, with the x-axis indicating the log2FC in gene expression and the y-axis showing the statistical significance (−log10padj). (C) The predicted expression of CcWRKY24, CcWRKY42, and CcWRKY70 at three stages. TPM values for CcWRKY genes were log-transformed. The heatmap corresponds to gene expression levels. (D) Relative expression levels of three CcWRKY genes quantified by qPCR. Error bars indicate standard errors of three biological replicates. Asterisks indicate the significance of the expression level difference among stages (ns p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001).
Using these transcriptomic data, we conducted differential gene expression (DGE) analyses between S1 and S2 and between S1 and S3 (Figure 4B). In total, 1062 up-regulated genes were found between S1 and S2, which were enriched in 51 gene ontology (GO) terms relating to ‘Biological processes’; meanwhile, 1279 up-regulated genes were found between S1 and S3, which were enriched to 66 GO terms (Table S5) relating to ‘Biological processes’. Notably, 34 enriched GO terms were present between both S1-S2 and S1-S3, some of which were related to the response to temperature, abscisic acid, light stimulus, and cell wall development (Table S5). Specifically, we found three (CcWRKY24, CcWRKY42, CcWRKY70) and five (CcWRKY24, CcWRKY42, CcWRKY48, CcWRKY58, CcWRKY70) CcWRKYs genes that were up-regulated from the S1 to the S2 stage and from the S1 to the S3 stage, respectively (Tables S6 and S7). We focused the subsequent validation analysis on CcWRKY24, CcWRKY42, and CcWRKY70 genes, considering that these three genes exhibited up-regulation from S1 to S2 as well as from S1 to S3 (log2[Fold Change (FC)] > 1; padj < 0.05) (Figure 4B).
The heatmap results based on the transcripts per kilobase per million mapped reads (TPM) profile showed that these three genes exhibited relatively low expression levels in the S1 stage. The expression level of CcWRKY42 was higher in the S2 and S3 stages compared with the S1 stage, while the expression level of CcWRKY24 gradually increased from the S1 to the S3 stage. The expression level of CcWRKY70 increased from the S1 stage to the S2 stage and was similar between the S2 and the S3 stage (Figure 4C, Table S8).
Furthermore, the qPCR experiments showed that the expression profiles of these three genes were consistent with those predicted by transcriptome data. The one-way ANOVA indicated that the expression levels of CcWRKY24 (F = 1790, df = 8, p < 0.001), CcWRKY42 (F = 46.35, df = 8, p = 0.001), and CcWRKY70 (F = 11.07, df = 8, p < 0.01) among the three time periods were significantly different (Table S9). Multiple comparisons (Tukey’s post-hoc analysis) indicated that the relative expression level of CcWRKY24 and CcWRKY42 increased sequentially from S1 to S3 (p < 0.05), while that of CcWRKY70 was higher in S2 and S3 compared to S1 (p < 0.05) and no significant difference was found between the latter two stages (Figure 4D).
3. Discussion
The WRKY TF family, one of the essential classes of TFs in plants, plays an indispensable role in plant growth, development, and stress response [4,44]. With the continuous expansion of relevant research, the genome-wide analysis of the WRKY gene family has been conducted in numerous species, among which the number of identified WRKY proteins differed. For example, there are more than 70 WRKY proteins in Arabidopsis [45], 58 in eggplant [46], 79 in potato [47], 109 in rice [48], and 185 in soybean [49]. In this study, 72 WRKY proteins were identified in C. camphora, which was comparable to those identified in Arabidopsis and potato but less than that in rice and soybean.
The prediction of physicochemical properties for 72 CcWRKY TFs indicated that 40 proteins were acidic (pI < 7), 32 were alkaline (pI > 7), and most (67 out of 72) were unstable (Table 1). Furthermore, there were also differences in the length, molecular weight, and aliphatic index of the 72 CcWRKY TFs identified (Table 1). Despite the variation in the physicochemical properties of these proteins, they were determined to be hydrophilic, which is similar to what was found by Guo et al. [50]. The observed variation in the physicochemical properties of these CcWRKYs indicates the structural complexity and functional divergence among them, which is consistent with the literature [51]. The structural complexity and functional diversity among CcWRKYs imply their versatile physiological roles and regulatory functions within the plant. Therefore, a deeper understanding of the structures and functions of CcWRKYs can help us gain better insights into the growth, development, and environmental stress response mechanisms of the camphor tree, as suggested by the study on tobacco. Additionally, the analysis of conserved domains revealed that almost all the CcWRKY genes possessed motif 1 and motif 2, which together formed a single WRKY conserved domain (Figure 3, Figures S1 and S3). This reflects the structural conservatism of CcWRKY TFs. Meanwhile, the same clade or closely related clades shared motif similarities, while possessing subgroup-specific motifs (Figure 3). This finding is similar to those reported by Guo et al. [50] and Hu et al. [52], which could partly explain the pattern of functional diversification in WRKY TFs. The majority of CcWRKY TFs were located at the nucleus, as subcellular localization predictions showed (Table 1), suggesting that CcWRKY TFs likely play a primary role in transcriptional regulation within the nucleus. Yang et al. [41] identified 3 TPS (CcTPS1, CcTPS3, CcTPS4) and 37 other genes (including 5 WRKY, 15 MYB, 10 ERF/AP2, 5 bZIP, and 2 BHLH) that may be crucially involved in the biosynthesis and regulation of monoterpenes in C. camphora. Chen et al. [53] discovered that AtGPPS11 and AtGGPPS12 interacted to regulate monoterpene biosynthesis in Arabidopsis flowers. Meanwhile, AtWRKY12 and AtWRKY13 interacted with SPL10 to modulate the age-mediated flowering process [27]. These findings imply putative interactions between CcWRKY and other TFs as well as within the CcWRKYs.
In our study, the phylogenetic analysis revealed that the members of CcWRKYs clustered with their corresponding Arabidopsis subfamilies (Figure 2). Specifically, CcWRKYs were categorized into three groups (groups I, II, and III). Group II can be further divided into five subgroups (IIa, IIb, IIc, IId, IIe). Among these five subgroups, group IIa was sister to group IIb, group IId was sister to group IIe, group IIc exhibited an orphan clade, while group III was nested within group II. This is consistent with the classification of A. thaliana [4,5,6], indicating that the identification and classification of the 72 CcWRKYs are reliable and that the WRKY TF families in C. camphora and A. thaliana share similar evolutionary processes and a high degree of conservatism.
The literature shows that group IIc harbors a greater sequence diversity than other subgroups in group II [3]. Similarly, their functions are also diverse. In our study, the phylogenetic analysis showed that both CcWRKY24 and CcWRKY42 belong to group IIc, which are orthologs of AtWRKY12 (Figure 2). Functionally, AtWRKY12 and AtWRKY13, a paralogous pair of TFs, likely play a role in the transition from a vegetative phase to a reproduction phase [2,27]. For example, the knockout of the AtWRKY12 gene could delay the flowering time, while the loss of the AtWRKY13 function would accelerate the flowering process [27,28]. Meanwhile, they also play different roles in stem development [2]. In our study, as the leaves developed from S1 to S3, the floral organs continued to develop at the same time (Figure 4A). Therefore, CcWRKY24 and CcWRKY42 were presumed to be involved in the regulation of plant growth associated with the development of both vegetative and reproductive organs. Additionally, phylogenetic analysis showed that CcWRKY70 was close to AtWRKY14, AtWRKY35, AtWRKY65, and AtWRKY69, all of which belong to subgroup IIe (Figure 2). Previous research indicates that WRKY14 (ABT1) is an important regulator of thermomorphogenesis in Arabidopsis, and its close homologs WRKY35 (ABT2), WRKY65 (ABT3), and WRKY69 (ABT4) also play critical roles in this responding process [54]. Therefore, we speculate that CcWRKY70 could also play a key role in plant thermomorphogenesis (which is also tightly linked to leaf expansion), enabling C. camphora to achieve coordinated growth under various combinations of temperature, light, and other environmental conditions. However, whether CcWRKY24/42/70 possesses these functions remains to be functionally validated. Functional analysis methods such as gene knockout and overexpression can be considered in future work.
The roots, bark, and leaves of C. camphora are rich in essential oils, which mainly comprise monoterpenes and sesquiterpenes [55]. During the processes of leaf growth and floral organ development in the camphor tree, there would be an accumulation of terpenoids and aromatic compounds. It is widely acknowledged that terpenoids and aromatic compounds, as important constitutes of plant secondary metabolites, have significant implications for the economic value and ecological functions of plants, including the camphor tree [56], the Arabian jasmine (Jasminum sambac) [57], the Chinese mahogany (Toona sinensis) [58], and the sweet osmanthus (Osmanthus fragrans) [59]. These compounds not only endow plants with unique aromas and flavors but also play a crucial role in plant stress resistance, defense mechanisms, and interactions with other organisms [60,61,62]. It is acknowledged that the WRKY TFs play pivotal roles in regulating these secondary metabolites. For instance, studies have indicated that WRKY genes may be involved in aroma synthesis by regulating the production of terpenes and aromatic volatiles [63,64,65]. Lu et al. [64] suggested that overexpression of JsWRKY51 is a crucial factor in enhancing the accumulation of β-ocimene, an important aromatic terpene component in Jasminum sambac. Regarding the synthesis and accumulation of these compounds, several studies have revealed that a large number of volatile compounds are gradually synthesized and accumulated during the bud-to-full-blooming flowering stages [57,66,67]. Specifically, Lu et al. [64] discovered that some WRKY genes were highly expressed in full-blooming flowers, as well as being significantly associated with the accumulation of multiple terpenoid compounds at the blooming stage. Meanwhile, Ding et al. [63] showed that eight OfWRKYs participated in regulating flowering in the sweet osmanthus. Among these eight genes, the expression patterns of OfWRKY7/19/36/139 were correlated with specific monoterpenes. Therefore, we speculate that CcWRKY TFs may be involved in the development of both leaf and floral organs in the camphor tree, as well as the accumulation of secondary metabolites. Although this study did not include metabolomic data, we strongly recommend that future studies integrate metabolomic data to further investigate the functional dynamics of WRKY genes during the leaf expansion stage or other development periods. This approach will help provide a more comprehensive understanding of the role of WRKY genes in plant growth and offer deeper insights into their complex regulatory networks.
Moreover, we found that three genes (CcWRKY24, CcWRKY42, CcWRKY70) continuously exhibited up-regulation across three leaf expansion stages. Given that WRKY genes may also play roles in other regulation pathways related to photosynthesis, which could retain stable expressions during leaf expansion, the number of identified CcWRKY genes with differential expressions among conditions is reasonable and aligns with the biological context. Furthermore, the qPCR experiments confirmed that the expression level of CcWRKY24 increased gradually from S1 to S3, while CcWRKY42 and CcWRKY70 demonstrated higher expression levels in S2 and S3 than in S1. This suggests that these three genes may be involved in various regulation pathways during the expansion of leaves. Additionally, studies have shown that WRKY TFs mediate morphogenesis during the vegetative growth phase of plants. For example, AtWRKY44 is a crucial regulator of early trichome development [68] and AtWRKY71 plays an important role in shoot branching [69]. Research on the role of WRKY TFs in leaf expansion is extremely scarce, but our current study has provided a foundation for future work to build on. In summary, our study appears to be the first one to investigate WRKY TFs across the C. camphora genome, profiling their gene expression during the leaf expansion phase and setting the stage for functional analyses of these and other TFs in the camphor tree.
4. Materials and Methods
4.1. Identification and Physicochemical Properties Analysis
Available AtWRKY protein sequences were obtained from the Arabidopsis Information Resource database (TAIR, http://www.arabidopsis.org/, accessed on 27 February 2024). Then, we downloaded the Hidden Markov Model for WRKY TFs (PF03106) from the Pfam website (v. 36.0) (http://pfam.xfam.org/, accessed on 27 February 2024) [70]. Subsequently, we used the Blast program (a cutoff of E ≤ 1 × e−5) and the Simple HMM search program available from Tbtools software (v. 2.064) [71] to identify WRKY family members from the whole genome of C. camphora [34] (https://doi.org/10.6084/m9.figshare.20647452.v1, accessed on 27 February 2024). The search results were merged, and the intersection was taken as candidate proteins. These candidate proteins were submitted to the NCBI CDD database [72] and the Batch SMART program in Tbtools for validation of conserved domains. The physicochemical properties and the subcellular localization information of the WRKY TFs were analyzed by the Online Tool ExPASy [73] and the WOLF PSORT tool [74], respectively. TargetP-2.0 was used to predict the presence of targeting proteins [75]. The NLS motifs were predicted using the NLStradamus tool [76]. We used the Gene Location Visualize module from the GFF program in Tbtools to visualize the distribution of CcWRKY genes on chromosomes. Additionally, we performed a chi-square test to determine whether these WRKY genes were randomly distributed across these chromosomes with the chisq.test function from the R package stats (v. 4.1.2) [77].
4.2. Phylogenetic Tree Analysis
The WRKY protein sequences of C. camphora and A. thaliana were aligned using MAFFT (v. 7.508; --auto) [78] and then trimmed using trimAl (v. 1.4; -automated1) [79]. Next, the consensus maximum likelihood (ML) phylogenetic tree was inferred using IQ-TREE (v. 2.2.0.3) with 1000 ultra-fast bootstrap (BS) replicates [80,81]. The best-fit protein substitution model was determined automatically with ModelFinder as supported by IQ-TREE (JTT+F+I+R6) (-m MFP) [82]. The phylogenetic tree was visualized using the tvBOT Online Tool [83]. Furthermore, we performed a chi-square test to examine whether the numbers of WRKY TFs differed between the camphor tree and A. thaliana among groups with the chisq.test function from the R package stats.
4.3. Analysis of Conserved Motifs of CcWRKY TFs
The WebLogo tool (v. 2.8.2) [84] was used to visualize the sequences of the conserved CcWRKY domains. Later, these sequences were compared by the Jalview program (v. 2.11.4.1) [85] after being aligned with the Clustal Omega program (v. 1.2.4) available from the EMBL-EBI Job Dispatcher [86]. The neighbor-joining (NJ) tree of CcWKRY proteins was inferred using MEGA11 [87]. The conserved motifs of WRKY TFs were predicted by the MEME analysis suite (v. 5.5.5) [88] with the number of motifs set to 10. Subsequently, the results were visualized by tvBOT. In addition, the cis-acting regulatory elements (2000 bp upstream from the starting codon) of the CcWRKY genes were searched by the PlantCARE tool [89].
4.4. Sample Collection and Transcriptome Sequencing
To analyze the CcWRKY gene expression profiles during leaf expansion, we collected healthy leaves of C. camphora at three different stages (S1: 10 AM, 9 April 2024; S2: 10 AM, 24 April 2024; S3: 10 AM, 7 May 2024) at Taizhou University, Zhejiang Province. All these days were sunny days. Compared to the preceding week (2 April 2024; 17 °C), temperatures at the three sampling points progressively increased (S1: 19 °C; S2: 21 °C; S3: 23 °C). Three biological replicates for every stage were sampled. The three sampled trees were all planted in 2004, sourced from the same nursery, and propagated from the same cloned genotype. Currently, the trees are approximately eight meters tall. They were planted in an open, sun-exposed area on the campus, growing side by side with no shading between them. Branches with flowers from the uniformly sunlit side of the trees were collected, and adjacent leaves near the floral organs were harvested. The leaves were immediately stored in liquid nitrogen upon collection and were kept frozen until RNA extraction. These leaf samples were sent to a commercial sequencing institute (Annoroad Gene Technology Co., Ltd., Beijing, China) for RNA extraction and transcriptome sequencing. The total RNA was extracted based on the pBIOZOL methods (https://www.protocols.io/view/rna-isolation-from-plant-tissue-protocol-7-pbiozol-81wgb1e9yvpk/v1). The purity, concentration, and integrity of RNA products were determined with a Nanodrop2000 microspectrophotometer and a Labchip GX touch microfluidic capillary system. Following the manufacturer’s protocol and the literature, RNA samples with OD260/OD280 ratio of ca. 2.0, total RNA amount > 1 μg, and RNA integrity number (RIN) > 6 were considered qualified [90]. Different RNA libraries were pooled according to the effective concentration and the target downstream data volume and then sequenced on the DNBSEQTM-T7 instrument to generate 150 bp paired-end reads. Adapter reads, reads containing ambiguous bases, and low-quality reads (bases with Q ≤ 20 accounted for more than 50% of the entire read length) were removed from the raw sequencing data with in-house scripts available in the sequencing institute.
4.5. Differential Expression Patterns of CcWRKY Genes
To investigate DEGs between these leaf expansion stages (S1-S3), we mapped clean transcriptomic reads to the C. camphora genome using HISAT2 (v. 2.2.1) [91]. Subsequently, we analyzed the expression levels of genes using StringTie (v. 2.2.1) [92]. The read count information was extracted using the prepDE.py script available in StringTie (Table S10). Based on the count information file, the DEGs were identified using the R package DESeq2 (v. 1.34.0) [93] based on the criteria of |log2FC| > 1 and padj < 0.05. Next, we annotated these DEGs by querying against A. thaliana and then enriched them to test whether these genes were statistically strongly associated with the GO terms. Annotation and enrichment analysis were conducted using KOBAS-i [94]. The GO terms under the ‘biological process’ category were focused on, and only those with Fisher’s exact test (with Benjamini and Hochberg FDR correction) p-value < 0.05 were kept. Meanwhile, we plotted a volcano plot and identified the upregulated WRKY genes between stages based on the results of our DEG analysis. To investigate the differences in expression levels of these upregulated genes during these stages, we utilized the getTPM.py script available in StringTie to perform normalization of read counts and gene lengths, thereby eliminating the impacts of sequencing depth and gene length biases, resulting in a TPM profile. The heatmap based on the TPM profile and the volcano plots were generated using the RNAdiff module (v1.1.0) (https://github.com/nongxinshengxin/RNAdiffAPP, accessed on 20 July 2024) available from TBtools.
4.6. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
To verify the expression differences of three CcWRKY genes that were up-regulated between both S1-S2 and S1-S3, we conducted a qPCR analysis using the mRNA dye-based method (with the intercalating of SYBR Green) [95]. The RNA was extracted from leaves that were used for the RNA-seq as described above. We used the FastKing RT Kit (with gDNase) (TIANGEN Biotech, Co., Ltd., Beijing, China) to remove gDNA and synthesize cDNA by following the manufacturer’s protocol. The CcActin gene was used as the reference gene for qPCR [41]. The primers for the selected CcWRKY genes were designed using the online software primer3 (v. 0.4.0) (Table S11) [96].
The qPCR was performed on a CFX96TM Real-Time System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using the ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech, Co., Ltd., Nanjing, China). The qPCR conditions consisted of initial denaturation at 95 °C for 10 s, followed by 40 cycles of denaturation at 95 °C for 10 s, and then annealing and extension at 60 °C for 30 s. Then, we performed a melting curve analysis to determine the specificity of the reactions. The relative expression of each gene was calculated by the 2−△△Ct method [97]. The expression levels of genes in S1 were set as the reference. Finally, we tested whether gene expression levels differed among the three stages with one-way ANOVA and Tukey’s post-hoc analysis using R package stats.
5. Conclusions
Through a comprehensive genome-wide study of the WRKY gene family in the camphor tree, we developed a fundamental understanding of their physicochemical properties and structural characteristics, phylogenetic relationships, and gene expression profiles during leaf expansion. The transcriptional up-regulation of certain CcWRKY genes during leaf expansion suggests their regulatory role in this important developmental phase. In the future, functional analyses of these WRKY TFs in C. camphora should be conducted to better understand their roles in leaf expansion and additional developmental phases. Despite some limitations, this study lays a solid foundation for further investigations into the regulatory mechanisms of WRKY TFs in the camphor tree.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f16020266/s1, Table S1: Results of conserved domain validation against the NCBI CDD database; Table S2: Results of conserved domain validation from the SMART program; Table S3 Information of cis-elements present in CcWRKY genes. Table S4: A summary of RNA samples and sequencing data; Table S5: Significantly enriched Gene Ontology terms (biological process) of DEGs between different stages; Table S6: The change of gene expression level between S1 and S2; Table S7: The change of gene expression level between S1 and S3; Table S8: TPM matrix of assembled transcripts among three stages; Table S9: Quantification Cq values from the qPCR experiments; Table S10: Gene count information at different stages; Table S11: Primer information of three targeted genes in the qPCR validation; Figure S1: Conserved domain logo of CcWRKY TFs; Figure S2: Comparison of WRKY domain sequences among 72 CcWRKY TFs; Figure S3: Ten types of conserved motifs in CcWRKY TFs. Figure S4: The cis-acting regulatory elements in CcWKRY genes.
Author Contributions
Conceptualization, H.L.; methodology, S.L. and J.J.; software, S.L.; validation, S.L. and H.L.; formal analysis, S.L. and J.J.; investigation, S.L. and H.L.; resources, H.L.; data curation, S.L.; writing—original draft preparation, S.L. and J.J.; writing—review and editing, S.L. and H.L.; visualization, S.L.; supervision, H.L.; project administration, H.L.; funding acquisition, H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 32100164.
Data Availability Statement
The data presented in this study are openly available in GenBank under the BioProject accession number PRJNA1164943.
Acknowledgments
The authors thank Junmin Li for the assistance in computational resources.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ishiguro, S.; Nakamura, K. Characterization of a CDNA Encoding a Novel DNA-Binding Protein, SPF1, That Recognizes SP8 Sequences in the 5′ Upstream Regions of Genes Coding for Sporamin and β-Amylase from Sweet Potato. Mol. Gen. Genet. MGG 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Wang, H.; Chen, W.; Xu, Z.; Chen, M.; Yu, D. Functions of WRKYs in Plant Growth and Development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef]
- Song, H.; Cao, Y.; Zhao, L.; Zhang, J.; Li, S. Review: WRKY Transcription Factors: Understanding the Functional Divergence. Plant Sci. 2023, 334, 111770. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY Superfamily of Plant Transcription Factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY Transcription Factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. The WRKY Transcription Factor Superfamily: Its Origin in Eukaryotes and Expansion in Plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar] [CrossRef]
- Rogers, H.J. From Models to Ornamentals: How Is Flower Senescence Regulated? Plant Mol. Biol. 2013, 82, 563–574. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. CRC Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY Family Gene, and HAIKU2 (IKU2), a Leucine-Rich Repeat (LRR) KINASE Gene, Are Regulators of Seed Size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Gu, Y.; Li, W.; Jiang, H.; Wang, Y.; Gao, H.; Liu, M.; Chen, Q.; Lai, Y.; He, C. Differential Expression of a WRKY Gene between Wild and Cultivated Soybeans Correlates to Seed Size. J. Exp. Bot. 2017, 68, 2717–2729. [Google Scholar] [CrossRef]
- Lei, R.; Li, X.; Ma, Z.; Lv, Y.; Hu, Y.; Yu, D. Arabidopsis WRKY2 and WRKY34 Transcription Factors Interact with VQ20 Protein to Modulate Pollen Development and Function. Plant J. 2017, 91, 962–976. [Google Scholar] [CrossRef]
- Yang, Y.; Chi, Y.; Wang, Z.; Zhou, Y.; Fan, B.; Chen, Z. Functional Analysis of Structurally Related Soybean GmWRKY58 and GmWRKY76 in Plant Growth and Development. J. Exp. Bot. 2016, 67, 4727–4742. [Google Scholar] [CrossRef]
- Cheng, Y.; JalalAhammed, G.; Yu, J.; Yao, Z.; Ruan, M.; Ye, Q.; Li, Z.; Wang, R.; Feng, K.; Zhou, G.; et al. Putative WRKYs Associated with Regulation of Fruit Ripening Revealed by Detailed Expression Analysis of the WRKY Gene Family in Pepper. Sci. Rep. 2016, 6, 39000. [Google Scholar] [CrossRef]
- Tiika, R.J.; Wei, J.; Ma, R.; Yang, H.; Cui, G.; Duan, H.; Ma, Y. Identification and Expression Analysis of the WRKY Gene Family during Different Developmental Stages in Lycium ruthenicum Murr. Fruit. PeerJ 2020, 8, e10207. [Google Scholar] [CrossRef]
- Ay, N.; Irmler, K.; Fischer, A.; Uhlemann, R.; Reuter, G.; Humbeck, K. Epigenetic Programming via Histone Methylation at WRKY53 Controls Leaf Senescence in Arabidopsis Thaliana. Plant J. 2009, 58, 333–346. [Google Scholar] [CrossRef]
- Brusslan, J.A.; Rus Alvarez-Canterbury, A.M.; Nair, N.U.; Rice, J.C.; Hitchler, M.J.; Pellegrini, M. Genome-Wide Evaluation of Histone Methylation Changes Associated with Leaf Senescence in Arabidopsis. PLoS ONE 2012, 7, e33151. [Google Scholar] [CrossRef]
- Fan, Z.; Tan, X.-L.; Shan, W.; Kuang, J.; Lu, W.; Chen, J. Characterization of a Transcriptional Regulator, BrWRKY6, Associated with Gibberellin-Suppressed Leaf Senescence of Chinese Flowering Cabbage. J. Agric. Food Chem. 2018, 66, 1791–1799. [Google Scholar] [CrossRef]
- Price, A.M.; Aros Orellana, D.F.; Salleh, F.M.; Stevens, R.; Acock, R.; Buchanan-Wollaston, V.; Stead, A.D.; Rogers, H.J. A Comparison of Leaf and Petal Senescence in Wallflower Reveals Common and Distinct Patterns of Gene Expression and Physiology. Plant Physiol. 2008, 147, 1898–1912. [Google Scholar] [CrossRef]
- Meng, L.; Yang, H.; Yang, J.; Wang, Y.; Ye, T.; Xiang, L.; Chan, Z.; Wang, Y. Tulip Transcription Factor TgWRKY75 Activates Salicylic Acid and Abscisic Acid Biosynthesis to Synergistically Promote Petal Senescence. J. Exp. Bot. 2024, 75, 2435–2450. [Google Scholar] [CrossRef]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced Heat and Drought Tolerance in Transgenic Rice Seedlings Overexpressing OsWRKY11 under the Control of HSP101 Promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef]
- Ma, P.; Guo, G.; Xu, X.; Luo, T.; Sun, Y.; Tang, X.; Heng, W.; Jia, B.; Liu, L. Transcriptome Analysis Reveals Key Genes Involved in the Response of Pyrus betuleafolia to Drought and High-Temperature Stress. Plants 2024, 13, 309. [Google Scholar] [CrossRef]
- Ge, M.; Tang, Y.; Guan, Y.; Lv, M.; Zhou, C.; Ma, H.; Lv, J. TaWRKY31, a Novel WRKY Transcription Factor in Wheat, Participates in Regulation of Plant Drought Stress Tolerance. BMC Plant Biol. 2024, 24, 27. [Google Scholar] [CrossRef]
- Luo, D.; Xian, C.; Zhang, W.; Qin, Y.; Li, Q.; Usman, M.; Sun, S.; Xing, Y.; Dong, D. Physiological and Transcriptomic Analyses Reveal Commonalities and Specificities in Wheat in Response to Aluminum and Manganese. Curr. Issues Mol. Biol. 2024, 46, 367–397. [Google Scholar] [CrossRef]
- Gonzalez, A.; Brown, M.; Hatlestad, G.; Akhavan, N.; Smith, T.; Hembd, A.; Moore, J.; Montes, D.; Mosley, T.; Resendez, J.; et al. TTG2 Controls the Developmental Regulation of Seed Coat Tannins in Arabidopsis by Regulating Vacuolar Transport Steps in the Proanthocyanidin Pathway. Dev. Biol. 2016, 419, 54–63. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, G.; Yu, D. Activated Expression of WRKY57 Confers Drought Tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef]
- Li, W.; Wang, H.; Yu, D. Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 Oppositely Regulate Flowering under Short-Day Conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar] [CrossRef]
- Ma, Z.; Li, W.; Wang, H.; Yu, D. WRKY Transcription Factors WRKY12 and WRKY13 Interact with SPL10 to Modulate Age-mediated Flowering. J. Integr. Plant Biol. 2020, 62, 1659–1673. [Google Scholar] [CrossRef]
- Chen, W.; Vermaak, I.; Viljoen, A. Camphor—A Fumigant during the Black Death and a Coveted Fragrant Wood in Ancient Egypt and Babylon—A Review. Molecules 2013, 18, 5434–5454. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, W. Conservation and Applications of Camphor Tree (Cinnamomum camphora) in China: Ethnobotany and Genetic Resources. Genet. Resour. Crop Evol. 2016, 63, 1049–1061. [Google Scholar] [CrossRef]
- Fazmiya, M.J.A.; Sultana, A.; Rahman, K.; Heyat, M.B.B.; Sumbul; Akhtar, F.; Khan, S.; Appiah, S.C.Y. Current Insights on Bioactive Molecules, Antioxidant, Anti-Inflammatory, and Other Pharmacological Activities of Cinnamomum camphora Linn. Oxid. Med. Cell. Longev. 2022, 2022, 9354555. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Chen, X.; Liao, X.; Peng, D.; Han, X.; Zhu, C.; Wang, P.; Hufnagel, D.E.; Wang, L.; Li, K.; et al. A Chromosome-Level Genome of the Camphor Tree and the Underlying Genetic and Climatic Factors for Its Top-Geoherbalism. Front. Plant Sci. 2022, 13, 827890. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Kim, D.-S.; Park, S.-H.; Park, H. Phytochemistry and Applications of Cinnamomum camphora Essential Oils. Molecules 2022, 27, 2695. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lin, H.-Y.; Wang, X.; Bi, B.; Gao, Y.; Shao, L.; Zhang, R.; Liang, Y.; Xia, Y.; Zhao, Y.-P.; et al. Genome and Whole-Genome Resequencing of Cinnamomum camphora Elucidate Its Dominance in Subtropical Urban Landscapes. BMC Biol. 2023, 21, 192. [Google Scholar] [CrossRef]
- Wu, Z.; Raven, P.H. (Eds.) Cinnamomum camphora. In Flora of China 7, 102; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2008. [Google Scholar]
- Lee, H.J.; Hyun, E.-A.; Yoon, W.J.; Kim, B.H.; Rhee, M.H.; Kang, H.K.; Cho, J.Y.; Yoo, E.S. In Vitro Anti-Inflammatory and Anti-Oxidative Effects of Cinnamomum camphora Extracts. J. Ethnopharmacol. 2006, 103, 208–216. [Google Scholar] [CrossRef]
- Marouf, A.; Harras, F.; Shehata, E.; Abd-Allah, G. Efficacy of Camphor Oil and Its Nano Emulsion on The Cotton Leafworm, Spodoptera Littoralis. Egypt. Acad. J. Biol. Sci. F Toxicol. Pest Control 2021, 13, 103–108. [Google Scholar] [CrossRef]
- Zhou, L.; Dong, L.; Huang, Y.; Shi, S.; Zhang, L.; Zhang, X.; Yang, W.; Li, L. Spatial Distribution and Source Apportionment of Polycyclic Aromatic Hydrocarbons (PAHs) in Camphor (Cinnamomum camphora) Tree Bark from Southern Jiangsu, China. Chemosphere 2014, 107, 297–303. [Google Scholar] [CrossRef]
- Zhang, L.; Jing, Z.; Li, Z.; Liu, Y.; Fang, S. Predictive Modeling of Suitable Habitats for Cinnamomum camphora (L.) Presl Using Maxent Model under Climate Change in China. Int. J. Environ. Res. Public Health 2019, 16, 3185. [Google Scholar] [CrossRef]
- Xiang, Z.; Zhao, M.; Ogbodo, U.S. Accumulation of Urban Insect Pests in China: 50 Years’ Observations on Camphor Tree (Cinnamomum Camphora). Sustainability 2020, 12, 1582. [Google Scholar] [CrossRef]
- Yang, Z.; Xie, C.; Huang, Y.; An, W.; Liu, S.; Huang, S.; Zheng, X. Metabolism and Transcriptome Profiling Provides Insight into the Genes and Transcription Factors Involved in Monoterpene Biosynthesis of Borneol Chemotype of Cinnamomum camphora Induced by Mechanical Damage. PeerJ 2021, 9, e11465. [Google Scholar] [CrossRef]
- Luan, X.; Xu, W.; Zhang, J.; Shen, T.; Chen, C.; Xi, M.; Zhong, Y.; Xu, M. Genome-Scale Identification, Classification, and Expression Profiling of MYB Transcription Factor Genes in Cinnamomum camphora. Int. J. Mol. Sci. 2022, 23, 14279. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Luan, X.; Chen, C.; Gong, X.; Li, X.; Li, H.; Wu, Z.; Liu, Q.; Xu, M.; Zhong, Y. A Systematic Genome-Wide Analysis and Screening of the NAC Family Genes Related to Wood Formation in Cinnamomum camphora. Genomics 2023, 115, 110631. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Chen, Z. Identification of Genes Encoding Receptor-like Protein Kinases as Possible Targets of Pathogen- and Salicylic Acid-Induced WRKY DNA-Binding Proteins in Arabidopsis. Plant J. 2000, 24, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Chen, C.; Chen, Z. Expression Profiles of the Arabidopsis WRKY Gene Superfamily during Plant Defense Response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y. Identification of WRKY Gene Family and Characterization of Cold Stress-Responsive WRKY Genes in Eggplant. PeerJ 2020, 8, e8777. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Zhao, P.; Nan, Y.; Nie, T.; Wang, R.; Ma, H.; et al. Genome-Wide Identification of the Potato WRKY Transcription Factor Family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef]
- Dai, X.; Wang, Y.; Zhang, W.-H. OsWRKY74, a WRKY Transcription Factor, Modulates Tolerance to Phosphate Starvation in Rice. J. Exp. Bot. 2016, 67, 947–960. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, R.; Su, W.; Sun, T.; Qi, M.; Zhang, X.; Wei, F.; Yu, Z.; Xiao, F.; Yan, L.; et al. A Comprehensive Analysis of the WRKY Family in Soybean and Functional Analysis of GmWRKY164-GmGSL7c in Resistance to Soybean Mosaic Virus. BMC Genom. 2024, 25, 620. [Google Scholar] [CrossRef]
- Guo, X.; Yan, X.; Li, Y. Genome-Wide Identification and Expression Analysis of the WRKY Gene Family in Rhododendron henanense subsp. lingbaoense. PeerJ 2024, 12, e17435. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, B.; Gu, G.; Yuan, J.; Shen, S.; Jin, L.; Lin, Z.; Lin, J.; Xie, X. Genome-Wide Identification and Expression Analysis of the R2R3-MYB Gene Family in Tobacco (Nicotiana Tabacum L.). BMC Genom. 2022, 23, 432. [Google Scholar] [CrossRef]
- Hu, W.; Ren, Q.; Chen, Y.; Xu, G.; Qian, Y. Genome-Wide Identification and Analysis of WRKY Gene Family in Maize Provide Insights into Regulatory Network in Response to Abiotic Stresses. BMC Plant Biol. 2021, 21, 427. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Fan, D.; Wang, G. Heteromeric Geranyl (Geranyl) Diphosphate Synthase Is Involved in Monoterpene Biosynthesis in Arabidopsis Flowers. Mol. Plant 2015, 8, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Wang, N.; Yin, Q.; Li, H.; Wu, A.-M.; Qin, G. Activation Tagging Identifies WRKY14 as a Repressor of Plant Thermomorphogenesis in Arabidopsis. Mol. Plant 2022, 15, 1725–1743. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhang, J.; Zhang, B.; Jin, X.; Zhang, H.; Jin, Z. Transcriptional Analysis of Metabolic Pathways and Regulatory Mechanisms of Essential Oil Biosynthesis in the Leaves of Cinnamomum Camphora (L.). Presl. Front. Genet. 2020, 11, 598714. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Y.; Fu, C.; Yang, H.; Liu, X.; Qiu, F.; Wang, X.; Wang, Z. Chemical Variation and Environmental Influence on Essential Oil of Cinnamomum camphora. Molecules 2023, 28, 973. [Google Scholar] [CrossRef]
- Chen, G.; Mostafa, S.; Lu, Z.; Du, R.; Cui, J.; Wang, Y.; Liao, Q.; Lu, J.; Mao, X.; Chang, B.; et al. The Jasmine (Jasminum sambac) Genome Provides Insight into the Biosynthesis of Flower Fragrances and Jasmonates. Genom. Proteom. Bioinform. 2023, 21, 127–149. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhong, X.-L.; Zhu, S.-H.; Wang, K.; Tan, G.-F.; Meng, P.-H.; Zhang, J. Research Advances in Toona Sinensis, a Traditional Chinese Medicinal Plant and Popular Vegetable in China. Diversity 2022, 14, 572. [Google Scholar] [CrossRef]
- Wu, L.; Liu, J.; Huang, W.; Wang, Y.; Chen, Q.; Lu, B. Exploration of Osmanthus Fragrans Lour.’s Composition, Nutraceutical Functions and Applications. Food Chem. 2022, 377, 131853. [Google Scholar] [CrossRef]
- Dudareva, N.; Pichersky, E. Biochemical and Molecular Genetic Aspects of Floral Scents. Plant Physiol. 2000, 122, 627–634. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, Function and Metabolic Engineering of Plant Volatile Organic Compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.; Yu, R.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile Terpenoids: Multiple Functions, Biosynthesis, Modulation and Manipulation by Genetic Engineering. Planta 2017, 246, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Ouyang, Q.; Li, Y.; Shi, T.; Li, L.; Yang, X.; Ji, K.; Wang, L.; Yue, Y. Genome-Wide Investigation of WRKY Transcription Factors in Sweet Osmanthus and Their Potential Regulation of Aroma Synthesis. Tree Physiol. 2020, 40, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Wang, X.; Mostafa, S.; Noor, I.; Lin, X.; Ren, S.; Cui, J.; Jin, B. WRKY Transcription Factors in Jasminum sambac: An Insight into the Regulation of Aroma Synthesis. Biomolecules 2023, 13, 1679. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Wan, W.; Yin, D.; Deng, X.; Ma, Z.; Gao, T.; Cao, X. Genome-Wide Analysis of WRKY Transcription Factor Genes in Toona sinensis: An Insight into Evolutionary Characteristics and Terpene Synthesis. Front. Plant Sci. 2023, 13, 1063850. [Google Scholar] [CrossRef]
- Yu, Y.; Lyu, S.; Chen, D.; Lin, Y.; Chen, J.; Chen, G.; Ye, N. Volatiles Emitted at Different Flowering Stages of Jasminum sambac and Expression of Genes Related to α-Farnesene Biosynthesis. Molecules 2017, 22, 546. [Google Scholar] [CrossRef]
- Barman, M.; Mitra, A. Temporal Relationship between Emitted and Endogenous Floral Scent Volatiles in Summer- and Winter-blooming Jasminum Species. Physiol. Plant. 2019, 166, 946–959. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a WRKY Transcription Factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Guo, D.; Zhang, J.; Wang, X.; Han, X.; Wei, B.; Wang, J.; Li, B.; Yu, H.; Huang, Q.; Gu, H.; et al. The WRKY Transcription Factor WRKY71/EXB1 Controls Shoot Branching by Transcriptionally Regulating RAX Genes in Arabidopsis. Plant Cell 2015, 27, 3112–3127. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting Sequence Signals in Targeting Peptides Using Deep Learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A Simple Hidden Markov Model for Nuclear Localization Signal Prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (TvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA Integrity Number for Assigning Integrity Values to RNA Measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Medrano, J.F. Real-Time PCR for MRNA Quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and Modifications of Primer Design Program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).