Optimal Phenology Windows for Discriminating Populus euphratica and Tamarix chinensis in the Tarim River Desert Riparian Forests with PlanetScope Data
Abstract
1. Introduction
2. Materials and Methods
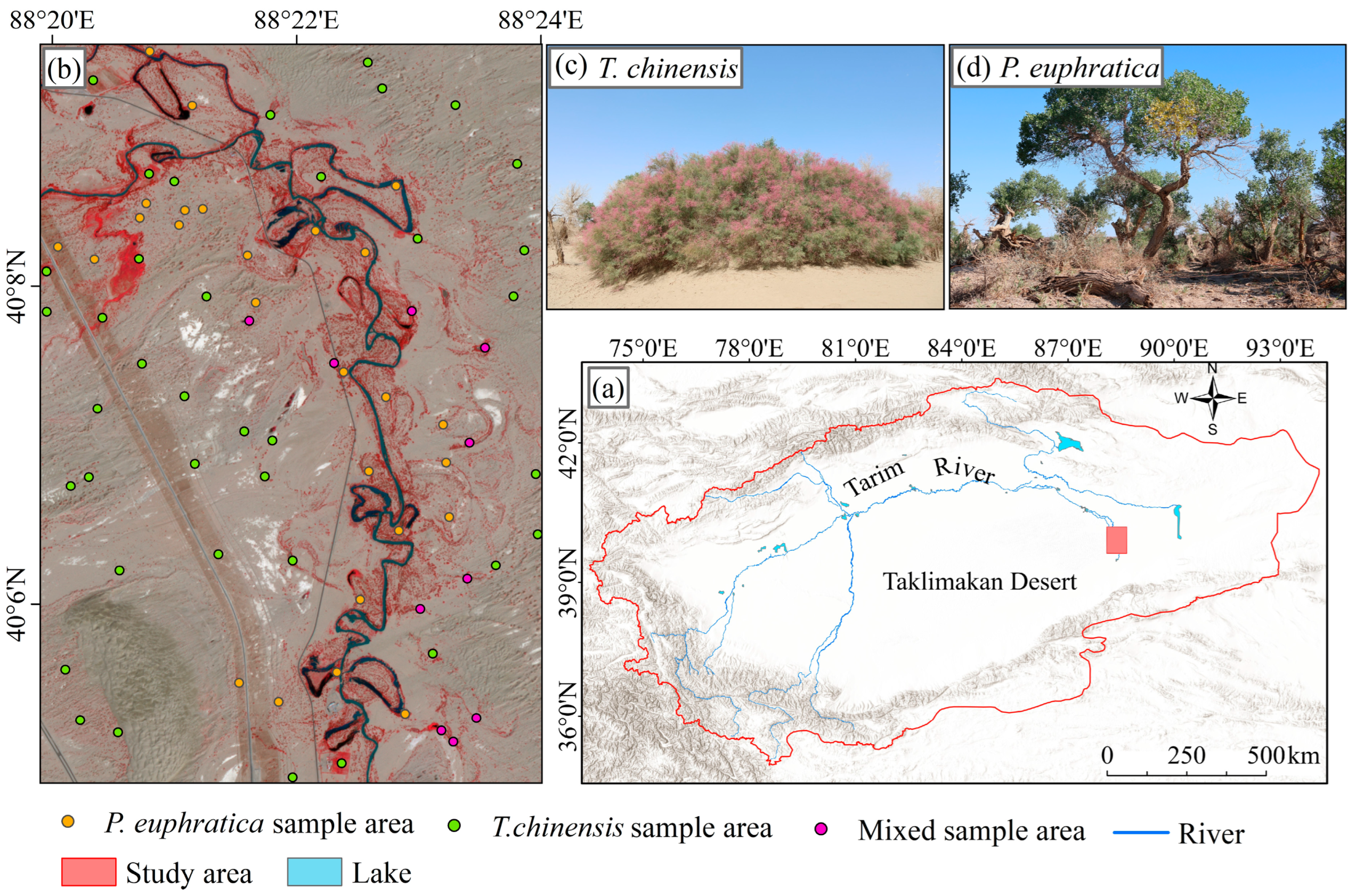
2.1. Study Sites
2.2. Materials
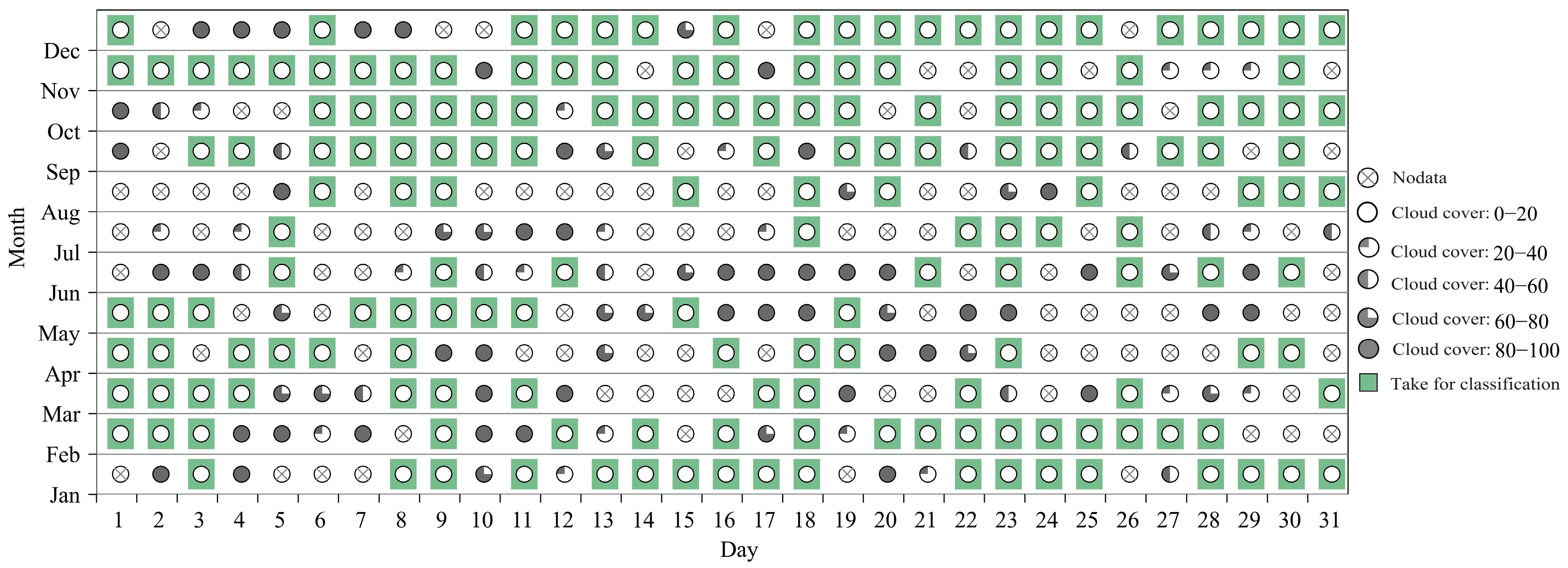
2.2.1. PlanetScope Images
2.2.2. Reference Imagery and Processing
2.2.3. Vegetation Indices
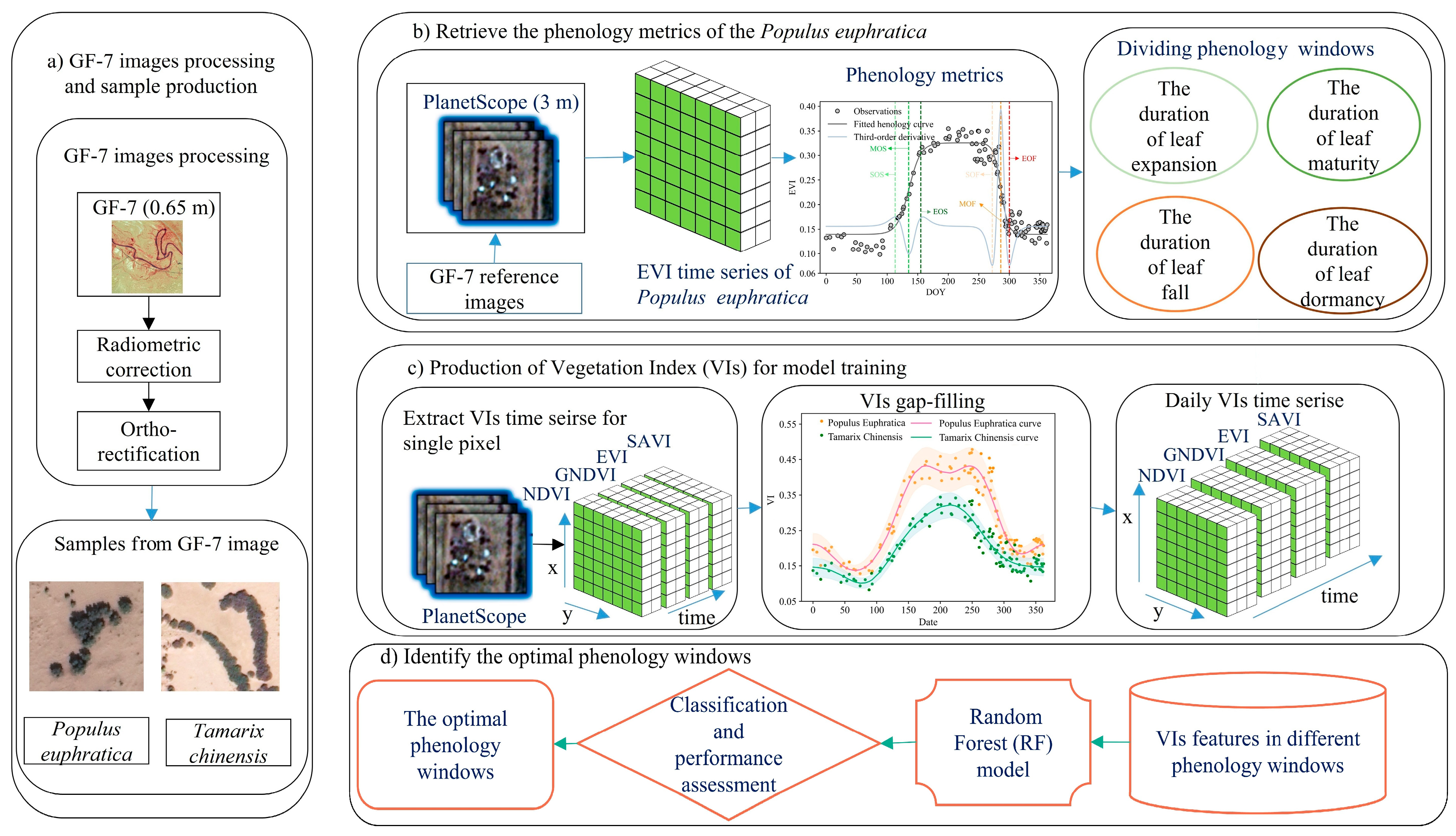
2.3. Methods
2.3.1. Time-Series VIs Reconstruction and Gap-Filling
2.3.2. Extracting Key Metrics of Phenology from PlanetScope
2.3.3. Classification and Performance Assessment
2.3.4. Selection of Optimal Phenology Windows
3. Results
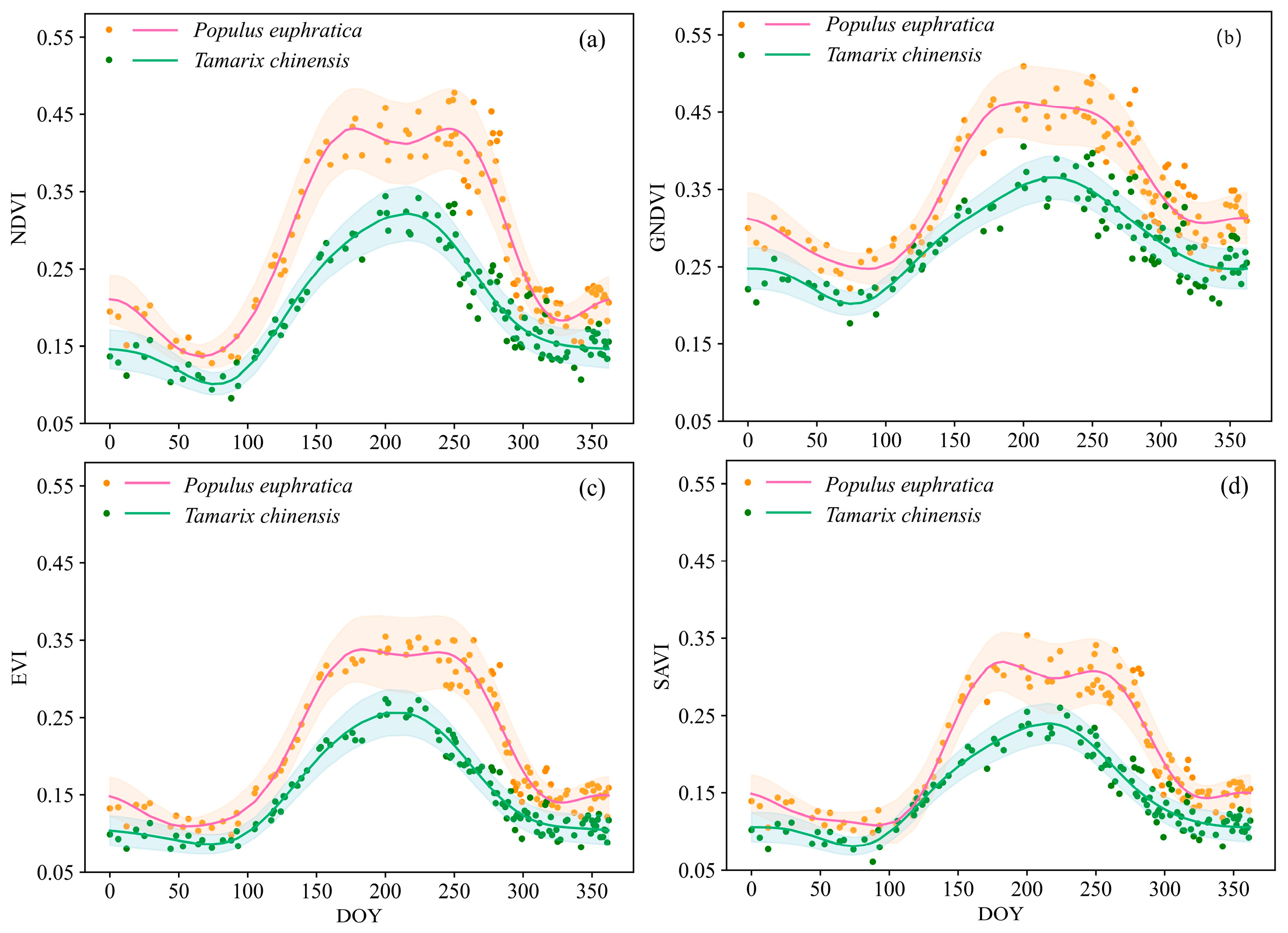
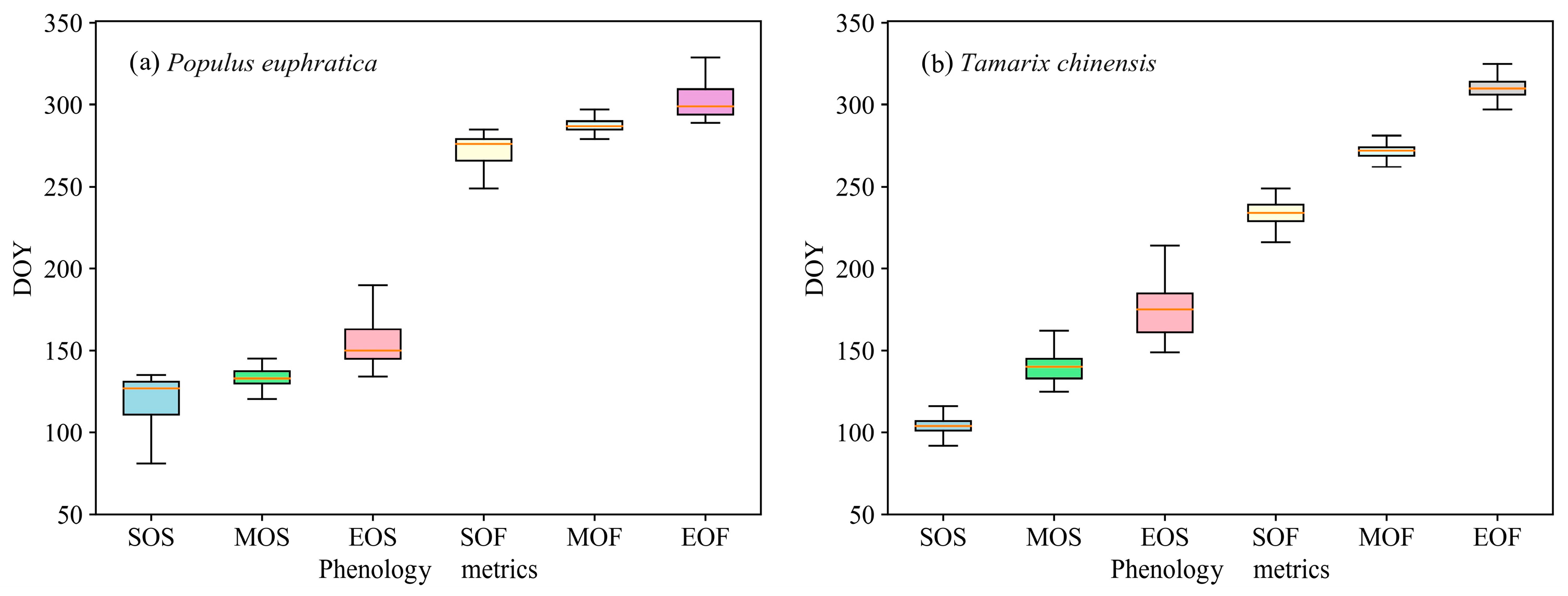
3.1. Vegetation Indices Profiles and Phenology Metrics
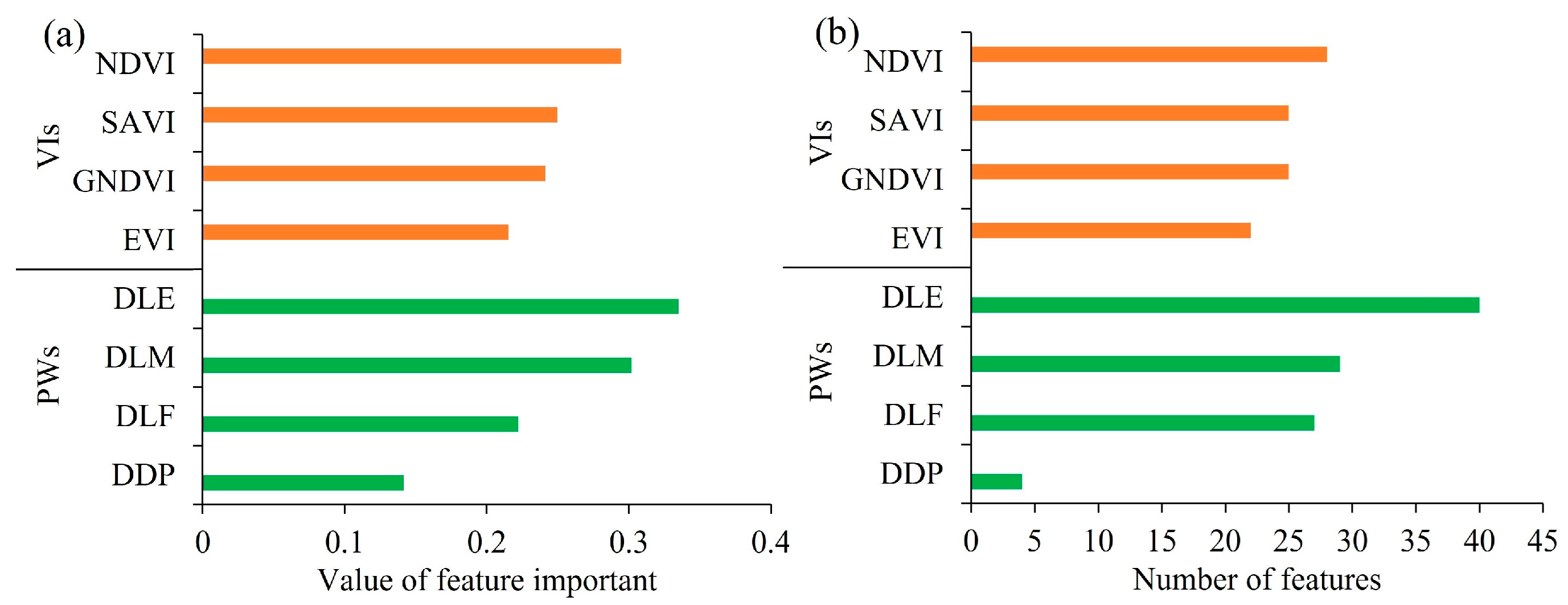
3.2. Model Training and Feature Importance
3.3. Performance of RF Model in Different Phenology Windows
3.4. Classification Performance Between Phenology Asynchrony and Synchrony Windows
3.5. Classification Results
4. Discussion
4.1. Optimal Phenology Windows for Discriminating P. euphratica and T. chinensis
4.2. Phenology Asynchrony Windows as Critical Time Windows for Tree Species Discrimination
4.3. Future Perspectives and Uncertainty Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SOS | Start of Spring |
| MOS | Middle of Spring |
| EOS | End of Spring |
| SOF | Start of Fall |
| MOF | Middle of Fall |
| EOF | End of Fall |
| DLE | Duration of Leaf Expansion |
| DLM | Duration of Leaf Maturity |
| DLF | Duration of Leaf Fall |
| DDP | Duration of the Dormancy Period |
| PAW | Phenology Asynchrony Window |
| PSW | Phenological Synchrony Window |
| RF | Random Forest |
References
- Guirado, E.; Delgado-Baquerizo, M.; Martínez-Valderrama, J.; Tabik, S.; Alcaraz-Segura, D.; Maestre, F.T. Climate legacies drive the distribution and future restoration potential of dryland forests. Nat. Plants 2022, 8, 879–886. [Google Scholar] [CrossRef]
- Wang, L.; Song, X.; Liu, Y.; Li, L.; Zhao, X.; Meng, P.; Fu, C.; Wei, W.; Wang, X.; Li, H. Effects of Long-Term Vegetation Restoration on Green Water Utilization Heterogeneity in the Loess Plateau Based on Field Experiments and Modeling. Plants 2025, 14, 644. [Google Scholar] [CrossRef] [PubMed]
- Keram, A.; Halik, Ü.; Keyimu, M.; Aishan, T.; Mamat, Z.; Rouzi, A. Gap dynamics of natural Populus euphratica floodplain forests affected by hydrological alteration along the Tarim River: Implications for restoration of the riparian forests. For. Ecol. Manag. 2019, 438, 103–113. [Google Scholar] [CrossRef]
- Peng, Y.; He, G.; Wang, G. Spatial-temporal analysis of the changes in Populus euphratica distribution in the Tarim National Nature Reserve over the past 60 years. Int. J. Appl. Earth Obs. Geoinf. 2022, 113, 103000. [Google Scholar] [CrossRef]
- Wang, J.; Han, P.; Zhang, Y.; Li, J.; Xu, L.; Shen, X.; Yang, Z.; Xu, S.; Li, G.; Chen, F. Analysis on ecological status and spatial–temporal variation of Tamarix chinensis forest based on spectral characteristics and remote sensing vegetation indices. Environ. Sci. Pollut. Res. 2022, 29, 37315–37326. [Google Scholar] [CrossRef]
- Ling, H.; Xu, H.; Guo, B.; Deng, X.; Zhang, P.; Wang, X. Regulating water disturbance for mitigating drought stress to conserve and restore a desert riparian forest ecosystem. J. Hydrol. 2019, 572, 659–670. [Google Scholar] [CrossRef]
- Zhang, P.; Deng, X.; Long, A.; Xu, H.; Ye, M.; Li, J. Change in Spatial Distribution Patterns and Regeneration of Populus euphratica under Different Surface Soil Salinity Conditions. Sci. Rep. 2019, 9, 9123. [Google Scholar] [CrossRef]
- Keyimu, M.; Halik, Ü.; Betz, F.; Dulamsuren, C. Vitality variation and population structure of a riparian forest in the lower reaches of the Tarim River, NW China. J. For. Res. 2018, 29, 749–760. [Google Scholar] [CrossRef]
- Deng, X.; Xu, H.; Ye, M.; Li, B.; Fu, J.; Yang, Z. Impact of long-term zero-flow and ecological water conveyance on the radial increment of Populus euphratica in the lower reaches of the Tarim River, Xinjiang, China. Reg. Environ. Change 2015, 15, 13–23. [Google Scholar] [CrossRef]
- Halik, Ü.; Aishan, T.; Betz, F.; Kurban, A.; Rouzi, A. Effectiveness and challenges of ecological engineering for desert riparian forest restoration along China’s largest inland river. Ecol. Eng. 2019, 127, 11–22. [Google Scholar] [CrossRef]
- Ling, H.; Zhang, P.; Xu, H.; Zhao, X. How to Regenerate and Protect Desert Riparian Populus euphratica Forest in Arid Areas. Sci. Rep. 2015, 5, 15418. [Google Scholar] [CrossRef]
- Yang, Y.; Zou, J.; Huang, Y.; Wu, Z.; Fang, T.; Xue, J.; Wang, D.; Wang, Y.; Wang, J.; Yang, X.; et al. Investigating the Earliest Identifiable Timing of Sugarcane at Early Season Based on Optical and SAR Time-Series Data. Remote Sens. 2025, 17, 2773. [Google Scholar] [CrossRef]
- Reiner, F.; Gominski, D.; Fensholt, R.; Brandt, M. An Operational Framework to Track Individual Farmland Trees Over Time at National Scales Using PlanetScope. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2025, 18, 1109–1121. [Google Scholar] [CrossRef]
- Mikołajczyk, Ł.; Hawryło, P.; Netzel, P.; Talaga, J.; Zdunek, N.; Socha, J. Classification of Tree Species in Poland Using CNNs Tabular-to-Pseudo Image Approach Based on Sentinel-2 Annual Seasonality Data. Forests 2025, 16, 1039. [Google Scholar] [CrossRef]
- Liu, H.; An, H. Analysis of the importance of five new spectral indices from WorldView-2 in tree species classification. J. Spat. Sci. 2020, 65, 455–466. [Google Scholar] [CrossRef]
- Yan, S.; Jing, L.; Wang, H. A New Individual Tree Species Recognition Method Based on a Convolutional Neural Network and High-Spatial Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 479. [Google Scholar] [CrossRef]
- Gazzea, M.; Kristensen, L.M.; Pirotti, F.; Ozguven, E.E.; Arghandeh, R. Tree Species Classification Using High-Resolution Satellite Imagery and Weakly Supervised Learning. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–11. [Google Scholar] [CrossRef]
- Adelabu, S.; Dube, T. Employing ground and satellite-based QuickBird data and random forest to discriminate five tree species in a Southern African Woodland. Geocarto Int. 2015, 30, 457–471. [Google Scholar] [CrossRef]
- Agarwal, S.; Vailshery, L.S.; Jaganmohan, M.; Nagendra, H. Mapping Urban Tree Species Using Very High Resolution Satellite Imagery: Comparing Pixel-Based and Object-Based Approaches. ISPRS Int. J. Geo-Inf. 2013, 2, 220–236. [Google Scholar] [CrossRef]
- Huang, Y.; Wen, X.; Gao, Y.; Zhang, Y.; Lin, G. Tree Species Classification in UAV Remote Sensing Images Based on Super-Resolution Reconstruction and Deep Learning. Remote Sens. 2023, 15, 2942. [Google Scholar] [CrossRef]
- Polyakova, A.; Mukharamova, S.; Yermolaev, O.; Shaykhutdinova, G. Automated Recognition of Tree Species Composition of Forest Communities Using Sentinel-2 Satellite Data. Remote Sens. 2023, 15, 329. [Google Scholar] [CrossRef]
- Grabska, E.; Frantz, D.; Ostapowicz, K. Evaluation of machine learning algorithms for forest stand species mapping using Sentinel-2 imagery and environmental data in the Polish Carpathians. Remote Sens. Environ. 2020, 251, 112103. [Google Scholar] [CrossRef]
- Kollert, A.; Bremer, M.; Löw, M.; Rutzinger, M. Exploring the potential of land surface phenology and seasonal cloud free composites of one year of Sentinel-2 imagery for tree species mapping in a mountainous region. Int. J. Appl. Earth Obs. Geoinf. 2021, 94, 102208. [Google Scholar] [CrossRef]
- Qiu, T.; Song, C.H.; Clark, J.S.; Seyednasrollah, B.; Rathnayaka, N.; Li, J.X. Understanding the continuous phenological development at daily time step with a Bayesian hierarchical space-time model: Impacts of climate change and extreme weather events. Remote Sens. Environ. 2020, 247, 111956. [Google Scholar] [CrossRef]
- Moon, M.; Seyednasrollah, B.; Richardson, A.D.; Friedl, M.A. Using time series of MODIS land surface phenology to model temperature and photoperiod controls on spring greenup in North American deciduous forests. Remote Sens. Environ. 2021, 260, 112466. [Google Scholar] [CrossRef]
- Liu, X.; Frey, J.; Munteanu, C.; Still, N.; Koch, B. Mapping tree species diversity in temperate montane forests using Sentinel-1 and Sentinel-2 imagery and topography data. Remote Sens. Environ. 2023, 292, 113576. [Google Scholar] [CrossRef]
- Lechner, M.; Dostálová, A.; Hollaus, M.; Atzberger, C.; Immitzer, M. Combination of Sentinel-1 and Sentinel-2 Data for Tree Species Classification in a Central European Biosphere Reserve. Remote Sens. 2022, 14, 2687. [Google Scholar] [CrossRef]
- Li, H.; Jia, M.; Zhang, R.; Ren, Y.; Wen, X. Incorporating the Plant Phenological Trajectory into Mangrove Species Mapping with Dense Time Series Sentinel-2 Imagery and the Google Earth Engine Platform. Remote Sens. 2019, 11, 2479. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Henebry, G.M.; Gao, F. Development and evaluation of a new algorithm for detecting 30 m land surface phenology from VIIRS and HLS time series. ISPRS J. Photogramm. Remote Sens. 2020, 161, 37–51. [Google Scholar] [CrossRef]
- Zhao, Y.; Lee, C.K.F.; Wang, Z.; Wang, J.; Gu, Y.; Xie, J.; Law, Y.K.; Song, G.; Bonebrake, T.C.; Yang, X.; et al. Evaluating fine-scale phenology from PlanetScope satellites with ground observations across temperate forests in eastern North America. Remote Sens. Environ. 2022, 283, 113310. [Google Scholar] [CrossRef]
- Peng, Y.; He, G.; Wang, G.; Zhang, Z. Large-Scale Populus euphratica Distribution Mapping Using Time-Series Sentinel-1/2 Data in Google Earth Engine. Remote Sens. 2023, 15, 1585. [Google Scholar] [CrossRef]
- Li, H.; Shi, Q.; Wan, Y.; Shi, H.; Imin, B. Using Sentinel-2 Images to Map the Populus euphratica Distribution Based on the Spectral Difference Acquired at the Key Phenological Stage. Forests 2021, 12, 147. [Google Scholar] [CrossRef]
- You, H.; Tian, S.; Yu, L.; Lv, Y. Pixel-Level Remote Sensing Image Recognition Based on Bidirectional Word Vectors. IEEE Trans. Geosci. Remote Sens. 2020, 58, 1281–1293. [Google Scholar] [CrossRef]
- Li, D.; Shi, Q.; Peng, L.; Wan, Y. Plant Population Classification Based on PointCNN in the Daliyabuyi Oasis, China. Forests 2023, 14, 1943. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, H.; Alifu, K.; Lv, Y. Remote sensing image description based on word embedding and end-to-end deep learning. Sci. Rep. 2021, 11, 3162. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Li, J.L.; Voorde, T.V.D.; Zhou, C.H.; Maeyer, P.D.; Ma, Y. Individual Populus euphratica Tree Detection in Sparse Desert Forests Based on Constrained 2-D Bin Packing. IEEE Trans. Geosci. Remote Sens. 2024, 62, 4407219. [Google Scholar] [CrossRef]
- Sadeh, Y.; Zhu, X.; Dunkerley, D.; Walker, J.P.; Zhang, Y.; Rozenstein, O.; Manivasagam, V.S.; Chenu, K. Fusion of Sentinel-2 and PlanetScope time-series data into daily 3 m surface reflectance and wheat LAI monitoring. Int. J. Appl. Earth Obs. Geoinf. 2021, 96, 102260. [Google Scholar] [CrossRef]
- Valderrama-Landeros, L.; Flores-Verdugo, F.; Rodríguez-Sobreyra, R.; Kovacs, J.M.; Flores-de-Santiago, F. Extrapolating canopy phenology information using Sentinel-2 data and the Google Earth Engine platform to identify the optimal dates for remotely sensed image acquisition of semiarid mangroves. J. Environ. Manag. 2021, 279, 111617. [Google Scholar] [CrossRef]
- Cheng, Y.; Vrieling, A.; Fava, F.; Meroni, M.; Marshall, M.; Gachoki, S. Phenology of short vegetation cycles in a Kenyan rangeland from PlanetScope and Sentinel-2. Remote Sens. Environ. 2020, 248, 112004. [Google Scholar] [CrossRef]
- Zhao, R.; Chen, Y.; Shi, P.; Zhang, L.; Pan, J.; Zhao, H. Land use and land cover change and driving mechanism in the arid inland river basin: A case study of Tarim River, Xinjiang, China. Environ. Earth Sci. 2013, 68, 591–604. [Google Scholar] [CrossRef]
- Liu, G.; Kurban, A.; Duan, H.; Halik, U.; Ablekim, A.; Zhang, L. Desert riparian forest colonization in the lower reaches of Tarim River based on remote sensing analysis. Environ. Earth Sci. 2014, 71, 4579–4589. [Google Scholar] [CrossRef]
- Wu, S.; Wang, J.; Yan, Z.; Song, G.; Chen, Y.; Ma, Q.; Deng, M.; Wu, Y.; Zhao, Y.; Guo, Z.; et al. Monitoring tree-crown scale autumn leaf phenology in a temperate forest with an integration of PlanetScope and drone remote sensing observations. ISPRS J. Photogramm. Remote Sens. 2021, 171, 36–48. [Google Scholar] [CrossRef]
- Tang, H.; Xie, J.; Tang, X.; Chen, W.; Li, Q. On-Orbit Radiometric Performance of GF-7 Satellite Multispectral Imagery. Remote Sens. 2022, 14, 886. [Google Scholar] [CrossRef]
- Xie, J.; Liu, R.; Tang, X.; Yang, X.; Zeng, J.; Mo, F.; Mei, Y. A Geometric Calibration Method Without a Field Site of the GF-7 Satellite Laser Relying on a Surface Mathematical Model. IEEE Trans. Geosci. Remote Sens. 2023, 61, 1–14. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Campos-Taberner, M.; Moreno-Martínez, Á.; Walther, S.; Duveiller, G.; Cescatti, A.; Mahecha, M.D.; Muñoz-Marí, J.; García-Haro, F.J.; Guanter, L.; et al. A unified vegetation index for quantifying the terrestrial biosphere. Sci. Adv. 2021, 7, eabc7447. [Google Scholar] [CrossRef] [PubMed]
- Graesser, J.; Stanimirova, R.; Friedl, M.A. Reconstruction of Satellite Time Series with a Dynamic Smoother. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 1803–1813. [Google Scholar] [CrossRef]
- Farmonov, N.; Amankulova, K.; Szatmári, J.; Urinov, J.; Narmanov, Z.; Nosirov, J.; Mucsi, L. Combining PlanetScope and Sentinel-2 images with environmental data for improved wheat yield estimation. Int. J. Digit. Earth 2023, 16, 847–867. [Google Scholar] [CrossRef]
- Liu, R.; Shang, R.; Liu, Y.; Lu, X. Global evaluation of gap-filling approaches for seasonal NDVI with considering vegetation growth trajectory, protection of key point, noise resistance and curve stability. Remote Sens. Environ. 2017, 189, 164–179. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Y.; Xu, W.; Wu, M.; Wang, L.; Lu, N.; Ou, G. A Seasonal Fresh Tea Yield Estimation Method with Machine Learning Algorithms at Field Scale Integrating UAV RGB and Sentinel-2 Imagery. Plants 2025, 14, 373. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. Available online: https://scikit-learn.sourceforge.net (accessed on 29 November 2023).
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Grybas, H.; Congalton, R.G. A Comparison of Multi-Temporal RGB and Multispectral UAS Imagery for Tree Species Classification in Heterogeneous New Hampshire Forests. Remote Sens. 2021, 13, 2631. [Google Scholar] [CrossRef]
- Osińska-Skotak, K.; Radecka, A.; Piórkowski, H.; Michalska-Hejduk, D.; Kopeć, D.; Tokarska-Guzik, B.; Ostrowski, W.; Kania, A.; Niedzielko, J. Mapping Succession in Non-Forest Habitats by Means of Remote Sensing: Is the Data Acquisition Time Critical for Species Discrimination? Remote Sens. 2019, 11, 2629. [Google Scholar] [CrossRef]
- Zhang, C.; Xia, K.; Feng, H.; Yang, Y.; Du, X. Tree species classification using deep learning and RGB optical images obtained by an unmanned aerial vehicle. J. For. Res. 2021, 32, 1879–1888. [Google Scholar] [CrossRef]
- Fang, F.; McNeil, B.E.; Warner, T.A.; Maxwell, A.E.; Dahle, G.A.; Eutsler, E.; Li, J. Discriminating tree species at different taxonomic levels using multi-temporal WorldView-3 imagery in Washington D.C., USA. Remote Sens. Environ. 2020, 246, 111811. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef]
- Weil, G.; Lensky, I.M.; Levin, N. Using ground observations of a digital camera in the VIS-NIR range for quantifying the phenology of Mediterranean woody species. Int. J. Appl. Earth Obs. Geoinf. 2017, 62, 88–101. [Google Scholar] [CrossRef]
- Cristani, M.; Lisein, J.; Michez, A.; Claessens, H.; Lejeune, P. Discrimination of Deciduous Tree Species from Time Series of Unmanned Aerial System Imagery. PLoS ONE 2015, 10, e0141006. [Google Scholar] [CrossRef]
- Wan, Y.; Shi, Q.; Dai, Y.; Marhaba, N.; Peng, L.; Peng, L.; Shi, H. Water Use Characteristics of Populus euphratica Oliv. and Tamarix chinensis Lour. at Different Growth Stages in a Desert Oasis. Forests 2022, 13, 236. [Google Scholar] [CrossRef]
- Dong, C.; Zhao, G.; Meng, Y.; Li, B.; Peng, B. The Effect of Topographic Correction on Forest Tree Species Classification Accuracy. Remote Sens. 2020, 12, 787. [Google Scholar] [CrossRef]
- Wang, M.; Li, M.; Wang, F.; Ji, X. Exploring the Optimal Feature Combination of Tree Species Classification by Fusing Multi-Feature and Multi-Temporal Sentinel-2 Data in Changbai Mountain. Forests 2022, 13, 1058. [Google Scholar] [CrossRef]




| Vegetation Indices | Equation |
|---|---|
| Normalized Difference Vegetation Index (NDVI) | |
| Enhanced Vegetation Index (EVI) | |
| Soil-Adjusted Vegetation Index (SAVI) | |
| Green Normalized Difference Vegetation Index (GNDVI) |
| Metric | P. euphratica | T. chinensis | Average F1 Score | ||||
|---|---|---|---|---|---|---|---|
| PA | UA | F1 Score | PA | UA | F1 Score | ||
| VIs (not filling-gap) | 0.9165 | 0.8662 | 0.8910 | 0.9002 | 0.8824 | 0.8912 | 0.8911 |
| VIs (gap-filling) | 0.9210 | 0.8925 | 0.9071 | 0.9222 | 0.8941 | 0.9079 | 0.9075 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Chen, X.; Zou, S. Optimal Phenology Windows for Discriminating Populus euphratica and Tamarix chinensis in the Tarim River Desert Riparian Forests with PlanetScope Data. Forests 2025, 16, 1560. https://doi.org/10.3390/f16101560
Wang Z, Chen X, Zou S. Optimal Phenology Windows for Discriminating Populus euphratica and Tamarix chinensis in the Tarim River Desert Riparian Forests with PlanetScope Data. Forests. 2025; 16(10):1560. https://doi.org/10.3390/f16101560
Chicago/Turabian StyleWang, Zhen, Xiang Chen, and Shuai Zou. 2025. "Optimal Phenology Windows for Discriminating Populus euphratica and Tamarix chinensis in the Tarim River Desert Riparian Forests with PlanetScope Data" Forests 16, no. 10: 1560. https://doi.org/10.3390/f16101560
APA StyleWang, Z., Chen, X., & Zou, S. (2025). Optimal Phenology Windows for Discriminating Populus euphratica and Tamarix chinensis in the Tarim River Desert Riparian Forests with PlanetScope Data. Forests, 16(10), 1560. https://doi.org/10.3390/f16101560






