Abstract
High-quality genetic maps are effective tools for elucidating the genetic mechanisms of complex quantitative traits and facilitating marker-assisted breeding. Species within the genus Liquidambar (commonly called sweetgum), particularly Liquidambar styraciflua and Liquidambar formosana, are significant forest resources worldwide. These sweetgum trees have been extensively utilized in medical and cosmetic applications for centuries as they contain large amounts of valuable secondary metabolites. Among these, shikimic acid is a notable metabolite with significant pharmaceutical applications. Despite advances in conventional breeding and propagation techniques for sweetgum, the genetic basis and regulatory mechanisms of valuable traits remain largely unexplored. In this study, we constructed the first high-density genetic map for sweetgum using whole-genome resequencing (WGR) of 220 progeny individuals derived from a cross of L. styraciflua × L. formosana. The genetic map spanned a total distance of 1428.51 centimorgans (cM) with an average inter-marker distance of 0.33 cM, incorporating 4268 bin markers across 16 linkage groups. To identify the genetic loci controlling the shikimic acid content, quantitative trait locus (QTL) mapping was carried out based on the genetic map. Two QTLs located on linkage group (LG) 12 were detected, encompassing a total of 213 genes within the QTL interval. Some of these genes are closely related to secondary metabolism in plants, including YUCCA and DXS genes. This study presents the first high-quality genetic map of sweetgum and provides a preliminary QTL analysis for shikimic acid content. Our findings establish a foundational framework for the genetic improvement of sweetgum through marker-assisted breeding and offer valuable insights for further research in sweetgum genetics.
1. Introduction
Liquidambar, commonly called sweetgum, belongs to the Hamamelidaceae family and comprises three major species: L. styraciflua (North America), L. formosana (Eastern Asia) and L. orientalis (Asia Minor). It is a relatively small genus with a remarkable distribution [1,2]. Among these, L. styraciflua is a prominent hardwood tree species native to eastern North America and northern Central America. This species is extensively utilized in the paper industry and for wood products such as furniture, plywood, and particle board. Additionally, it holds significant horticultural value [3,4]. L. formosana, known as Chinese sweetgum, is widely distributed in southern China and exhibits an exceptional adaptability, relatively fast growth, and high ecological value. As one of the main afforestation species in the subtropical regions of China, it provides economic and ecological benefits and has been widely used for urban landscaping, medicinal applications and ornamental purposes [5,6,7]. Diverse compounds extracted from its leaves possess numerous applications in multiple fields [7,8,9]. Furthermore, sweetgum trees serve as a source of valuable raw materials that can be used for medicinal purposes, which are derived primarily from their resinous sap and have been used to treat common ailments for centuries [10]. The leaves also contain beneficial compounds such as essential oils and shikimic acid [11,12]. Shikimic acid, integral to the biosynthesis of plant secondary metabolites, is synthesized through the shikimic acid pathway [13]. It has been utilized as a precursor in the pharmaceutical industry for the production of the antiviral drug Tamiflu, which is used for the effective treatment of influenza viruses [10]. However, research on shikimic acid in sweetgum trees remains limited, with previous studies primarily focusing on detecting its content [14,15]. Comprehensive molecular biology research on shikimic acid in this species is still lacking.
The combined economic and ecological benefits of sweetgum have driven breeders to implement genetic improvement measures aimed at developing a new germplasm with rapid growth, high timber quality, and increased secondary metabolites. However, the lengthy growth cycle of sweetgum, similar to other tree species, hampers the effectiveness of conventional breeding. Marker-assisted selection (MAS) breeding presents a more efficient breeding method, shortening cycles and improving breeding process [16,17]. High-density genetic linkage maps are a valuable genomic tool for MAS, particularly crucial for the analysis of economic traits, which are often controlled by polygenes with minor individual effects. The use of high-density genetic linkage maps is indispensable in sweetgum, facilitating quantitative trait locus (QTL) mapping, map-based cloning, and MAS breeding.
Significant advancements have been made in the past three decades in the development of genetic linkage maps for forestry species including Pinus [18,19,20], Eucalyptus [21,22], and Populus [23,24]. Early genetic maps were constructed using first-generation and second-generation molecular markers such as RAPD, RFLP, AFLP, or SSR, which were limited by the constraints of sequencing technology, resulting in an insufficient map density. Despite these limitations, foundational genetic resources and valuable methodologies were established, preparing the groundwork for the subsequent era of high-throughput genomic sequencing.
Single nucleotide polymorphisms (SNPs), considered as the third generation of molecular markers, offer distinct advantages due to their high abundance and uniform distribution throughout the genome [25]. The advent of next-generation sequencing tools, characterized by a high throughput and low cost, has made SNPs particularly valuable for constructing high-quality genetic maps. In recent years, numerous genetic maps for important trees have been constructed based on SNP markers. In conifers, SNP-based genetic maps for Larch and Chinese fir were constructed using specific-locus-amplified fragment (SLAF) sequencing [26,27]. Similarly, some high-density genetic maps based on SNP markers have been reported in Populus [28,29]. In economic trees, such as Ziziphus jujuba Mill., Camellia sinensis, Eriobotrya japonica Lindl and Chinese chestnut, genetic maps based on SNP makers play a crucial role in elucidating the formation mechanisms of fruit- and leaf-related traits [30,31,32,33].
Two principal sequencing strategies are commonly employed for detecting SNP loci to construct high-density genetic maps: reduced representation genome sequencing (RRGS) and whole-genome resequencing (WGR). RRGS, which does not depend on the availability of reference genomic sequences, typically involves the selective sequencing of specific DNA fragments obtained through enzymatic or mechanical fragmentation [34]. This strategy has been widely used in recent years due to its relatively low cost. On the other hand, the increasing availability of whole-genome sequences has facilitated the use of WGR, which allows for comprehensive variation detection and SNP identification throughout the genome, thereby improving the resolution and utility of genetic maps. WGR has become a pivotal tool in forest genetics, being used extensively for constructing high-density genetic maps in species such as Populus, Camellia sinensis, and Myrica rubra [28,30,35]. Nevertheless, a genetic map for sweetgum remains unachieved. The construction of a high-resolution genetic map by WGR would facilitate further genetic studies and breeding operations in sweetgum.
In this study, we established an F1 full-sibling segregating population through interspecific hybridization between L. styraciflua (female) and L. formosana (male). A total of 220 hybrid progenies were randomly selected for genome-wide genotyping using WGR technology, along with the two parents. This effort resulted in the construction of the first high-density genetic map for sweetgum, which was then utilized to conduct QTL mapping for shikimic acid content. This genetic map is a pioneering achievement in sweetgum research, providing a robust framework for further QTL mapping of valuable traits and functional genetic studies, thus significantly advancing the genetic improvement of this economically and ecologically important species.
2. Materials and Methods
2.1. Plant Materials and Shikimic Acid Content Detection
The F1 mapping population originated from a cross between L. styraciflua (female parent) and L. formosana (male parent) conducted in 2019. The female parent, selected for its elegant tree form and straight trunk, was located at our research base in Yanling County, Henan Province. The male parent, planted in Qingdao, Shandong Province, was selected in this study to cultivate more adaptable varieties for future selection. L. formosana is naturally distributed in the subtropical areas of southern China, with its northern boundary reaching the Qinling Mountains and the Dabie Mountains [6]. Since Qingdao is situated in the warm temperate zone, north of its natural distribution area, the male parent can be considered to have a relatively high cold tolerance.
Hybrid seeds were sown in a greenhouse at Beijing Forestry University in 2020, yielding over 5000 full-sib progenies. To minimize the influence of the environment on the experiment, all the seedlings were cultivated in a uniform environment, and the seedlings were planted in pots of the identical specifications, with the same temperature and humidity. This reduces the environmental impact to the minimum. From these, 220 individuals were randomly selected to form the mapping population. Before sequencing, four SSR primers were used to confirm the authenticity of the hybrid population (Table S1) [36]. Shikimic acid content measurements were conducted during the early growth stage to enhance seedling production efficiency. In August 2021, when the hybrid sweetgums exhibited vigorous growth and a high leaf biomass, fully expanded leaf samples were collected and subsequently oven-dried for shikimic acid content detection. Shikimic acid was extracted from dried leaves in a water bath at 70 °C for 2 h, and its content was quantified using High-Performance Liquid Chromatography (HPLC) with a detection wavelength of 213 nm, employing a mobile phase of acetonitrile and 1% phosphoric acid solution [37].
2.2. DNA Extraction, Library Construction and Sequencing
Healthy young leaves were collected from both parental and offspring plants, immediately frozen, and stored at −80 °C for DNA extraction. Genomic DNA was extracted from all samples using the CTAB method and assessed for concentration and quality with a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing libraries were prepared from qualified DNA samples, and whole-genome resequencing was performed using the Illumina Hiseq 2500 sequencing platform with a paired-end 150 bp sequencing strategy.
2.3. Data Analysis and Linkage Map Construction
The raw sequencing reads were filtered to generate clean reads, which were then aligned to the reference genome assembled by our group (unpublished) using BWA software (v 0.7.11) [38]. SNP calling between two parents and F1 progeny was performed with GATK software (v 4.2.0.0) [39], with subsequent stringent filtering to derive final SNP clusters for genetic map construction. The markers were further filtered based on several criteria: (1) the removal of markers homozygous in both parents; (2) a minimum 4× depth for parental markers; (3) the exclusion of markers that were not located on chromosomes. According to the genotyping information of parents, the genotype of each SNP in the offspring was determined. HighMap software was employed to construct the genetic map using a Bin-mapping method [40]. SNPs were assigned to linkage groups based on their physical positions, and recombination analysis was performed for each marker. Non-recombinant SNPs were grouped into bins to serve as markers on the genetic map.
2.4. QTL Mapping
QTL mapping for shikimic acid content was carried out using the interval mapping method in the MapQTL 6.0 software [41]. A threshold for logarithm of odds (LODs) values was established using 1000 permutation tests. If no mapping interval was identified at LODs thresholds corresponding to 0.995 and 0.95 confidence levels, the threshold was manually adjusted to 3.0. QTLs were named according to trait names and linkage group locations. Based on the positions of flanking markers, all genes within the interval were identified as candidates. Gene annotations from the reference hybrid sweetgum genome were utilized to analyze the candidate genes in the target intervals.
3. Results
3.1. Preliminary Identification of Hybrid Populations
The capillary electrophoresis results of four pairs of SSR primers showed clear amplification in both parents, all within the targeted fragment sizes. Specifically, the PCR products of primer L37 displayed homozygosity in both parents. In contrast, primers C30 and C10 were heterozygous in the paternal parent and homozygous in the maternal parent. Primer C67 exhibited heterozygosity in both parents (Figure S1). The amplification patterns of these four primers in 220 progenies conformed to Mendelian inheritance, with each progeny inheriting one allele from L. styraciflua and one from L. formosana, thereby confirming the authenticity of the hybrid offspring (Figure S2). These amplification results indicated that all progenies were full-sib hybrids.
3.2. Analysis of WGR Data
High-throughput sequencing was employed to perform whole-genome resequencing on the 2 parent samples and 220 progeny individuals, resulting in the generation of substantial clean base pairs: 17.59 Gb in the female, 20.16 Gb in the male, and 765.86 Gb in the offspring (Table 1). The sequencing efforts yielded 58,761,436 and 67,321,840 clean reads in the maternal and paternal parents, respectively, and a total of 2,559,946,028 clean reads across the progeny. Alignment with a reference genome showed high mapping efficiencies of 93.28% for the female, 90.83% for the male, and 94.86% for the offspring. The average sequencing depths achieved were 23× for the female, 25× for the male, and 4.26× across the progeny. These high-quality sequencing data are instrumental in constructing a high-density genetic map for sweetgum, essential for subsequent genetic analyses and breeding advancements.

Table 1.
Information from sequencing data of 220 offspring and their parents.
3.3. Construction of Genetic Linkage Map
In this study, a comprehensive analysis of SNP loci was conducted, identifying a total of 5,943,582 SNPs through mapping to the reference genome and subsequent SNP calling. Of these, 3,741,944 polymorphic SNP markers were discerned between the two parental genomes, exhibiting a Transition/Transversion (Ti/Tv) ratio of 1.80. Following rigorous marker screening and filtering, which included the removal of markers with homozygous genotypes and those with insufficient sequencing depth, a bin mapping method was employed to construct the genetic map. The resultant high-density genetic map spans a total genetic distance of 1428.51 cM and comprises 4268 bin markers distributed across 16 linkage groups (LGs), with an average inter-marker distance of 0.33 cM (Figure 1 and Table 2).
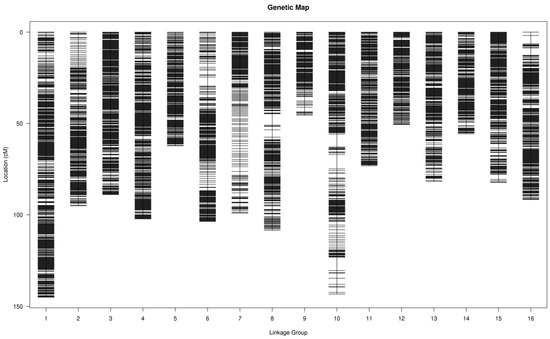
Figure 1.
High-density genetic map of sweetgum constructed by bin markers.

Table 2.
Information from the constructed genetic map.
The number of bin markers in each LG ranged from 140 (LG09) to 470 (LG01), with genetic distances ranging from 45.51 cM (LG09) to 144.97 cM (LG01). The maximum and minimum average marker intervals appeared in LG 7, 3, 5 and 12, which were 0.48 cM for LG07 and 0.28 cM for LG3, LG5, and LG12, respectively. Notably, three LGs exhibited gaps between adjacent bins exceeding 5 cM, the largest of which was 7.86 cM in LG10. A collinearity analysis was performed to evaluate the map’s quality by comparing the genetic positions of all bin markers to their physical positions on the reference genome. This analysis demonstrated a high level of consistency between the linkage groups and the physical map (Figure S3), confirming the high quality of the constructed sweetgum genetic map, which is suitable for further genetic studies and QTL mapping research.
3.4. QTL Mapping for Shikimic Acid Content
Shikimic acid, a pivotal raw material for the pharmaceutical industry, is notably abundant in the leaves of sweetgum. Our quantitative analysis of this compound revealed that the average content of shikimic acid in the leaf dry weight was 82.7 mg/g, exhibiting a variability ranging from 41.1 mg/g to 127.8 mg/g, with a coefficient of variation of 23.8%. This variability suggests that the shikimic acid content displays a normal distribution, aligning with the expected genetic distribution law of quantitative traits (Figure S4).
In this study, our genetic analysis identified two significant quantitative trait loci (QTLs) on LG12 that are associated with variations in shikimic acid content, each surpassing an LODs score greater than 3 (Figure 2 and Table 3). The identified QTLs, designated as qSK-12.21 and qSK-12.2, account for 7.5% and 8.2% of the phenotypic variance in shikimic acid content, respectively. Specifically, The QTL qSK-12.2 spans a genetic interval from 13.912 cM to 25.995 cM, covering a distance of 12.083 cM. Meanwhile, QTL qSK-12.1 is located between 12.543 cM and 13.228 cM. These findings highlight the genetic regions contributing to the biosynthesis and accumulation of shikimic acid in sweetgum, offering potential targets for future breeding programs aimed at enhancing shikimic acid production.
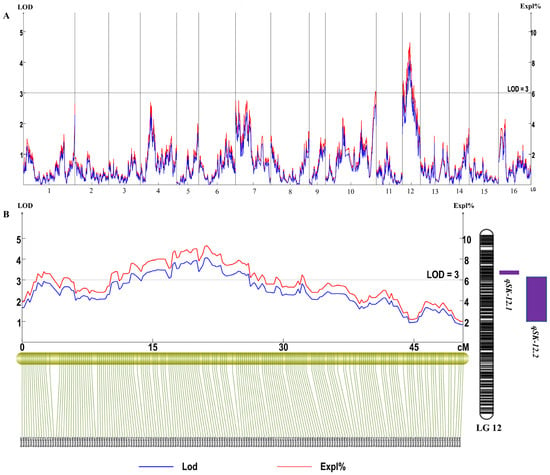
Figure 2.
Result of QTL mapping for shikimic acid content in sweetgum. (A) LODs score and phenotypic variation explained (%) of markers for shikimic acid content on 16 LGs. (B) Distributions of QTLs on LG12.

Table 3.
Information on QTLs for shikimic acid content.
3.5. Potential Candidate Gene Mining
The high-density genetic map of sweetgum constructed in this study leveraged re-sequencing technology, demonstrating a good collinearity and high consistency with the reference genome. The availability of the reference genome significantly enhanced our capacity for gene mining within QTL intervals and facilitated the prediction of candidate genes.
The analysis revealed that the region of QTL qSK-12.1 spans from 17,037,373 bp to 17,204,647 bp on chromosome 12. Due to its relatively short interval, no genes were detected within the QTL qSK-12.1 that were associated with shikimic acid content. In contrast, the QTL qSK-12.2 covers a more extensive interval, ranging from 17,220,616 bp to 21,068,059 bp on chromosome 12. This interval housed 213 genes, with 205 functionally annotated through a database comparison, identifying numerous enzymes related to secondary metabolism (Tables S2 and S3). Gene Ontology (GO) enrichment analysis indicated significant enrichment of these genes in biochemical processes, with a total of 350 genes distributed across 15 secondary annotations of biochemical processes (Figure 3). Notably, the ‘metabolic process’ category encompassed the highest number of genes, totaling 92, underscoring the substantial influence of plant metabolism on shikimic acid content.
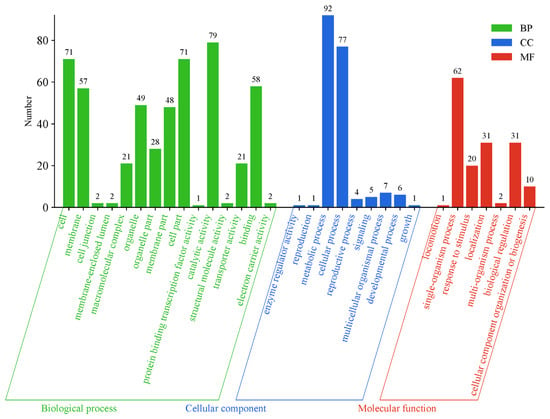
Figure 3.
GO function analysis of the genes in the QTL interval.
Shikimic acid, an intermediate in the shikimic acid pathway, serves as a bridge between primary and secondary metabolism. The shikimic acid pathway utilizes phosphoenolpyruvate from primary metabolism and erythritol-4-phosphate to synthesize chorismite, a precursor for the biosynthesis of aromatic amino acids [13,42]. The dynamics of shikimic acid content are influenced by multiple factors, including the metabolism of upstream substrates and the metabolic consumption of downstream products.
Within the QTL interval, the genes Lsf12G005840.1, Lsf12G005830.1, and Lsf12G005440.1 were identified as YUCCA genes, which play a pivotal role in auxin biosynthesis. Additionally, two genes were identified as tocopherol cyclase genes, impacting the efficiency of aromatic amino acid utilization in the downstream shikimic acid pathway. Other key genes include 1-deoxyxylulose-5-phosphate synthase (Lsf12G005220.1), zeaxanthin cyclase (Lsf12G005310.1, Lsf12G005320.1), and carotenoid cleavage dioxygenase (Lsf12G006480.1, Lsf12G006460.1, Lsf12G006470.1), pivotal in the upstream metabolic pathways crucial for terpenoid synthesis, such as plant hormone ABA and carotenoids.
4. Discussion
High-density genetic maps are pivotal in the QTLs mapping of crucial agronomic traits. High-density genetic maps are pivotal in the QTLs mapping of crucial agronomic traits and in advancing molecular breeding [30,43]. In this study, we constructed the first high-density genetic map of sweetgum using recombination bin makers, based on an F1 interspecific population (L. styraciflua × L. formosana) comprising 220 individuals. The hybrid population not only supported the construction of genetic maps but also facilitated the cultivation of superior germplasm through crossbreeding. In woody plants, the selection of a mapping population is often constrained by factors such as long growth cycles, extended juvenile periods, and self-incompatibility. Nevertheless, the F1 population is considered optimal for constructing genetic maps in such species. The size of the mapping population critically influences the quality of the map and the precision of QTLs analysis. Populations exceeding 150 individuals are generally deemed ideal for mapping purposes, and those comprising over 200 individuals are sufficient to construct high-quality genetic maps and perform robust QTLs mapping analyses [44,45]. Additionally, the selection of parental genotypes is crucial, as significant genetic diversity between parents ensures the development of numerous polymorphic markers, as is considering fertility factors. The high combining ability between L. formosana and L. styraciflua greatly facilitates the construction of the genetic map of sweetgum. The careful selection of the mapping population and parental genotypes is thus fundamental to ensuring the high quality of the resultant genetic map.
Woody plants are essential resources globally and play a critical role in terrestrial ecosystems. Genetic maps are fundamental tools that enhance in-depth research into woody plants and accelerate the breeding process. Historically, the genetic maps constructed in early research were constrained by molecular marker sequencing techniques, and an insufficient marker density limited their practical research. SNPs are considered ideal markers for constructing genetic linkage maps due to their high number and extensive genome coverage [25]. Ongoing advancements in sequencing technologies and bioinformatics have enabled the large-scale development of SNP markers, significantly enhancing map resolution. RRGS techniques are commonly employed for the development of SNP markers in forestry genetics. Meanwhile, the trend towards whole-genome assembly in various species has positioned WGR as the most efficient and precise method for developing SNP markers. This technique significantly improves the accuracy of genetic mapping and greatly enhances the efficiency of locus identification, making it an optimal method for constructing genetic maps for plants. This approach has been successfully applied in several crops including cantaloupe [46], rice [47], and sesame [48], and has been increasingly used in the construction of genetic maps for economically important forest species [32,49,50].
The range of studies employing resequencing technology for genetic mapping in forest and horticultural plants demonstrates that genetic maps constructed using this technology typically feature an average density of less than 1 cM, underscoring the effectiveness of the WGR strategy in SNP discovery and high-density linkage map construction (Table 4). With the complete sweetgum genome sequence assembled by us, employing the WGR method for genome-wide SNP discovery and the construction of a high-density genetic map in sweetgum became feasible. In this study, we selected 220 offspring, a number exceeding that used in most comparable studies, thereby laying a solid foundation for enhancing map quality. The resulting high-density genetic map of sweetgum spans 1428.51 cM, with an average marker distance of 0.33 cM and includes a total of 4268 bin markers. Compared to other studies (Table 4), our map exhibits an advantage in terms of map density and population size, which significantly contribute to the enhanced accuracy of QTL mapping.

Table 4.
Summary of reported genetic maps by WGR in forest and horticultural plants.
Most economically significant traits in trees are quantitative, and controlled by multiple genes. The primary goal of QTL mapping is to elucidate the genetic mechanisms underlying these valuable traits and to support breeding practices to improve the efficiency of genetic improvement. Sweetgum, noted for its unique volatile substances, aromatic compounds, and medicinal secondary metabolites, is a valuable tree species in which such traits are particularly relevant. Shikimic acid, a valuable metabolite abundant in sweetgum leaves, is utilized within industrial applications to synthesize the antiviral drug Oseltamivir, which combats the H5N1 influenza virus [12]. Conducting QTL mapping for shikimic acid content in sweetgum is thus a strategic approach to accelerate the molecular breeding process in sweetgum.
Shikimic acid is biosynthesized via the shikimate pathway, which serves as a crucial junction between primary and secondary metabolism, playing a pivotal role in various plant physiological processes. Many valuable metabolites are synthesized through the shikimic acid pathway and its downstream secondary metabolic pathways. It is estimated that, in higher plants, 20% or more of the total fixed carbon flows through the shikimate pathway, predominantly for the synthesis of the various secondary metabolites [42,55]. The aromatic amino acids produced by this pathway are pivotal not only for protein synthesis, but also as precursors for numerous significant metabolites such as flavonoids, phenols, and phenylpropanoids [56]. Additionally, the synthesis of lignin, essential for wood formation, is also derived from the shikimic acid pathway [57].
In this study, HPLC technology was utilized to quantify the shikimic acid content in sweetgum leaves. Combined with the high-quality genetic map we constructed, two QTLs on the LG 12 were identified, and extensive gene mining within these intervals was performed. A total of 213 genes were located in the QTL intervals, with 205 having functional annotations. The majority of these genes are associated with plant secondary metabolism, indicating the substantial influence of plant metabolism on shikimic acid content.
A notable proportion of the secondary metabolites in sweetgum leaves are derivatives of the shikimic acid pathway. The downstream metabolism of these compounds may provide feedback that regulates shikimic acid content. The rate of downstream secondary metabolism may affect the consumption and retention of shikimic acid, thereby affecting changes in its content. The downstream metabolism-related genes identified in the interval are particularly important. Among the genes in the interval, three genes (Lsf12G005840.1, Lsf12G005830.1, and Lsf12G005440.1) were annotated as YUCCA genes encoding YUCCA (YUC) flavin monooxygenase-like proteins crucial for auxin synthesis. Indole-3-acetic acid (IAA), the main form of auxin produced by plants, is synthesized predominantly through the tryptophan-dependent pathway, which involves the conversion of tryptophan to indole-3-pyruvate (IPA) by aminotransferases and the subsequent conversion of IPA to IAA by YUCCA flavin monooxygenases [58,59,60]. We postulated that these genes modulated the utilization of tryptophan derived from the shikimate pathway, thereby influencing shikimic acid content. Additionally, two genes in the interval encode tocopherol cyclase (TC), a key enzyme in the vitamin E biosynthesis pathway, also known as “tocopherol” [61]. This pathway utilizes homogentisate acid produced from tyrosine (synthesized by the shikimic acid pathway) as one of two precursors for tocopherol synthesis [62]. This connection underscores the integration of the tocopherols synthesis pathway with the shikimic acid pathway.
Furthermore, upstream metabolic pathways also impact the shikimic acid pathway. Phosphoenolpyruvate, one of the two precursor compounds in the shikimic acid pathway, is catalyzed to produce pyruvate, an essential precursor in the methylerythritol phosphate (MEP) pathway for synthesizing terpenoid compounds like plant hormone abscisic acid and carotenoids [63]. Notably, Lsf12G005220.1 was annotated as 1-deoxyxylulose-5-phosphate synthase (DXS), a rate-limiting enzyme in the MEP pathway [64]. In addition, three genes (Lsf12G006480.1, Lsf12G006460.1, Lsf12G006470.1) and two genes (Lsf12G005310.1 and Lsf12G005320.1) detected in the interval encode key enzymes involved in secondary metabolism, associated with abscisic acid synthesis and carotenoid degradation, both of which are downstream metabolic pathways of the MEP pathway. Furthermore, several transcription factors were found in the QTL interval. Five genes were annotated as ERF transcription factors belonging to the AP2/ERF gene family, which play a significant role in regulating plant growth and development processes as well as responses to abiotic stresses [65]. These findings not only enhance our understanding of the complex genetic factors influencing secondary metabolite production in sweetgum but also provide foundational support for marker-assisted breeding programs.
The resources and knowledge generated here lay a solid groundwork for future research into valuable quantitative traits in sweetgum. The 205 annotated genes from the shikimic acid content QTL intervals identified in this study provide valuable genetic resources for advancing molecular breeding in sweetgum. In the future work, we will undertake in-depth research on these genes, concentrating on a specific gene from a particular interval. We will focus on conducting comprehensive investigations into gene function validation though genetic transformation, gene expression, functional interactions, and revealing molecular regulatory mechanisms involving these critical genes, thereby enhancing our understanding and manipulation of these economically and ecologically important pathways. We will also explore the impacts of environmental factors, abiotic stress, and the interactions between the environment and genes on shikimic acid content, which will potentially lead to the development of sweetgum varieties with optimized traits for industrial and medicinal use.
5. Conclusions
In this study, whole-genome resequencing was performed on 220 progenies derived from an interspecific cross between L. styraciflua and L. formosana, along with their parents. Finally, the first high-density bin-based genetic map of sweetgum was constructed after comprehensive genomic analysis. The map covered a total distance of 1428.51 cM with an average inter-marker distance of 0.33 cM, containing 4268 markers across 16 linkage groups. This map of sweetgum facilitated detailed QTL analysis for shikimic acid content, leading to the identification of two significant QTLs on the LG12. Within these QTL intervals, a total of 213 candidate genes were identified, with a notable number of these genes being closely associated with secondary metabolism according to their functional annotations. Overall, the genetic map and QTL mapping results provide abundant and critical insights into the genetic basis of the shikimic acid content in this important tree species, and they provide basic support for the molecular breeding of and future valuable quantitative trait research into sweetgum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f15091662/s1, Figure S1: Capillary electrophoresis pattern of four pairs of SSR primers amplified in the parent of sweetgum; Figure S2: Capillary electrophoresis of two offspring as example; Figure S3: Collinearity analysis of the linkage map and genome based on the arrangement order of markers on genetic map; Figure S4: Distribution chart of shikimic acid content. Table S1: Primers for detecting the authenticity of hybrid progeny; Table S2: Result of database comparison of candidate genes from QTL interval; Table S3: Functional annotation of genes in QTL interval by database comparison.
Author Contributions
Conceptualization, J.Z. (Jinfeng Zhang); formal analysis, Y.F., Y.L. and H.L.; resources, Y.F., F.B., D.Z. and Z.P.; data curation, Y.F.; writing—original draft preparation, Y.F. and H.L.; writing—review and editing, Y.F. and J.Z. (Jian Zhao); visualization, Y.F.; supervision, J.Z. (Jinfeng Zhang) and J.Z. (Jian Zhao); project administration, J.Z. (Jinfeng Zhang); funding acquisition, Y.F. and J.Z. (Jinfeng Zhang). All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Fundamental Research Funds for the Central Universities (2019ZY39), National Key R&D Program of China (2023YFD2200602), the National Natural Science Foundation of China (No. 32271836), National Forestry and Grassland Administration Promotion Project of China (2020133102), and Special Funds for Laboratory Construction and Safe Operation of Beijing Forestry University (BJFUSY20230008).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
Dingju Zhan and Zhenwu Pang is employed by Guangxi Bagui Forest and Flowers Seedlings Co., Ltd., their employer’s company was not involved in this study, and there is no relevance between this research and their company.
Abbreviations
| QTL | Quantitative trait locus |
| cM | Centimorgan |
| LG | Linkage group |
| SNP | Single nucleotide polymorphism |
| MAS | Molecular marker-assisted selection |
| RAPD | Random amplified polymorphic DNA |
| RFLP | Restriction fragment length polymorphism |
| AFLP | Amplified fragment length polymorphism |
| SSR | Simple sequence repeats |
| SLAF | Specific-locus amplified fragment |
| RRGS | Reduced Representation Genome Sequencing |
| WGR | Whole-Genome Resequencing |
| CTAB | Cetyltrimethylammonium bromide |
| HPLC | High-performance liquid chromatography |
| LOD | Logarithm of odds |
| GO | Gene ontology |
| MEP | Methylerythritol phosphate |
| DXS | 1-deoxyxylulose-5-phosphate synthase |
References
- Brand, M.H.; Lineberger, R.D. Micropropagation of American Sweetgum (Liquidambar styraciflua L.). In High-Tech and Micropropagation II. Biotechnology in Agriculture and Forestry; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1992; Volume 18, pp. 3–24. [Google Scholar]
- Ickert-Bond, S.; Pigg, K.; Wen, J. Comparative infructescence morphology in Liquidambar (Altingiaceae) and its evolutionary significance. Am. J. Bot. 2005, 92, 1234–1255. [Google Scholar] [CrossRef] [PubMed]
- Sutter, E.G. Sweetgum (Liquidambar styraciflua L.). In Trees II. Biotechnology in Agriculture and Forestry; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1989; Volume 5, pp. 287–299. [Google Scholar]
- Ďurkovič, J.; Lux, A. Micropropagation with a novel pattern of adventitious rooting in American sweetgum (Liquidambar styraciflua L.). Trees 2010, 24, 491–497. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Y.; Chen, H.; Chen, C.; Liu, Z.; Han, C.; Wu, Q.; Yu, F. Transcriptomic Analyses Reveal Key Genes Involved in Pigment Biosynthesis Related to Leaf Color Change of Liquidambar formosana Hance. Molecules 2022, 27, 5433. [Google Scholar] [CrossRef]
- Sun, R.; Lin, F.; Huang, P.; Zheng, Y. Moderate Genetic Diversity and Genetic Differentiation in the Relict Tree Liquidambar formosana Hance Revealed by Genic Simple Sequence Repeat Markers. Front. Plant Sci. 2016, 7, 1411. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Guan, Y.-J.; Chen, Q.-Z.; Yuan, L.-H.; Xu, Q.-Q.; Zhou, M.-L.; Liu, H.; Lin, W.; Zhang, Z.-D.; Zhou, Z.-L.; et al. Pentacyclic Triterpenes from the resin of Liquidambar formosana have anti-angiogenic properties. Phytochemistry 2021, 184, 112676. [Google Scholar] [CrossRef]
- DeCarlo, A.; Zeng, T.; Dosoky, N.S.; Satyal, P.; Setzer, W.N. The Essential Oil Composition and Antimicrobial Activity of Liquidambar formosana Oleoresin. Plants 2020, 9, 822. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Pan, Y.; Wang, H.; Zhang, Y.; Lei, Q.; Zhu, Z.; Li, H.; Liang, M. Antioxidant activities of Liquidambar formosana Hance leaf extracts. Med. Chem. Res. 2010, 19, 166–176. [Google Scholar] [CrossRef]
- Lingbeck, J.M.; O’Bryan, C.A.; Martin, E.M.; Adams, J.P.; Crandall, P.G. Sweetgum: An ancient source of beneficial compounds with modern benefits. Pharmacogn Rev. 2015, 9, 1–11. [Google Scholar] [CrossRef]
- El-Readi, M.Z.; Eid, H.H.; Ashour, M.L.; Eid, S.Y.; Labib, R.M.; Sporer, F.; Wink, M. Variations of the chemical composition and bioactivity of essential oils from leaves and stems of Liquidambar styraciflua (Altingiaceae). J. Pharm. Pharmacol. 2013, 65, 1653–1663. [Google Scholar] [CrossRef]
- Bochkov, D.V.; Sysolyatin, S.V.; Kalashnikov, A.I.; Surmacheva, I.A. Shikimic acid: Review of its analytical, isolation, and purification techniques from plant and microbial sources. J. Chem. Biol. 2012, 5, 5–17. [Google Scholar] [CrossRef]
- Herrmann, K.M.; Weaver, L.M. THE SHIKIMATE PATHWAY. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Enrich, L.B.; Scheuermann, M.L.; Mohadjer, A.; Matthias, K.R.; Eller, C.F.; Newman, M.S.; Fujinaka, M.; Poon, T. Liquidambar styraciflua: A renewable source of shikimic acid. Tetrahedron Lett. 2008, 49, 2503–2505. [Google Scholar] [CrossRef]
- Martin, E.; Duke, J.; Pelkki, M.; Clausen, E.C.; Carrier, D.J. Sweetgum (Liquidambar styraciflua L.): Extraction of shikimic acid coupled to dilute acid pretreatment. Appl. Biochem. Biotechnol. 2010, 162, 1660–1668. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, L.; Lu, W.; Zhong, J.; Du, H.; Liu, P.; Du, Q.; Du, L.; Qing, J. Construction of SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Growth Traits in Eucommia ulmoides Oliver. Forests 2022, 13, 1479. [Google Scholar] [CrossRef]
- Nelson, C.D.; Nance, W.L.; Doudrick, R.L. A partial genetic linkage map of slash pine (Pinus elliottii Engelm. var. elliottii) based on random amplified polymorphic DNAs. Theor. Appl. Genet. 1993, 87, 145–151. [Google Scholar] [CrossRef]
- Remington, D.L.; Whetten, R.W.; Liu, B.H.; O’Malley, D.M. Construction of an AFLP genetic map with nearly complete genome coverage in Pinus taeda. Theor. Appl. Genet. 1999, 98, 1279–1292. [Google Scholar] [CrossRef]
- Hirao, T.; Matsunaga, K.; Hirakawa, H.; Shirasawa, K.; Isoda, K.; Mishima, K.; Tamura, M.; Watanabe, A. Construction of genetic linkage map and identification of a novel major locus for resistance to pine wood nematode in Japanese black pine (Pinus thunbergii). BMC Plant Biol. 2019, 19, 424. [Google Scholar] [CrossRef] [PubMed]
- Thamarus, K.A.; Groom, K.; Murrell, J.; Byrne, M.; Moran, G.F. A genetic linkage map for Eucalyptus globulus with candidate loci for wood, fibre, and floral traits. Theor. Appl. Genet. 2002, 104, 379–387. [Google Scholar] [CrossRef]
- Sumathi, M.; Bachpai, V.K.W.; Deeparaj, B.; Mayavel, A.; Dasgupta, M.G.; Nagarajan, B.; Rajasugunasekar, D.; Sivakumar, V.; Yasodha, R. Quantitative trait loci mapping for stomatal traits in interspecific hybrids of Eucalyptus. J. Genet. 2018, 97, 323–329. [Google Scholar] [CrossRef]
- Cervera, M.T.; Storme, V.; Ivens, B.; Gusmão, J.; Liu, B.H.; Hostyn, V.; Van Slycken, J.; Van Montagu, M.; Boerjan, W. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics 2001, 158, 787–809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, Z.; Yang, K.; Li, B. Genetic mapping in (Populus tomentosa x Populus bolleana) and P. tomentosa Carr. using AFLP markers. Theor. Appl. Genet. 2004, 108, 657–662. [Google Scholar] [CrossRef]
- Lander, E.S. The new genomics: Global views of biology. Science 1996, 274, 536–539. [Google Scholar] [CrossRef]
- Dong, M.; He, Q.; Zhao, J.; Zhang, Y.; Yuan, D.; Zhang, A.J. Genetic Mapping of Prince Rupprecht’s Larch (Larix principis-rupprechtii Mayr) by Specific-Locus Amplified Fragment Sequencing. Genes 2019, 10, 583. [Google Scholar] [CrossRef]
- Chen, X.; Xiong, C.; Lou, Y.; Xu, H.; Cheng, Q.; Sun, S.; Xiao, F. High-Density Genetic Map and QTL Analysis in Cunninghamia lanceolate: Insights into Growth and Wood-Color Traits. Forests 2023, 14, 1591. [Google Scholar] [CrossRef]
- Sun, P.; Jia, H.; Cheng, X.; Zhang, Y.; Li, J.; Zhang, L.; Lu, M.; Zhang, J.; Hu, J. Genetic architecture of leaf morphological and physiological traits in a Populus deltoides ‘Danhong’ × P. simonii ‘Tongliao1’ pedigree revealed by quantitative trait locus analysis. Tree Genet. Genomes 2020, 16, 45. [Google Scholar] [CrossRef]
- Tong, C.; Li, H.; Wang, Y.; Li, X.; Ou, J.; Wang, D.; Xu, H.; Ma, C.; Lang, X.; Liu, G.; et al. Construction of High-Density Linkage Maps of Populus deltoides × P. simonii Using Restriction-Site Associated DNA Sequencing. PLoS ONE 2016, 11, e0150692. [Google Scholar] [CrossRef][Green Version]
- Peng, Z.; Zhao, C.; Li, S.; Guo, Y.; Xu, H.; Hu, G.; Liu, Z.; Chen, X.; Chen, J.; Lin, S.; et al. Integration of genomics, transcriptomics and metabolomics identifies candidate loci underlying fruit weight in loquat. Hortic. Res. 2022, 9, uhac037. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Z.; Tang, H.; Zhang, Q.; Zhou, G.; Li, X. High-Density Genetic Map Construction and QTL Mapping of Leaf and Needling Traits in Ziziphus jujuba Mill. Front. Plant Sci. 2019, 10, 1424. [Google Scholar] [CrossRef]
- An, Y.; Chen, L.; Tao, L.; Liu, S.; Wei, C. QTL Mapping for Leaf Area of Tea Plants (Camellia sinensis) Based on a High-Quality Genetic Map Constructed by Whole Genome Resequencing. Front. Plant Sci. 2021, 12, 705285. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, Y.; Lai, J.; Wu, J.; Wu, C.; Hu, W.; Wu, X.; Gong, B. The Construction of a High-Density Genetic Map for the Interspecific Cross of Castanea mollissima × C. henryi and the Identification of QTLs for Leaf Traits. Forests 2023, 14, 1684. [Google Scholar] [CrossRef]
- Miller, M.R.; Dunham, J.P.; Amores, A.; Cresko, W.A.; Johnson, E.A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007, 17, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yu, Z.; Qi, X.; Wang, Z.; Zheng, Y.; Ren, H.; Liang, S.; Zheng, X. Construction of a High-Density Genetic Map and Identification of Leaf Trait-Related QTLs in Chinese Bayberry (Myrica rubra). Front. Plant Sci. 2021, 12, 675855. [Google Scholar] [CrossRef]
- Chen, S.; Dong, M.; Zhang, Y.; Qi, S.; Liu, X.; Zhang, J.; Zhao, J. Development and Characterization of Simple Sequence Repeat Markers for, and Genetic Diversity Analysis of Liquidambar formosana. Forests 2020, 11, 203. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, G.; Chen, K.; Chen, X.; Hong, Q.; Kan, J. Assessment of fresh star anise (Illicium verum Hook.f.) drying methods for influencing drying characteristics, color, flavor, volatile oil and shikimic acid. Food Chem. 2021, 342, 128359. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ma, C.; Hong, W.; Huang, L.; Liu, M.; Liu, H.; Zeng, H.; Deng, D.; Xin, H.; Song, J.; et al. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS ONE 2014, 9, e98855. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.; Van Ooijen, J.W.; Ooijen, J.; Hoorn, J.; Duin, J.; Jw, V.T.V. MapQTL®6. Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma BV: Wageningen, The Netherlands, 2009. [Google Scholar]
- Herrmann, K.M. The Shikimate Pathway: Early Steps in the Biosynthesis of Aromatic Compounds. Plant Cell 1995, 7, 907–919. [Google Scholar] [CrossRef]
- Zhang, Z.; Xie, W.; Zhang, J.; Wang, N.; Zhao, Y.; Wang, Y.; Bai, S. Construction of the first high-density genetic linkage map and identification of seed yield-related QTLs and candidate genes in Elymus sibiricus, an important forage grass in Qinghai-Tibet Plateau. BMC Genom. 2019, 20, 861. [Google Scholar] [CrossRef]
- Ferreira, A.; Ferreira, M.; Silva, L.; Cruz, C. Estimating the effects of population size and type on the accuracy of genetic maps. Genet. Mol. Biol. 2006, 29, 187–192. [Google Scholar] [CrossRef]
- Li, H.; Hearne, S.; Bänziger, M.; Li, Z.; Wang, J. Statistical properties of QTL linkage mapping in biparental genetic populations. Heredity 2010, 105, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Deng, G.; Mou, H.; Xu, Y.; Chen, L.; Yang, J.; Zhang, M. A re-sequencing-based ultra-dense genetic map reveals a gummy stem blight resistance-associated gene in Cucumis melo. DNA Res. 2017, 25, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, L.; Wang, J.; Sun, J.; Xia, X.; Geng, X.; Wang, X.; Xu, Z.; Xu, Q. Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biol. 2018, 16, 102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Miao, H.; Li, C.; Wei, L.; Duan, Y.; Ma, Q.; Kong, J.; Xu, F.; Chang, S. Ultra-dense SNP genetic map construction and identification of SiDt gene controlling the determinate growth habit in Sesamum indicum L. Sci. Rep. 2016, 6, 31556. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Xu, Y.; Yin, Y.; Huang, T.; Zhang, B.; Wang, Y.; Li, Y.; Cao, Y.; An, W. A consensus and saturated genetic map provides insight into genome anchoring, synteny of Solanaceae and leaf- and fruit-related QTLs in wolfberry (Lycium Linn.). BMC Plant Biol. 2021, 21, 350. [Google Scholar] [CrossRef]
- Yan, F.; Luo, Y.; Bao, J.; Pan, Y.; Wang, J.; Wu, C.; Liu, M. Construction of a highly saturated genetic map and identification of quantitative trait loci for leaf traits in jujube. Front. Plant Sci. 2022, 13, 1001850. [Google Scholar] [CrossRef]
- Liu, D.; Ye, Y.; Tang, R.; Gong, Y.; Chen, S.; Zhang, C.; Mei, P.; Chen, J.; Chen, L.; Ma, C. High-density genetic map construction and QTL mapping of a zigzag-shaped stem trait in tea plant (Camellia sinensis). BMC Plant Biol. 2024, 24, 382. [Google Scholar] [CrossRef]
- Jiang, Y.; Luo, H.; Yu, B.; Ding, Y.; Kang, Y.; Huang, L.; Zhou, X.; Liu, N.; Chen, W.; Guo, J.; et al. High-Density Genetic Linkage Map Construction Using Whole-Genome Resequencing for Mapping QTLs of Resistance to Aspergillus flavus Infection in Peanut. Front. Plant Sci. 2021, 12, 745408. [Google Scholar] [CrossRef]
- Jiang, J.; Fan, X.; Zhang, Y.; Tang, X.; Li, X.; Liu, C.; Zhang, Z. Construction of a High-Density Genetic Map and Mapping of Firmness in Grapes (Vitis vinifera L.) Based on Whole-Genome Resequencing. Int. J. Mol. Sci. 2020, 21, 797. [Google Scholar] [CrossRef]
- Guan, W.; Ke, C.; Tang, W.; Jiang, J.; Xia, J.; Xie, X.; Yang, M.; Duan, C.; Wu, W.; Zheng, Y. Construction of a High-Density Recombination Bin-Based Genetic Map Facilitates High-Resolution Mapping of a Major QTL Underlying Anthocyanin Pigmentation in Eggplant. Int. J. Mol. Sci. 2022, 23, 10258. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Adams, Z.P.; Ehlting, J.; Edwards, R. The regulatory role of shikimate in plant phenylalanine metabolism. J. Theor. Biol. 2019, 462, 158–170. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef]
- Di, D.W.; Wu, L.; Zhang, L.; An, C.W.; Zhang, T.Z.; Luo, P.; Gao, H.H.; Kriechbaumer, V.; Guo, G.Q. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci. Rep. 2016, 6, 36866. [Google Scholar] [CrossRef] [PubMed]
- Dłużewska, J.; Szymańska, R.; Gabruk, M.; Kós, P.B.; Nowicka, B.; Kruk, J. Tocopherol Cyclases-Substrate Specificity and Phylogenetic Relations. PLoS ONE 2016, 11, e0159629. [Google Scholar] [CrossRef]
- DellaPenna, D.; Pogson, B.J. Vitamin synthesis in plants: Tocopherols and carotenoids. Annu. Rev. Plant Biol. 2006, 57, 711–738. [Google Scholar] [CrossRef]
- Nagegowda, D.A.; Gupta, P. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020, 294, 110457. [Google Scholar] [CrossRef]
- Tian, S.; Wang, D.; Yang, L.; Zhang, Z.; Liu, Y. A systematic review of 1-Deoxy-D-xylulose-5-phosphate synthase in terpenoid biosynthesis in plants. Plant Growth Regul. 2022, 96, 221–235. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).