Comparative Analysis of Alternative Splicing in Moso Bamboo and Its Dwarf Mutant, Phyllostachys edulis ‘Tubaeformis’
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and RNA Extraction
2.2. Observation Based on Scanning Electron Microscope
2.3. PacBio Single-Molecule Long-Read Sequencing Sequencing and Analysis
2.4. RNA-Sequencing Library Preparation, Sequencing, and Analysis
2.5. Functional Annotation of Genes and Isoforms
2.6. Alternative Splicing Events
3. Results
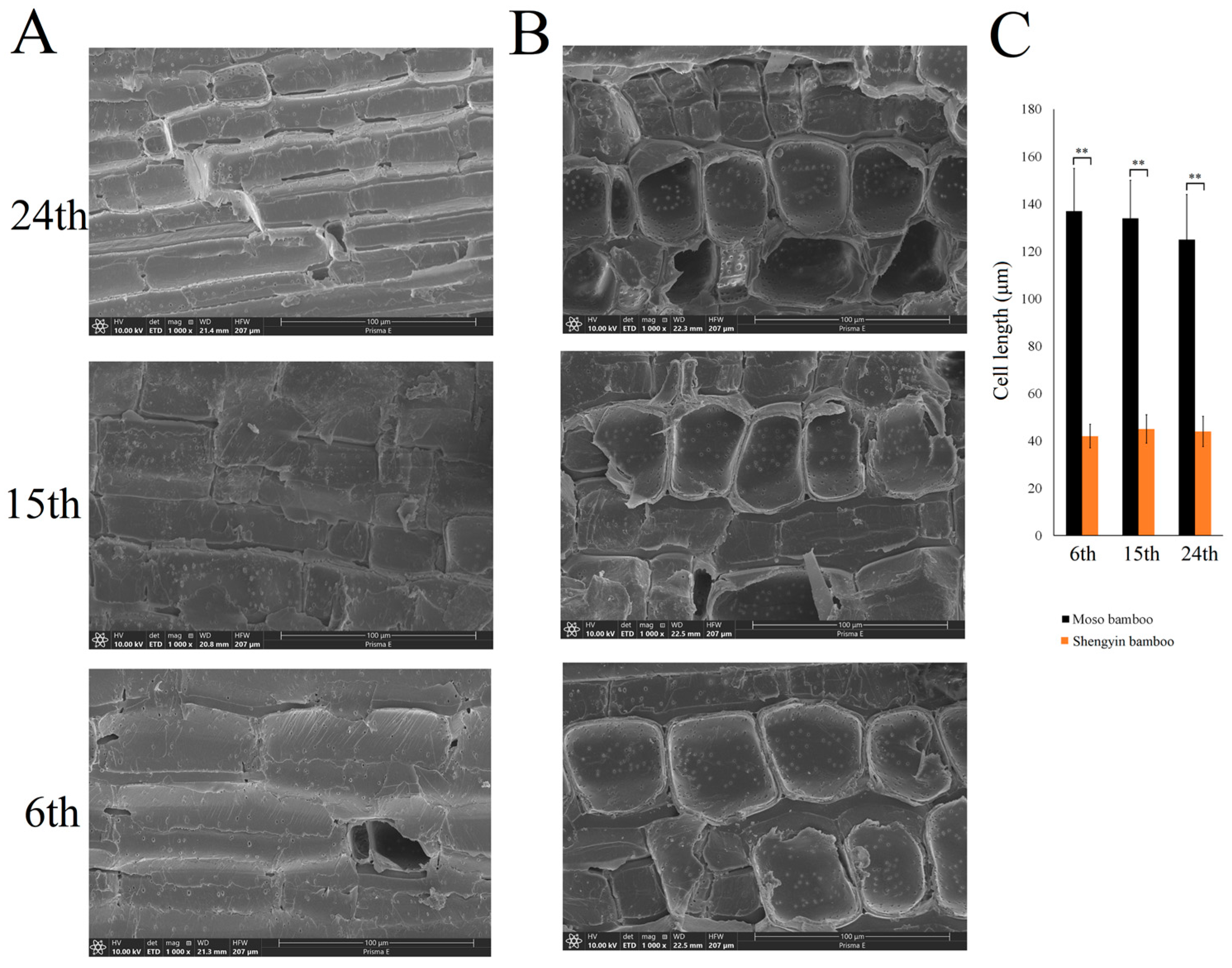
3.1. Observation of Mature Culm Internodes
3.2. Overview of Isoform Sequencing (Iso-Seq)
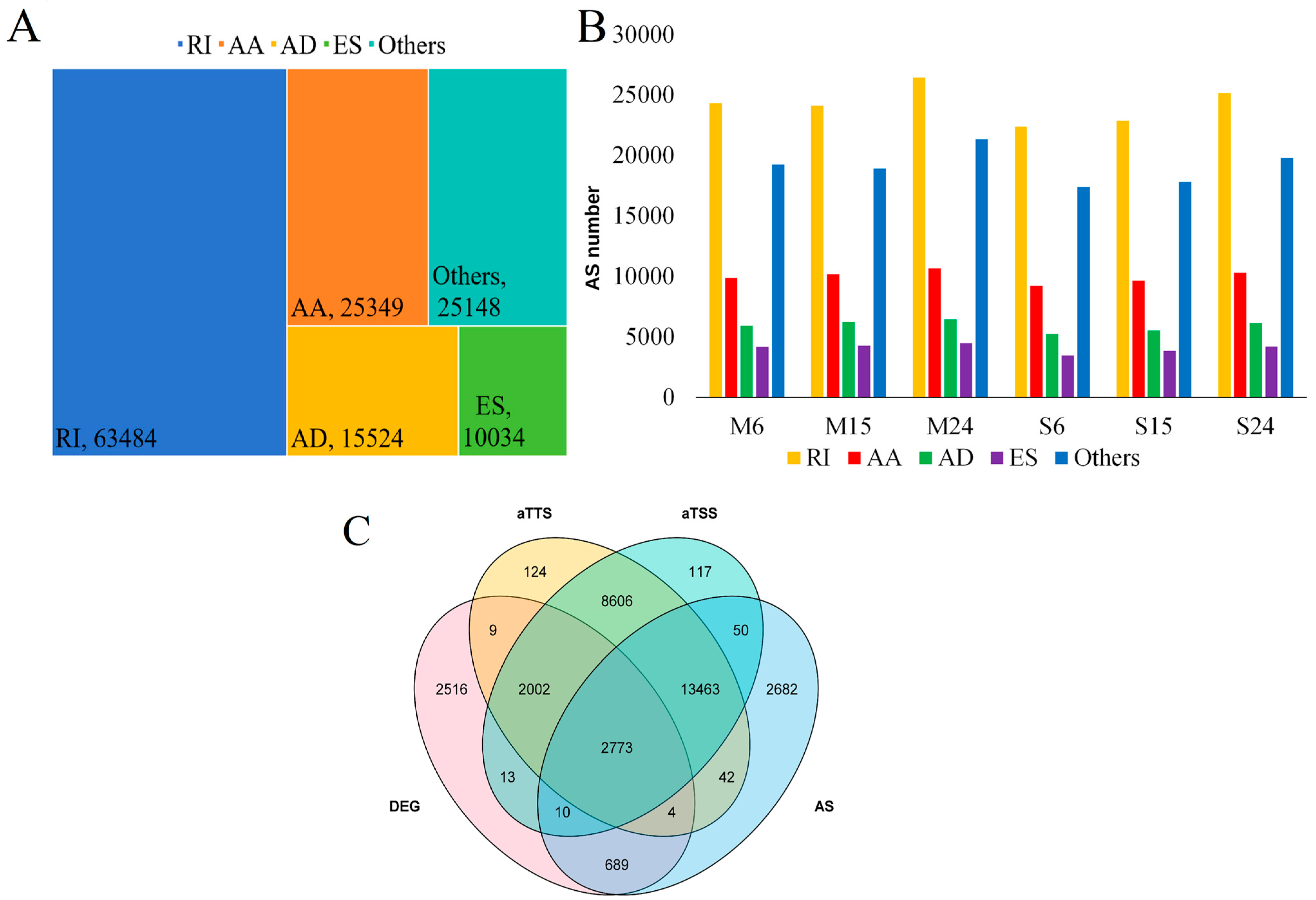
3.3. AS Increased Transcriptome Complexity in Moso Bamboo and Shengyin Bamboo Growing Culms
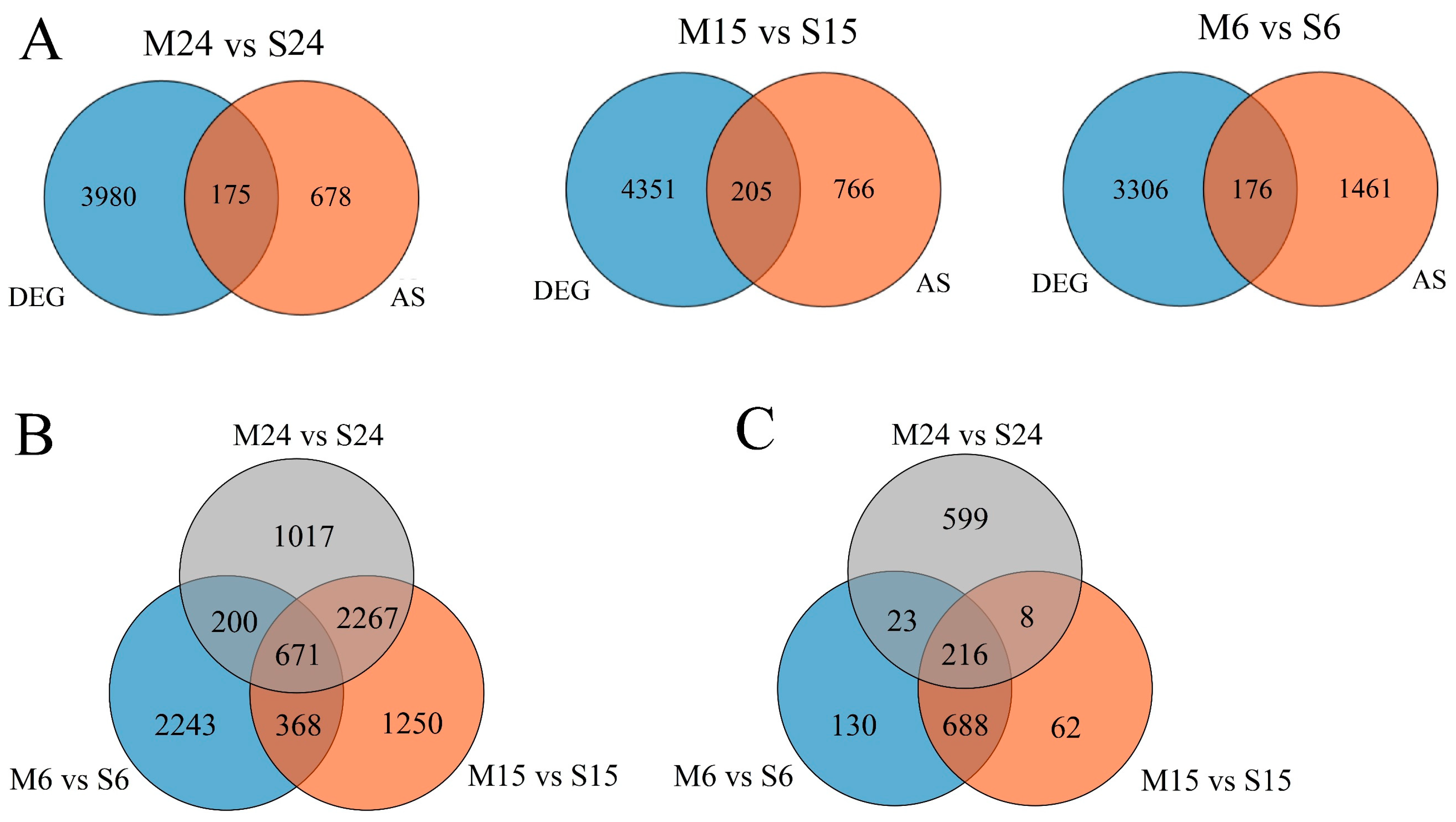
3.4. Analysis of Differences in Alternative Splicing Events between Moso Bamboo and Shengyin Bamboo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Peng, Z.H.; Lu, Y.L.; Li, L.B.; Zhao, Q.; Feng, Q. The draft genome of the fast-growing non-timber forest species Moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef]
- Li, L.; Cheng, Z.C.; Ma, Y.J.; Bai, Q.S.; Li, X.Y.; Cao, Z.H.; Wu, Z.N.; Gao, J. The association of hormone signaling, transcription and anatomy during shoot growth in Moso bamboo. Plant Biotechnol. J. 2018, 16, 72–85. [Google Scholar] [CrossRef]
- Wei, Q.; Jiao, C.; Guo, L. Exploring key cellular processes and candidate genes regulating the primary thickening growth of Moso underground shoots. New Phytol. 2017, 214, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.J. Study on the Regulation of Culm Type in Phyllostachys Edulis f. tubaeformis; Chinese Academy of Forestry: Beijing, China, 2017. [Google Scholar]
- Fiszbein, A.; Krick, K.S.; Begg, B.E.; Burge, C.B. Exon-Mediated Activation of Transcription Starts. Cell 2019, 179, 1551–1565. [Google Scholar] [CrossRef] [PubMed]
- Barash, Y.; Calarco, J.A.; Gao, W.; Pan, Q.; Wang, X.; Shai, O.; Blencowe, B.J.; Frey, B.J. Deciphering the splicing code. Nature 2010, 465, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Yang, W.; Liu, G.; Cui, X.; Meng, H.; Zhao, H.Y.; Li, J.; Liu, Z.; Zhang, M.Q.; Cai, L. Dynamic alternative splicing during mouse preimplantation embryo development. Front. Bioeng. Biotechnol. 2020, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Lin, X.; Tang, W.; Deng, Q.; Wang, Y.; Lin, Y.; He, W.; Zhang, Y.; Li, M.; Luo, Y.; et al. Transcriptomic Complexity in Strawberry fruit development and maturation revealed by nanopore sequencing. Front. Plant Sci. 2020, 13, 872054. [Google Scholar] [CrossRef]
- Chen, M.X.; Zhang, K.L.; Zhang, M.; Das, D.; Fang, Y.M.; Dai, L.; Zhang, J.; Zhu, F.Y. Alternative splicing and its regulatory role in woody plants. Tree Physiol. 2020, 40, 1475–1486. [Google Scholar] [CrossRef]
- Treutlein, B.; Gokce, O.; Quake, S.R.; Sudhof, T.C. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, E1291–E1299. [Google Scholar] [CrossRef]
- Lynch, K.W. Regulation of alternative splicing by signal transduction pathways. Adv. Exp. Med. Biol. 2007, 623, 161–174. [Google Scholar] [PubMed]
- Wang, T.; Liu, L.; Wang, X.; Liang, L.; Yue, J.; Li, L. Comparative Analyses of Anatomical Structure, Phytohormone Levels, and Gene Expression Profiles Reveal Potential Dwarfing Mechanisms in Shengyin Bamboo (Phyllostachys edulis f. tubaeformis). Int. J. Mol. Sci. 2018, 19, 1697. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Yu, J.; Xu, D.W.; Ai, K.Q.; Bao, E.; Zou, Z.R. Exposure to lower red to far-red light ratios improve tomato tolerance to salt stress. BMC Plant Biol. 2018, 18, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.S.; Gao, Z.M.; Wang, L.; Wang, J.L.; Wang, S.B.; Fei, B.H. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience 2018, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Foissac, S.; Sammeth, M. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007, 35, W297–W299. [Google Scholar] [CrossRef]
- Muller, I.B.; Meijers, S.; Kampstra, P.; van Dijk, S.; van Elswijk, M.; Lin, M.; Wojtuszkiewicz, A.M.; Jansen, G.; de Jonge, R.; Cloos, J. Computational comparison of common event-based differential splicing tools: Practical considerations for laboratory researchers. BMC Bioinform. 2021, 22, 347. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Schafer, S.L.; Miao, K.; Benson, C.C.; Heinig, M.; Cook, S.A.; Hubner, N. Alternative Splicing Signatures in RNA-seq Data: Percent Spliced in (PSI). Curr. Protoc. Hum. Genet. 2015, 87, 11–16. [Google Scholar] [CrossRef]
- Lazzara, F.E.; Rodriguez, R.E.; Palatnik, J.F. Molecular mechanisms regulating GRF activity in plant growth, development, and environmental responses. J. Exp. Bot. 2024, 26, erae179. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, P.; Wang, J.; Xu, Z.Y. Growth-regulating factors: Conserved and divergent roles in plant growth and development and potential value for crop improvement. Plant J. 2023, 113, 1122–1145. [Google Scholar] [CrossRef]
- Yan, X.; Bai, D.; Song, H.; Lin, K.; Pang, E. Alternative splicing during fruit development among fleshy fruits. BMC Genom. 2021, 22, 762. [Google Scholar] [CrossRef]
- Martin, G.; Marquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 35. [Google Scholar] [CrossRef]
- Ding, Y.Q.; Wang, Y.; Qiu, C.; Qian, W.J.; Xie, H.; Ding, Z.T. Alternative splicing in tea plants was extensively triggered by drought, heat and their combined stresses. PeerJ 2020, 8, e8258. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, J.L.; Tong, L.; Fang, Z.Y.; Yang, L.M.; Zhuang, M.; Zhang, Y.Y.; Lv, H.H. Global survey of the full-length cabbage transcriptome (Brassica oleracea var. capitata L.) reveals key alternative splicing events involved in growth and disease response. Int. J. Mol. Sci. 2021, 22, 10443. [Google Scholar] [CrossRef] [PubMed]
- Punzo, P.; Ruggiero, A.; Possenti, M.; Perrella, G.; Nurcato, R.; Costa, A.; Morelli, G.; Grillo, S.; Batelli, G. DRT111/SFPS splicing factor controls abscisic acid sensitivity during seed development and germination. Plant Physiol. 2020, 183, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Qin, Z.; Ren, G.; Deng, P.; Ji, W.; Jiao, C.; Wu, L. Complexity and regulation of age-dependent alternative splicing in Brachypodium distachyon. Plant Physiol. 2023, 192, 2703–2722. [Google Scholar] [CrossRef]
- Iida, K.; Go, M. Survey of conserved alternative splicing events of mRNAs encoding SR proteins in land plants. Mol. Biol. Evol. 2006, 23, 1085–1094. [Google Scholar] [CrossRef]
- Nicola, V.; Claudio, F.; Elisa, C.C.; Andrea, T.; Davide, C.; Michela, D.; Rosanna, Z.; Massimiliano, C.; Vannozzi, A.; ClaudioBonghi Margherita, L.; et al. Deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014, 14, 99. [Google Scholar]
- Meyer, K.; Koester, T.; Staiger, D. Pre-mRNA Splicing in Plants: In Vivo Functions of RNA-Binding Proteins Implicated in the Splicing Process. Biomolecules 2015, 5, 1717–1740. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, Q.; Jia, Y.; Deng, P.; Gao, J. Transcriptome and anatomical comparisons reveal the specific characteristics and genes involved in distinct types of growing culms. Ind. Crops Prod. 2021, 171, 113865. [Google Scholar] [CrossRef]
- Paula, D. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011, 6, 49–54. [Google Scholar]
- Hartmann, L.; Wießner, T.; Wachter, A. Subcellular Compartmentation of Alternatively Spliced Transcripts Defines SERINE/ARGININE-RICH PROTEIN30 Expression. Plant Physiol. 2018, 176, 2886–2903. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, Q.; Liu, P.; Huang, J.; Zhang, S.; Yang, G.; Wu, C.; Zheng, C.; Yan, K. Dual roles of the serine/arginine-rich splicing factor SR45a in promoting and interacting with nuclear cap-binding complex to modulate the salt-stress response in Arabidopsis. New Phytol. 2021, 230, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, J.; Bassham, D.C.; Howell, S.H. Daily temperature cycles promote alternative splicing of RNAs encoding SR45a, a splicing regulator in maize. Plant Physiol. 2021, 186, 1318–1335. [Google Scholar] [CrossRef]
- Tao, G.Y.; Ramakrishnan, M.; Vinod, K.K.; Yrjälä, K.; Satheesh, V.; Cho, J.; Fu, Y.; Zhou, M. Multi-omics analysis of cellular pathways involved in different rapid growth stages of moso bamboo. Tree Physiol. 2020, 40, 1487–1508. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hu, T.; Li, X.P.; Mu, S.H.; Cheng, Z.C.; Ge, W.; Gao, J. Genome-wide analysis of shoot growth-associated alternative splicing in moso bamboo. Mol. Genet. Genom. 2016, 5, 10. [Google Scholar] [CrossRef]
- Wei, Q.; Jiao, C.; Ding, Y.; Gao, S.; Guo, L.; Chen, M.; Hu, P.; Xia, S.; Ren, G.; Fei, Z. Cellular and molecular characterizations of a slow-growth variant provide insights into the fast growth of bamboo. Tree Physiol. 2018, 38, 641–654. [Google Scholar] [CrossRef]
- Gao, Z.; Guo, L.; Ramakrishnan, M.; Xiang, Y.; Jiao, C.; Jiang, J.; Vinod, K.K.; Fei, Z.; Que, F.; Ding, Y.; et al. Cellular and molecular characterizations of the irregular internode division zone formation of a slow-growing bamboo variant. Tree Physiol. 2022, 42, 570–584. [Google Scholar] [CrossRef]
- Yu, H.; Tian, C.; Yu, Y.; Jiao, Y. Transcriptome Survey of the Contribution of Alternative Splicing to Proteome Diversity in Arabidopsis thaliana. Mol. Plant. 2016, 9, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, J.; Wang, Q.; Lv, K.; Du, K.; Zhang, W.; Li, Q.; Kang, X.; Wei, H. Growth-Regulating Factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phytol. 2021, 230, 612–628. [Google Scholar] [CrossRef]
- Kuijt, S.J.; Greco, R.; Agalou, A.; Shao, J.; ‘t Hoen, C.C.; Övernäs, E.; Osnato, M.; Curiale, S.; Meynard, D.; van Gulik, R.; et al. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 2014, 164, 1952–1966. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zeng, J.; Ren, Y.; Chen, D.; Li, W.; Gao, F.; Cao, Y.; Luo, T.; Yuan, G.; Wu, X.; et al. OsGIF1 positively regulates the sizes of stems, leaves, and grains in rice. Front. Plant Sci. 2017, 8, 1730. [Google Scholar] [CrossRef] [PubMed]
- Erika, Ő.; Csaba, P.; Binish, M.; Aladár, P.; Tünde, L. E2FB interacts with RETINOBLASTOMA RELATED and regulates cell proliferation during leaf development. Plant Physiol. 2020, 182, 518–533. [Google Scholar]
- Horvath, B.M.; Kourova, H.; Nagy, S.; Nemeth, E.; Magyar, Z.; Papdi, C.; Ahmad, Z.; Sanchez-Perez, G.F.; Perilli, S.; Blilou, I. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017, 36, 1261–1278. [Google Scholar] [CrossRef]
- Cui, K.; He, C.Y.; Zhang, J.G.; Duan, A.G.; Zeng, Y.F. Temporal and spatial profiling of internode elongation-associated protein expression in rapidly growing culms of bamboo. J. Proteome Res. 2012, 11, 2492–2507. [Google Scholar] [CrossRef]
- Meng, L.; Chen, S.; Li, D.; Huang, M.; Zhu, S. Genome-Wide Characterization and Evolutionary Expansion of Poplar NAC Transcription Factors and Their Tissue-Specific Expression Profiles under Drought. Int. J. Mol. Sci. 2022, 24, 253. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Goue, N.; Igarashi, H.; Ohtani, M.; Nakano, Y.; Mortimer, J.C.; Nishikubo, N.; Kubo, M.; Katayama, Y.; Kakegawa, K.; et al. Vascular-Related Nac-Domain6 and Vascular-Related Nac-Domain7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010, 153, 906–914. [Google Scholar] [CrossRef]
- Chen, Z.; Peng, Z.; Liu, S.; Leng, H.; Luo, J.; Wang, F.; Yi, Y.; Resco de Dios, V.; Lucas, G.R.; Yao, Y.; et al. Overexpression of PeNAC122 gene promotes wood formation and tolerance to osmotic stress in poplars. Physiol. Plant 2022, 174, e13751. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, Q.; Li, Z.; Gao, J. Genome-wide identification and functional characterization of the PheE2F/DP gene family in Moso bamboo. BMC Plant Biol. 2021, 21, 158. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, S.; Martinez-Cortes, T. Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions. Int. J. Mol. Sci. 2018, 19, 2876. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, Z.; Sun, Y.; Su, Y.; Cheng, L.; Liu, D.; Lin, S.; Li, L. Comparative Analysis of Alternative Splicing in Moso Bamboo and Its Dwarf Mutant, Phyllostachys edulis ‘Tubaeformis’. Forests 2024, 15, 1233. https://doi.org/10.3390/f15071233
Qiu Z, Sun Y, Su Y, Cheng L, Liu D, Lin S, Li L. Comparative Analysis of Alternative Splicing in Moso Bamboo and Its Dwarf Mutant, Phyllostachys edulis ‘Tubaeformis’. Forests. 2024; 15(7):1233. https://doi.org/10.3390/f15071233
Chicago/Turabian StyleQiu, Zhenhua, Yuanyuan Sun, Yanhui Su, Long Cheng, Dong Liu, Shuyan Lin, and Long Li. 2024. "Comparative Analysis of Alternative Splicing in Moso Bamboo and Its Dwarf Mutant, Phyllostachys edulis ‘Tubaeformis’" Forests 15, no. 7: 1233. https://doi.org/10.3390/f15071233
APA StyleQiu, Z., Sun, Y., Su, Y., Cheng, L., Liu, D., Lin, S., & Li, L. (2024). Comparative Analysis of Alternative Splicing in Moso Bamboo and Its Dwarf Mutant, Phyllostachys edulis ‘Tubaeformis’. Forests, 15(7), 1233. https://doi.org/10.3390/f15071233



