Transcriptional Profiling of BpWRKY49 Reveals Its Role as a Master Regulator in Stress Signaling Pathways in Birch (Betula platyphylla)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants Growth and Treatment
2.2. RNA Isolation and RT-qPCR
2.3. Bioinformatic Analysis
2.4. Cloning of BpWRKY49
2.5. Subcellular Localization
2.6. Transactivation Assay
2.7. Binding Specificity Assay and Y1H
2.8. DNA Affinity Purification Sequencing
3. Results
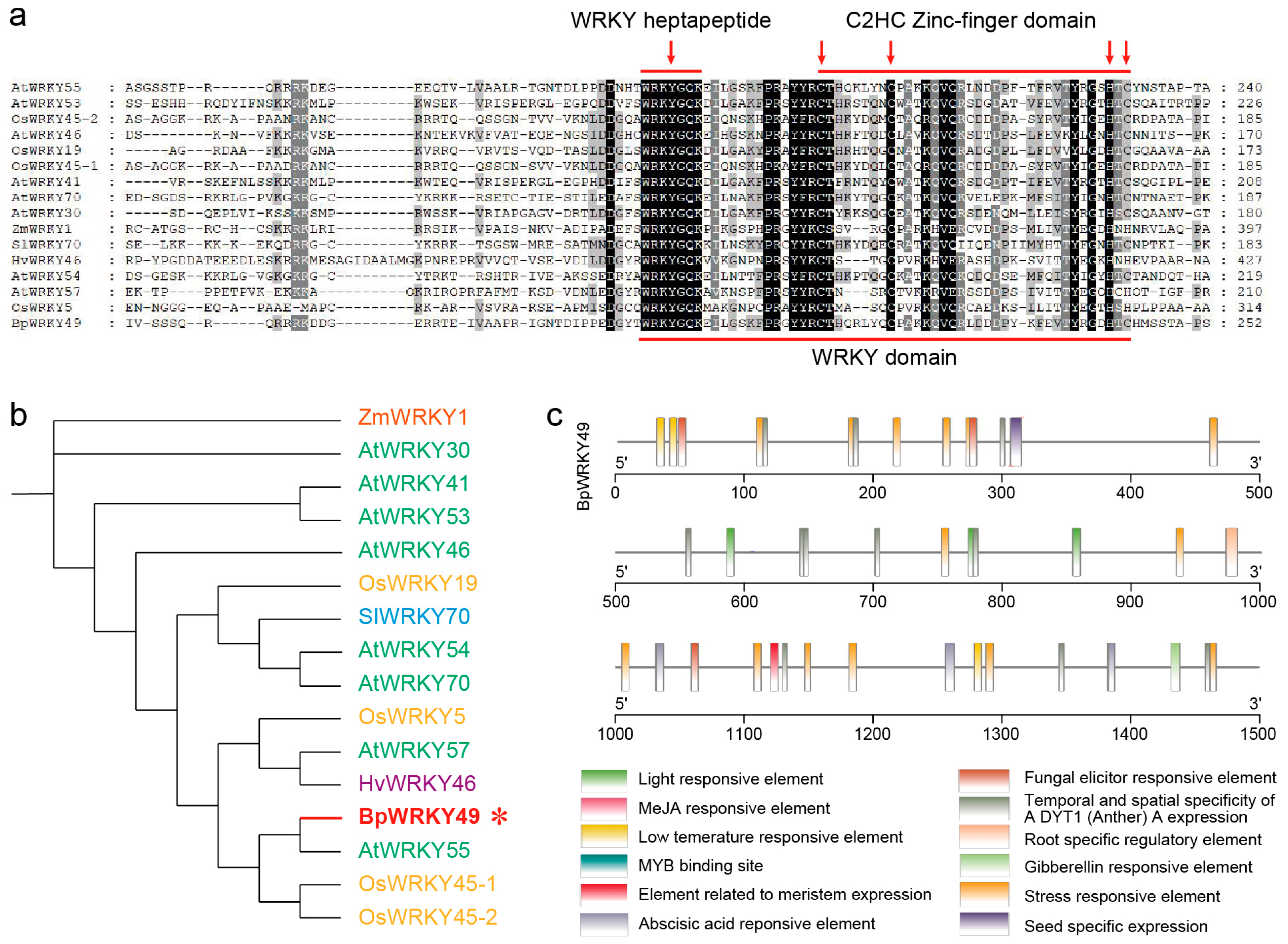
3.1. Phylogenetic and Promoter Analysis of BpWRKY49
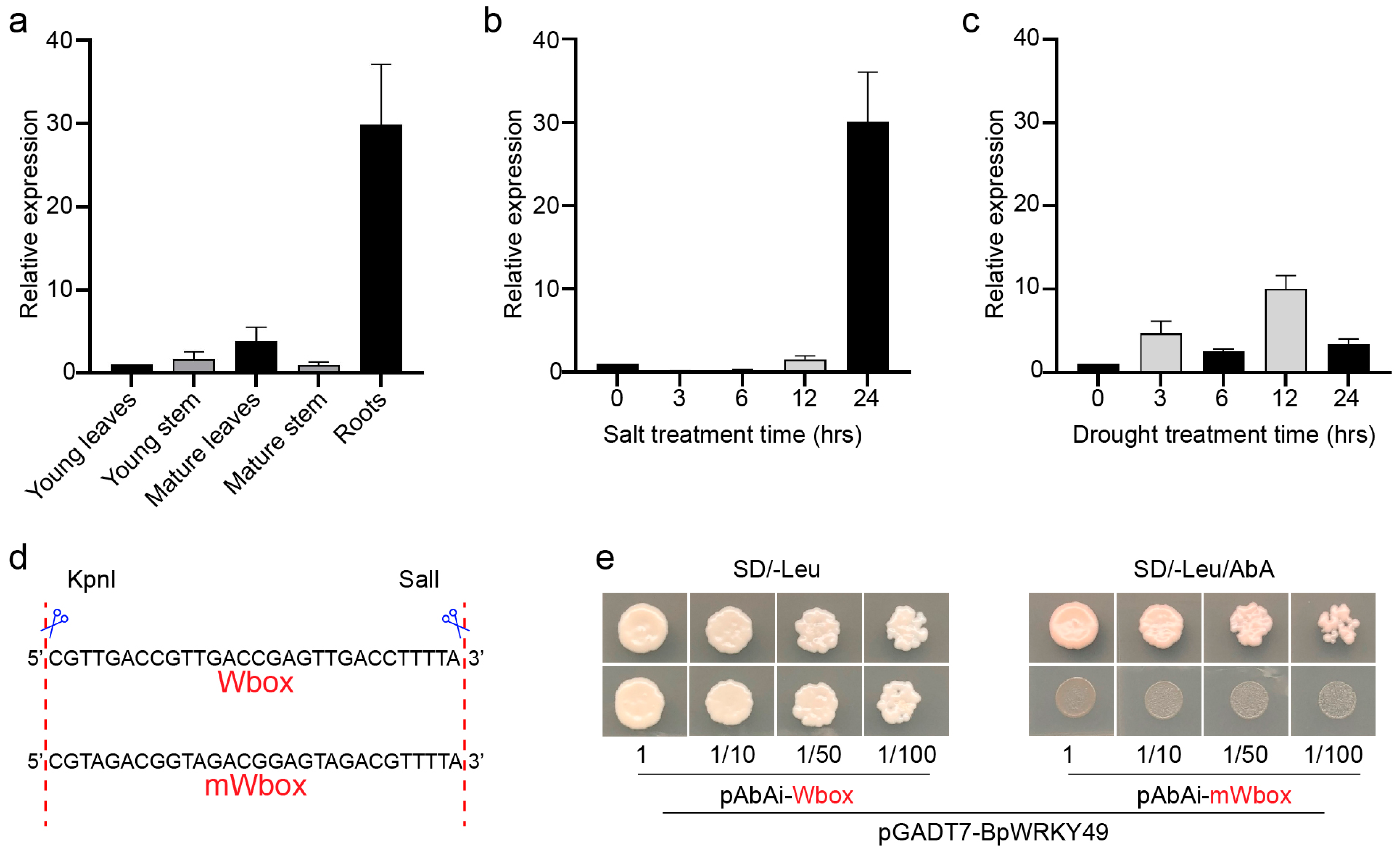
3.2. Expression Analysis of BpWRKY49 and Binding Specificity to W-Box
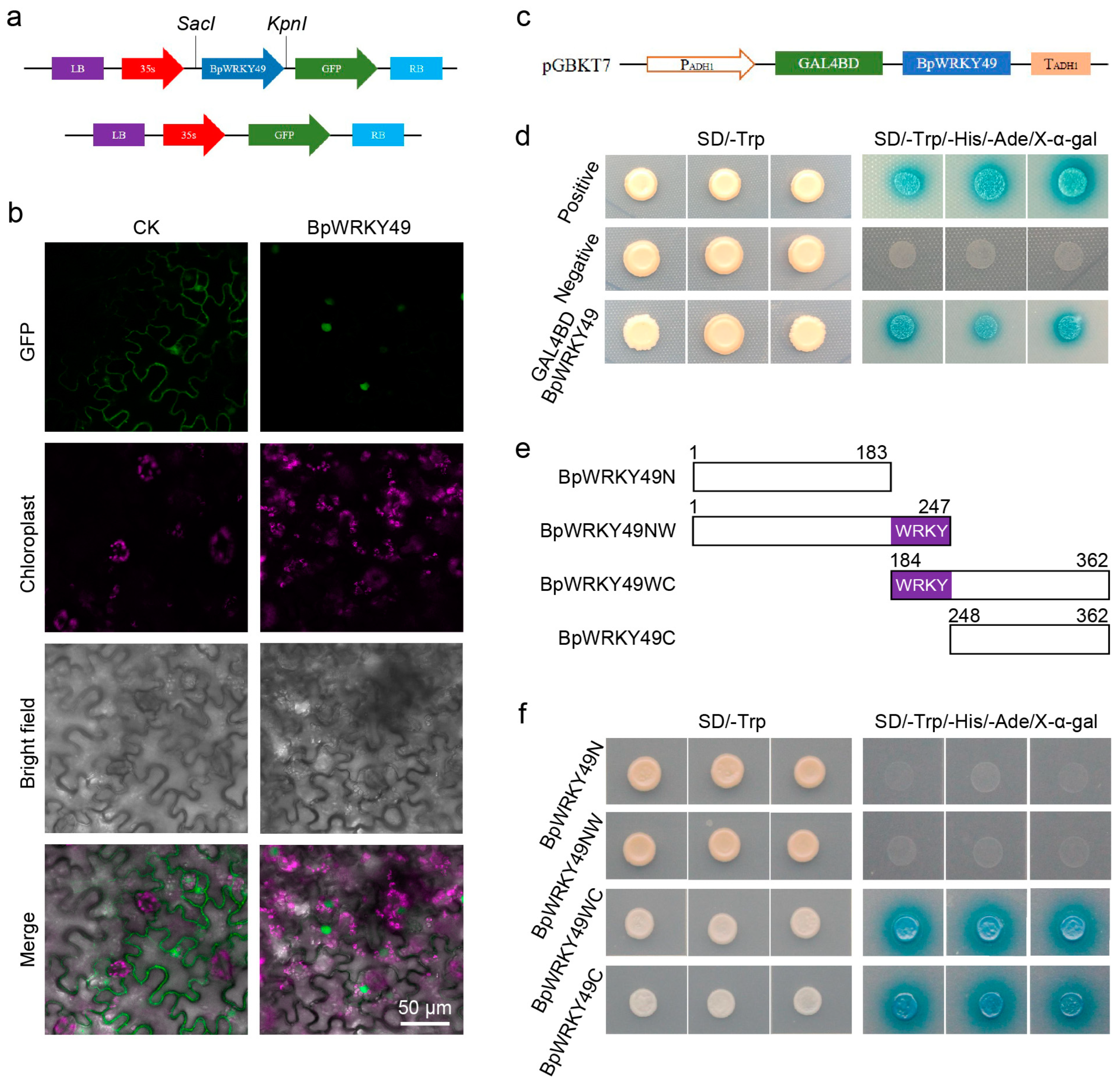
3.3. BpWRKY49 Was a Nuclear Localized Protein with Transcription Activity on C-Terminal in Yeast
3.4. Genome-Wide Identification of Binding Sites of BpWRKY49
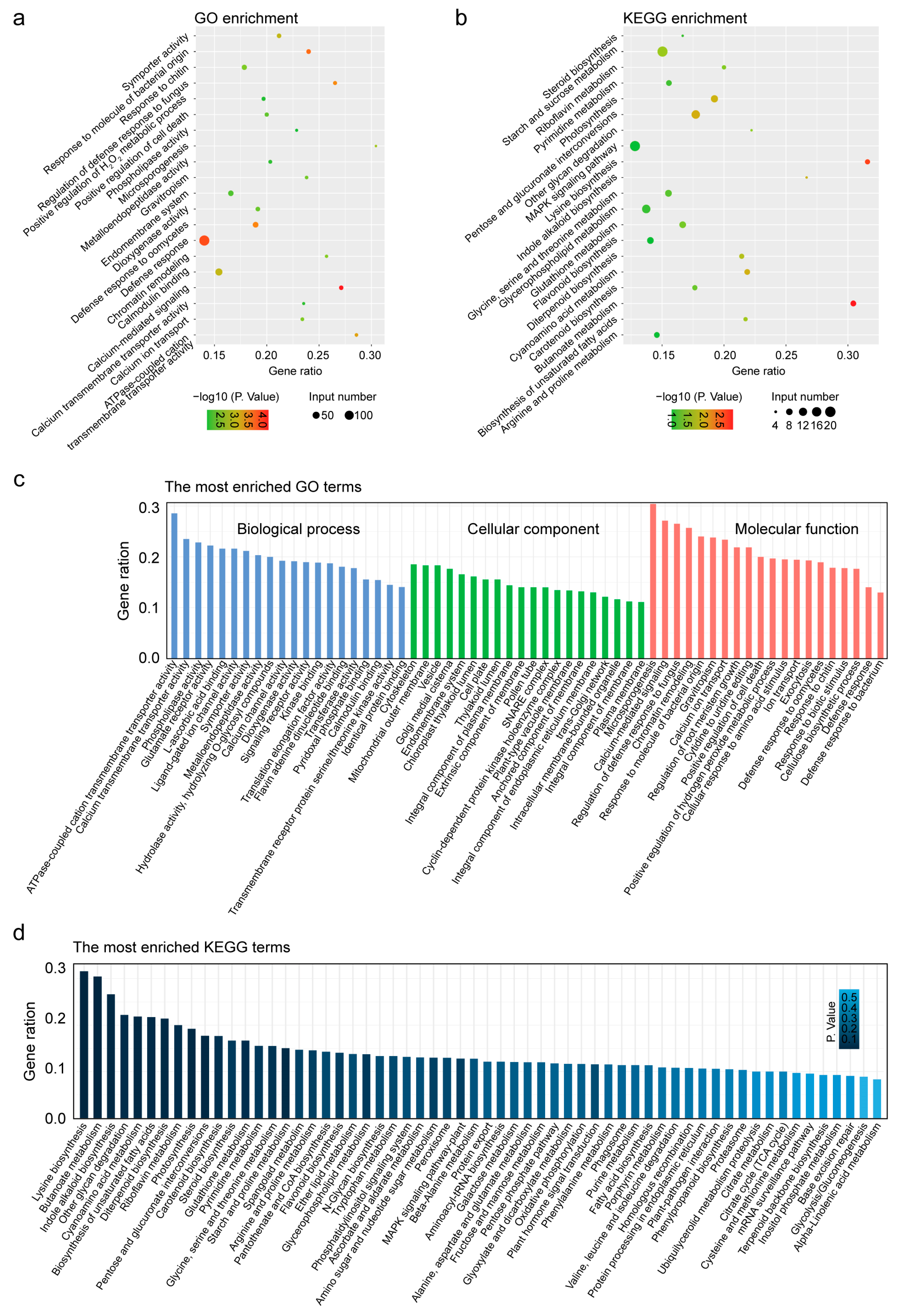
3.5. Defense Response and MAPK Signaling Pathway Were Most Enriched GO and KEGG Terms
3.6. Several Salt-Stress-Responsive Genes Were Targets of BpWRKY49
3.7. BpWRKY49 Bound to BpPUB21, BpBTL15, and BpHIP47 In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef] [PubMed]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. Int. 2015, 22, 4056–4075. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.S.; Tester, M. Salinity tolerance of Arabidopsis: A good model for cereals? Trends Plant Sci. 2007, 12, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Arif, M.S.; Ashraf, M.A.; Mahmood, R.; Yasmeen, T.; Shakoor, M.B.; Shahzad, S.M.; Ali, M.; Saleem, I.; Arif, M. A comprehensive review on rice responses and tolerance to salt stress. Adv. Rice Res. Abiotic Stress Toler. 2019, 133–158. [Google Scholar] [CrossRef]
- Omar, M.; Osman, M.; Kasim, W.; Abd El-Daim, I. Improvement of salt tolerance mechanisms of barley cultivated under salt stress using Azospirillum brasilense. In Salinity and Water Stress; Springer: Berlin/Heidelberg, Germany, 2009; Volume 44, pp. 133–147. [Google Scholar]
- Seleiman, M.F.; Aslam, M.T.; Alhammad, B.A.; Hassan, M.U.; Maqbool, R.; Chattha, M.U.; Khan, I.; Gitari, H.I.; Uslu, O.S.; Roy, R. Salinity stress in wheat: Effects, mechanisms and management strategies. Phyton 2022, 91, 668–694. [Google Scholar] [CrossRef]
- Li, C.; Li, D.; Shao, F.; Lu, S. Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genom. 2015, 16, 200. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C.; et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Xu, Z.S.; Tian, C.; Huang, Y.; Wang, F.; Xiong, A.S. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 2016, 6, 23101. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chi, Y.; Wang, Z.; Zhou, Y.; Fan, B.; Chen, Z. Functional analysis of structurally related soybean GmWRKY58 and GmWRKY76 in plant growth and development. J. Exp. Bot. 2016, 67, 4727–4742. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.A.S.; Cavalcanti, J.H.F.; Pimentel, K.G.; Medeiros, D.B.; Silva, J.C.F.; Condori-Apfata, J.A.; Lapidot-Cohen, T.; Brotman, Y.; Nunes-Nesi, A.; Fernie, A.R.; et al. The significance of WRKY45 transcription factor in metabolic adjustments during dark-induced leaf senescence. Plant Cell Environ. 2022, 45, 2682–2695. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, H.; Yu, D. Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 Oppositely Regulate Flowering under Short-Day Conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar] [CrossRef]
- Guan, Y.; Meng, X.; Khanna, R.; LaMontagne, E.; Liu, Y.; Zhang, S. Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in Arabidopsis. PLoS Genet. 2014, 10, e1004384. [Google Scholar] [CrossRef]
- Lippok, B.; Birkenbihl, R.P.; Rivory, G.; Brümmer, J.; Schmelzer, E.; Logemann, E.; Somssich, I.E. Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 2007, 20, 420–429. [Google Scholar] [CrossRef]
- Park, C.J.; Shin, Y.C.; Lee, B.J.; Kim, K.J.; Kim, J.K.; Paek, K.H. A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to Tobacco mosaic virus and Xanthomonas campestris. Planta 2006, 223, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Karim, A.; Jiang, Y.; Guo, L.; Ling, Z.; Ye, S.; Duan, Y.; Li, C.; Luo, K. Isolation and characterization of a subgroup IIa WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 2015, 35, 1129–1139. [Google Scholar] [CrossRef]
- Merz, P.R.; Moser, T.; Höll, J.; Kortekamp, A.; Buchholz, G.; Zyprian, E.; Bogs, J. The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiol. Plant 2015, 153, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cao, H.; Huang, B.; Zheng, X.; Liang, K.; Wang, G.L.; Sun, X. The WRKY10-VQ8 module safely and effectively regulates rice thermotolerance. Plant Cell Environ. 2022, 45, 2126–2144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tong, C.; Cao, L.; Zheng, P.; Tang, X.; Wang, L.; Miao, M.; Liu, Y.; Cao, S. Regulatory module WRKY33-ATL31-IRT1 mediates cadmium tolerance in Arabidopsis. Plant Cell Environ. 2023, 46, 1653–1670. [Google Scholar] [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, X.; Yang, B.; Xu, S.; Wei, X.; Zhao, P.; Niu, F.; Sun, M.; Wang, C.; Cheng, H.; et al. WRKY55 transcription factor positively regulates leaf senescence and the defense response by modulating the transcription of genes implicated in the biosynthesis of reactive oxygen species and salicylic acid in Arabidopsis. Development 2020, 147, dev189647. [Google Scholar] [CrossRef]
- Peng, X.; Hu, Y.; Tang, X.; Zhou, P.; Deng, X.; Wang, H.; Guo, Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 2012, 236, 1485–1498. [Google Scholar] [CrossRef]
- Han, M.; Ryu, H.-S.; Kim, C.-Y.; Park, D.-S.; Ahn, Y.-K.; Jeon, J.-S. OsWRKY30 is a transcription activator that enhances rice resistance to the Xanthomonas oryzae pathovar oryzae. J. Plant Biol. 2013, 56, 258–265. [Google Scholar] [CrossRef]
- Shen, H.; Liu, C.; Zhang, Y.; Meng, X.; Zhou, X.; Chu, C.; Wang, X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012, 80, 241–253. [Google Scholar] [CrossRef]
- Huangfu, J.; Li, J.; Li, R.; Ye, M.; Kuai, P.; Zhang, T.; Lou, Y. The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2016, 17, 697. [Google Scholar] [CrossRef]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 2011, 62, 4863–4874. [Google Scholar] [CrossRef]
- Hassan, S.; Lethin, J.; Blomberg, R.; Mousavi, H.; Aronsson, H. In silico based screening of WRKY genes for identifying functional genes regulated by WRKY under salt stress. Comput. Biol. Chem. 2019, 83, 107131. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, Y.; He, L. A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plant. Plant Mol. Biol. 2023, 113, 171–191. [Google Scholar] [CrossRef]
- Gu, J.; Lv, F.; Gao, L.; Jiang, S.; Wang, Q.; Li, S.; Yang, R.; Li, Y.; Li, S.; Wang, P. A WRKY Transcription Factor CbWRKY27 Negatively Regulates Salt Tolerance in Catalpa bungei. Forests 2023, 14, 486. [Google Scholar] [CrossRef]
- Ma, L.; Li, X.; Zhang, J.; Yi, D.; Li, F.; Wen, H.; Liu, W.; Wang, X. MsWRKY33 increases alfalfa (Medicago sativa L.) salt stress tolerance through altering the ROS scavenger via activating MsERF5 transcription. Plant Cell Environ. 2023, 46, 3887–3901. [Google Scholar] [CrossRef]
- Huang, J.; Liu, F.; Chao, D.; Xin, B.; Liu, K.; Cao, S.; Chen, X.; Peng, L.; Zhang, B.; Fu, S. The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 11999. [Google Scholar] [CrossRef]
- Shen, L.; Xia, X.; Zhang, L.; Yang, S.; Yang, X. SmWRKY11 acts as a positive regulator in eggplant response to salt stress. Plant Physiol. Biochem. 2023, 205, 108209. [Google Scholar] [CrossRef]
- Jiang, Y.; Tong, S.; Chen, N.; Liu, B.; Bai, Q.; Chen, Y.; Bi, H.; Zhang, Z.; Lou, S.; Tang, H. The PalWRKY77 transcription factor negatively regulates salt tolerance and abscisic acid signaling in Populus. Plant J. 2021, 105, 1258–1273. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H. Plant salt response: Perception, signaling, and tolerance. Front. Plant Sci. 2022, 13, 1053699. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and Drought Stress Signal Transduction in Plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Rai, G.K.; Mishra, S.; Chouhan, R.; Mushtaq, M.; Chowdhary, A.A.; Rai, P.K.; Kumar, R.R.; Kumar, P.; Perez-Alfocea, F.; Colla, G.; et al. Plant salinity stress, sensing, and its mitigation through WRKY. Front. Plant Sci. 2023, 14, 1238507. [Google Scholar] [CrossRef]
- Rastogi, S.; Pandey, M.M.; Kumar Singh Rawat, A. Medicinal plants of the genus Betula—Traditional uses and a phytochemical–pharmacological review. J. Ethnopharmacol. 2015, 159, 62–83. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Y.; Yu, L.; Zheng, T.; Wang, S.; Yue, Z.; Jiang, J.; Kumari, S.; Zheng, C.; Tang, H.; et al. Genome sequence and evolution of Betula platyphylla. Hortic. Res. 2021, 8, 37. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, X.; Cao, J.; Bai, H.; Wang, R.; Wang, C.; Xu, Z.; Li, C.; Liu, G. Genome-Wide Identification and Characterization of WRKY Transcription Factors in Betula platyphylla Suk. and Their Responses to Abiotic Stresses. Int. J. Mol. Sci. 2023, 24, 15000. [Google Scholar] [CrossRef]
- Bartlett, A.; O’Malley, R.C.; Huang, S.C.; Galli, M.; Nery, J.R.; Gallavotti, A.; Ecker, J.R. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 2017, 12, 1659–1672. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Vasimuddin, M.; Misra, S.; Li, H.; Aluru, S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019; pp. 314–324. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, Q.; Qin, L.; Li, L. A Cys2His2 Zinc Finger Transcription Factor BpSZA1 Positively Modulates Salt Stress in Betula platyphylla. Front. Plant Sci. 2022, 13, 823547. [Google Scholar] [CrossRef]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef]
- Turck, F.; Zhou, A.; Somssich, I.E. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in Parsley. Plant Cell 2004, 16, 2573–2585. [Google Scholar] [CrossRef]
- Wang, W.; Li, T.; Chen, J.; Zhang, X.; Wei, L.; Yao, S.; Zeng, K. A self-regulated transcription factor CsWRKY33 enhances resistance of citrus fruit to Penicillium digitatum. Postharvest Biol. Technol. 2023, 198, 112267. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, C.; Xie, F.; Hua, Q.; Zhang, Z.; Zhang, R.; Chen, J.; Zhao, J.; Hu, G.; Qin, Y. A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. Int. J. Mol. Sci. 2021, 22, 2171. [Google Scholar] [CrossRef]
- Lv, B.; Wu, Q.; Wang, A.; Li, Q.; Dong, Q.; Yang, J.; Zhao, H.; Wang, X.; Chen, H.; Li, C. A WRKY transcription factor, FtWRKY46, from Tartary buckwheat improves salt tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 147, 43–53. [Google Scholar] [CrossRef]
- Li, X.; Tang, Y.; Zhou, C.; Zhang, L.; Lv, J. A Wheat WRKY Transcription Factor TaWRKY46 Enhances Tolerance to Osmotic Stress in transgenic Arabidopsis Plants. Int. J. Mol. Sci. 2020, 21, 1321. [Google Scholar] [CrossRef]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef]
- Matsushita, A.; Inoue, H.; Goto, S.; Nakayama, A.; Sugano, S.; Hayashi, N.; Takatsuji, H. Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J. 2013, 73, 302–313. [Google Scholar] [CrossRef]
- Sun, J.; Hu, W.; Zhou, R.; Wang, L.; Wang, X.; Wang, Q.; Feng, Z.; Li, Y.; Qiu, D.; He, G.; et al. The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 2015, 34, 23–35. [Google Scholar] [CrossRef]
- Ma, J.; Li, C.; Sun, L.; Ma, X.; Qiao, H.; Zhao, W.; Yang, R.; Song, S.; Wang, S.; Huang, H. The SlWRKY57-SlVQ21/SlVQ16 module regulates salt stress in tomato. J. Integr. Plant Biol. 2023, 65, 2437–2455. [Google Scholar] [CrossRef]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019, 280, 132–142. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; He, S.P.; Gao, Y.; Wang, N.N.; Lu, R.; Li, X.B. A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol. Biochem. 2019, 141, 231–239. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, P.; Xiong, F.; Zhang, X.; Song, H. WRKY Genes Improve Drought Tolerance in Arachis duranensis. Front. Plant Sci. 2022, 13, 910408. [Google Scholar] [CrossRef]
- Yang, J.; Chen, H.; Yang, C.; Ding, Q.; Zhao, T.; Wang, D. A WRKY transcription factor WRKY184 from Brassica napus L. is involved in flowering and secondary wall development in transgenic Arabidopsis thaliana. Plant Growth Regul. 2020, 92, 427–440. [Google Scholar] [CrossRef]
- Han, P.-L.; Dong, Y.-H.; Jiang, H.; Hu, D.-G.; Hao, Y.-J. Molecular cloning and functional characterization of apple U-box E3 ubiquitin ligase gene MdPUB29 reveals its involvement in salt tolerance. J. Integr. Agric. 2019, 18, 1604–1612. [Google Scholar] [CrossRef]
- Kim, M.S.; Kim, J.H.; Amoah, J.N.; Seo, Y.W. Wheat (Triticum aestivum. L) Plant U-box E3 ligases TaPUB2 and TaPUB3 enhance ABA response and salt stress resistance in Arabidopsis. FEBS Lett. 2022, 596, 3037–3050. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, W.; Li, Q.; Zhang, G.; Zhao, X.; Li, G.; Li, Y.; Wang, Y.; Wang, W. The wheat E3 ligase TaPUB26 is a negative regulator in response to salt stress in transgenic Brachypodium distachyon. Plant Sci. 2020, 294, 110441. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Jiang, N.; Wang, M.; Du, Z.; Chen, J.; Huang, Y.; Li, M.; Jin, Y.; Li, J.; Wan, J.; et al. OsHIPP17 is involved in regulating the tolerance of rice to copper stress. Front. Plant Sci. 2023, 14, 1183445. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Peng, X.; Wang, X.; Wang, C. The heavy metal-associated isoprenylated plant protein (HIPP) gene family plays a crucial role in cadmium resistance and accumulation in the tea plant (Camellia sinensis L.). Ecotoxicol. Environ. Saf. 2023, 260, 115077. [Google Scholar] [CrossRef] [PubMed]
- Parasyri, A.; Barth, O.; Zschiesche, W.; Humbeck, K. The Barley Heavy Metal Associated Isoprenylated Plant Protein HvFP1 Is Involved in a Crosstalk between the Leaf Development and Abscisic Acid-Related Drought Stress Responses. Plants 2022, 11, 2851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Feng, H.; Feng, C.; Xu, H.; Huang, X.; Wang, Q.; Duan, X.; Wang, X.; Wei, G.; Huang, L.; et al. Isolation and characterisation of cDNA encoding a wheat heavy metal-associated isoprenylated protein involved in stress responses. Plant Biol. 2015, 17, 1176–1186. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, S.; Jing, R.; Abbas, M.; Hu, Z.; Kalsoom, R.; Hussain, S.S.; Du, L.; Lin, J.; Zhang, X. Transcriptional Profiling of BpWRKY49 Reveals Its Role as a Master Regulator in Stress Signaling Pathways in Birch (Betula platyphylla). Forests 2024, 15, 605. https://doi.org/10.3390/f15040605
Abbas S, Jing R, Abbas M, Hu Z, Kalsoom R, Hussain SS, Du L, Lin J, Zhang X. Transcriptional Profiling of BpWRKY49 Reveals Its Role as a Master Regulator in Stress Signaling Pathways in Birch (Betula platyphylla). Forests. 2024; 15(4):605. https://doi.org/10.3390/f15040605
Chicago/Turabian StyleAbbas, Sammar, Ruotong Jing, Manzar Abbas, Zijian Hu, Rabia Kalsoom, Syed Sarfaraz Hussain, Liang Du, Jinxing Lin, and Xi Zhang. 2024. "Transcriptional Profiling of BpWRKY49 Reveals Its Role as a Master Regulator in Stress Signaling Pathways in Birch (Betula platyphylla)" Forests 15, no. 4: 605. https://doi.org/10.3390/f15040605
APA StyleAbbas, S., Jing, R., Abbas, M., Hu, Z., Kalsoom, R., Hussain, S. S., Du, L., Lin, J., & Zhang, X. (2024). Transcriptional Profiling of BpWRKY49 Reveals Its Role as a Master Regulator in Stress Signaling Pathways in Birch (Betula platyphylla). Forests, 15(4), 605. https://doi.org/10.3390/f15040605







