Abstract
Anthracnose of Camellia plants is caused by the Colletotrichum species. The fungal pathogens mainly infect the leaves of plants and lead to serious economic losses. However, knowledge of Camellia phyllosphere microbial community after Colletotrichum infection has not been explored which limited our understanding of the relationship between the Camellia anthracnose outbreak and interacting microorganisms. In this study, three economically and ecologically important Camellia species with anthracnose symptoms were collected and subjected to bacterial and fungal composition analysis, diversity, co-occurrence characteristics, isolation of key strains, and tie-back pathogenicity test. The results indicated that Sphingomonas and Methylobacterium were the dominant bacterial genera over the three Camellia species and Pallidocercospora, Colletotrichum, and Pichia were the dominant fungal genera. The co-occurrence analysis showed that Methylobacterium, Sphingomonas, Massilia, and Allorhizobium were the key bacterial taxa and Colletotrichum, Pallidocercospora, Pichia, Septophoma, and Septoria were the key fungal taxa over the three infected plants. The hub taxa, including the species significantly associated with the Colletotrichum abundance, were mostly beneficial bacteria over the three Camellia species. Further co-culture and tie-back pathogenicity tests verified that the hub taxa associated with pathogenic Colletotrichum in the microbial networks may play promoting/inhibiting roles on Colletotrichum infection. The results highlight the importance of phytopathological conditions for the interactions between microbial members of foliar fungal and bacterial communities.
1. Introduction
A range of biotic and abiotic factors shapes plant microbial communities. Among them, the pathogen invasion plays the most influential role [1,2]. Soil-borne fungal pathogens such as Fusarium oxysporum and Rhizoctonia solani can impact plant microbiome assembly in the rhizosphere [3]. The wheat gathers family members of Chitinophagaceae and Flavobacteriaceae in the root endosphere that is invaded by R. solani [4]. Furthermore, plant diseases are often caused by co-infection of several pathogens [5]. Espinoza et al. disclosed that twig dieback disease is caused by one or more pathogens, and different pathogens can cause the same symptoms [6]. Other study showed that during Ca. sinensis anthracnose, Pseudopestalotiopsis increased significantly in the middle- and late-stages of anthracnose, which could lead to the expansion of disease caused by pathogenic Colletotrichum species [7]. Even at different stages of the disease outbreak, microbial community differed in its structural characteristics [8]. Therefore, the pathogen–host model study combined with the complexity of microbial community on different plant niches is conducive to the understanding of plant diseases.
Different niches of plant have complex and diverse microorganisms. They shape the community composition and structures of plant habitats through multiple interactions to influence the host plants’ physiological metabolism and health status [9]. The phyllosphere is defined as the collective microenvironment of the plant aerial parts such as the fruits, flowers and leaves. Specifically, leaves have been extensively studied considering their significance in plant growth and adaptability. Microorganisms with an average density of 106–107 cells/cm2 inhabit plant leave surfaces [10]. The foliar microbes can form colonization groups of specific phylogenetic types through long-term plant–microbial and microbial–microbial interactions under a variety of factors [11]. A recent study indicated that the chemical signaling interactions between a phyllosphere enriched with fungi and pathogenic bacteria significantly modulate rice disease resistance, underscoring the critical role of foliar microorganism interactions in effective disease management [12].
The plant Camellia species represented by Ca. sinensis, Ca oleifera, and Ca. japonica have important economic and ecological values especially in East Asia [13]. The fungal Colletotrichum genus consists of a large number of plant pathogens, and they can parasitize numerous economic crops worldwide, such as pear, citrus, and coffee [14,15,16]. In recent years, the anthracnose caused by Colletotrichum species has become the most destructive disease in Camellia plantations. [17,18]. Up till now, most studies of Camellia anthracnose have been focusing on pathogen characterization, pathogenicity, and disease mechanisms [14,15,16,17,18,19]. There are limited studies on the microbial community related to the plant leaf anthracnose disease. The purpose of this study was to gain more understanding on foliar microbial community structure upon Camellia anthracnose infection caused by Colletotrichum species using 16S rRNA and ITS amplicon sequencing. We further isolated the microbial species in the diseased lesions and performed co-culture and tie-back pathogenicity testing together with the pathogen to explore the role of the foliar microbiome members during infection, which could provide theoretical support for new strategies to control Camellia anthracnose.
2. Methods
2.1. Sample Collection
Plant leaves of Ca. oleifera (30°3′32″ N, 119°57′16″ E), Ca. sinensis (30°15′37″ N, 119°43°45″ E), and Ca. japonica (30°15′42″ N, 119°43′16″ E) were collected from plantations in Zhejiang Province, China. The plantations were in a natural state with no chemical spraying. Nine leaves with typical anthracnose symptoms of three Camellia plants from each area were randomly collected and mixed. The samples were placed in sterile bags at 4 °C in ice boxes and transported back to the lab. Under aseptic conditions, the diseased parts of leaves (0.8 mm diam) were drilled and transferred to 2 mL sterile tubes. They were stored at −80 °C for subsequent experiments.
2.2. Microbial DNA Extraction, PCR Amplification, and Illumina Sequencing
The microbial DNA extraction kit (UltraCleanR Microbial DNA Kit, MoBio, New York, NY, USA) was utilized for DNA extraction from plant anthracnose lesions. The DNA concentration and purity were assessed using 1% agarose gel electrophoresis and ultraviolet spectrophotometry. The regions of the bacterial 16S rRNA V4–V5 and fungal ITS were amplified. The primers F:5′-AACMGGATTAGATACCCKG-3′ and R:5′-ACGTCATCCCCACCTTCC-3′ were used for the bacterial community, while the universal primers F:5′-GGAAGTAAAAGTCGTAACAAGG-3′ and R:5′-GCTGCGTTCTTCATCGATGC-3 were used for the fungal community. A sequencing library was constructed by incorporating index codes into the product using the NEB Next® Ultra™ DNA library prep kit for Illumina (NEB, Ipswich, MA, USA). The library was subsequently sequenced on an Illumina HiSeq platform utilizing 250 bp paired-end reads via a cBot cluster generation system (Illumina, Westlake Village, CA, USA), following the manufacturer’s recommendations.
2.3. Bioinformatics Analysis
In order to generate amplicon sequence variants (ASVs), DADA2 [20] was utilized to eliminate low-quality reads, noise, and chimeras. The representative ASVs were annotated using the databases of SILVA for bacteria [21] and UNITE for fungi [22]. Taxonomic annotation of the detected communities was conducted at various levels of phylum, class, order, family, genus, and species. Microbial diversity analysis was conducted to calculate the Chao1, Shannon, and Simpson indices, and rarefaction curves were obtained using the Mothur software (version 1.30.1) [23]. The vegan R package was used to perform principal coordinates analysis (PCoA) based on the Bray–Curtis distances (version 2.5.3) [24]. PCoA was plotted with the ggplot2 R package (version 2.2.1) [25]. Coupled with effect size measurements (LEfSe) analysis, linear discriminant analysis (LDA) was used to identify marker species with significant differences between samples at all classification levels simultaneously with the LEfSe software (version 1.0) [26]. In addition, ANOSIM analysis of similarities test was carried out to statistically compare the sample mean distance of different groups based on the Bray–Curtis distance matrix. The correlation between the top 30 relative abundances of fungal and bacterial ASVs was analyzed using the following online tool: https://hiplot.com.cn/basic/cor-heatmap (accessed on 9 July 2022).
Co-occurrence patterns were reconstructed for network analysis. This was achieved by calculating multiple abundance correlations at the genus level. The R package “Psych” was utilized to analyze OTUs present in more than 2% of the samples in 3 groups and to calculate multiple abundance correlation matrices. Co-occurrence was considered robust if the Spearman’s correlation coefficient (ρ) was between >0.05 and p < 0.6. The interactive platform Gephi (v.0.9.2) was used to visualize the network [27]. The network node represents an individual OTU, while the edge signifies the correlation between pairs of nodes within the microbiome network. The topological characteristics of bacterial and fungal networks consist of average path length, clustering coefficient, and connectivity together with co-concurrent (positive) and mutually exclusive (negative) correlation numbers, modularity, and network diameter. The role of individual nodes were determined by considering their topological features of degree and closeness centrality.
2.4. Isolation and Culture of Microorganisms from Anthracnose Lesions
The remaining diseased leaves were used to isolate the fungal and bacterial taxa in the anthracnose lesions of the three plants. The diseased tissues were cut into 2–5 mm × 2–5 mm fragments at the junction of diseased and healthy tissue after being treated with 75% ethanol for 45 s and washed three times in sterile water. The tissues were transferred to a 2% malt extract agar (MEA) and Luria–Bertani (LB) medium at 25 °C. After 3–4 days of incubation, the fungi and bacteria growing from the edge of the diseased tissues were picked and placed on new media. The obtained pure cultures were stored in 25% glycerol at −80 °C for future use.
The Ezup Column Fungi Genomic DNA Purification Kit was employed for fungal genomic DNA extraction. The primers for ITS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) were used for amplification. The PCR reaction was carried out with an initial denaturation at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 52 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. Bacterial genomic DNA were extracted with the TIANamp Bacteria DNA Kit with primers for 16S_8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 16S_926R (5′-CCGTCAATTCCTTTAAGTTT-3′). The PCR reaction conditions were 5 min at 95 °C, followed by 40 cycles of 40 s at 94 °C, 50 s at 55 °C, 1 min at 72 °C, and a 10 min final extension at 72 °C.
The characterization of the Colletotrichum isolates were achieved based on comparison of multi-locus sequences and morphological features. PCRs were performed with primers for ITS4/ITS5, ACT-512F/ACT-783R, T1/Bt2b, and GDF1/GDR1 [28,29,30,31] (Table 1) to obtain partial regions of ITS, ACT, TUB2, and GAPDH gene sequences following Weir et al. [32]. The concatenated datasets of these four gene sequences were analyzed using the PhyloSuite v1.2.2 software via the maximum likelihood (ML) method.

Table 1.
Primers used in the study.
2.5. Co-Culture and Tie-Back Experiment
Based on the results of co-occurrence analysis, the bacterial and fungal taxa significantly associated with the relative abundance of Colletotrichum in the leaf lesions were identified. Isolates with positive and negative correlations obtained in the previous step were selected for co-culture with C. fructicola and C. camelliae. The culture media containing C. fructicola (LAJ-06) and C. camelliae (ZSJ-013) isolates were punched with a 5 mm sterile cork-borer and transported to the center of 9 cm MEA plates, respectively. The related bacterial and fungal cakes were inoculated 2 cm away from the Colletotrichum strains. The plates were incubated in the dark at 25 °C for a week to observe the growth of the strains. For each group, the experiment was repeated three times.
Healthy mature leaves of three Camellia plants were collected for the combined tie-back test. After being immersed in sterile water and dried on a filter paper under sterile conditions, they were placed in Petri dishes covered with wet filter papers and punctured with a sterilized needle (0.5 mm-diameter insect pin). The associated isolates were cultured in darkness at 25 °C on MEA plates for one week. The isolate cakes were obtained from the same diameter of cultures with a 6 mm sterile puncher. The Colletotrichum cake was inoculated on both the left and right sides of the leaves. The inoculated leaves were placed in a growth chamber with 85% relative humidity at 26 °C and subjected to a 12/12 h light/dark photoperiod. The lesions were measured on the 7th day following inoculation. The Colletotrichum cake on the left side of the leaves was removed and replaced with the cakes of related isolates with the same diameter. Each combination was set with eight replicates. After continuously moisturizing the culture for 7 days, the change value of the lesion diameters was calculated. Each experiment was performed with eight replicates. All statistical analyses were conducted utilizing GraphPad Prism (version 8.0.2), employing analysis of variance (ANOVA). Upon identifying significant treatment effects (p < 0.05), multiple comparisons were further examined using the least significant difference (LSD) test.
3. Results
3.1. Phyllosphere Fungal and Bacterial Community Diversity of Three Camellia Plants Under Anthracnose
Camellia leaves with anthracnose symptoms were collected from orchards of Ca. sinensis, Ca. oleifera, and Ca. japonica in January 2019 in the Zhejiang province, China to explore variation in the phyllosphere microbiota. Barcode-labeled sequences were used for samples identification. A total of 58,210, 74,751, and 63,118 high-quality bacterial 16S rRNA reads and 112,409, 91,745, and 108,157 fungal ITS reads were obtained from three samples with an average of 65,360 reads per sample. These reads were categorized into 7230 bacterial operational taxonomic units (OTUs) and 930 fungal OTUs with a sequence similarity level of 97%.
For the bacterial community, the Shannon and Chao1 indices of Ca. sinensis samples were higher than those of Ca. oleifera and Ca. japonica, and Ca. sinensis > Ca. japonica > Ca. oleifera. The fungal Shannon index followed the order Ca. sinensis > Ca. japonica > Ca. oleifera, which was similar to the bacterial community. The Chao1 index followed the order Ca. oleifera > Ca. sinensis > Ca. japonica. Generally, the Shannon index of the bacterial and fungal communities in Ca. sinensis exhibited higher values compared to those observed in the other two samples. It indicated that the microbial diversity and richness are highest in Ca. sinensis anthracnose lesions, while richness of the bacterial species is highest in Ca. oleifera (Figure 1a,c). In contrast, no significant differences were found between the three samples.
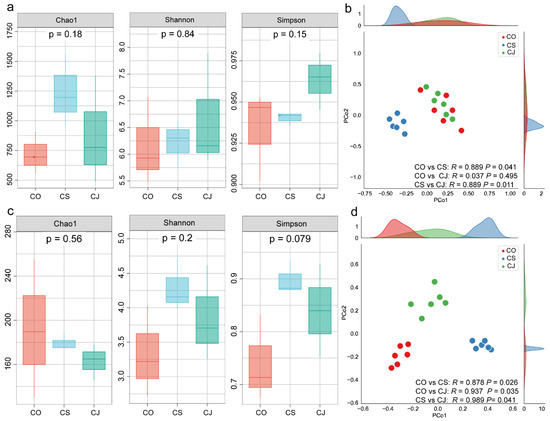
Figure 1.
Community structure and microbial diversity of anthracnose spot microbiotas of Ca. sinensis (CS), Ca. oleifera (CO), and Ca. japonica (CJ) samples. (a,b) Bacterial community diversity and PCoA scatter plot, respectively. (c,d) Fungal community. The Chao1, Shannon, and Simpson indices were included and PCoA was performed to characterize beta-diversity.
The beta-diversity of the microbial community was evaluated via PCoA analysis using the Bray–Curtis dissimilarity. For the bacterial community, the community structure from the Ca. oleifera and Ca. japonica samples clustered together, which was completely separated from that of Ca. sinensis. For the fungal community, the community structure in three Camellia anthracnose lesions were separated from each other and had significant differences (Figure 1b,d).
3.2. Phyllosphere Bacterial and Fungal Community Composition of Three Camellia Plants Under Anthracnose
The dominant bacterial taxa at the phylum level in all samples from the three plants were Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes. At the genus level, Sphingomonas (21.93%), Methylobacterium (9.97%), Massilia (5.54%), and Aquabacterium (4.08%) were the most abundant taxa in Ca. oleifera; Methylobacterium (34.21%), Rhizobium (15.21%), Curtobacterium (12.61%), and Sphingomonas (6.75%) were the most abundant taxa in Ca. sinensis, and Methylobacterium (22.45%), Sphingomonas (11.6%), Aquabacterium (6.9%), and Kineococcus (4.96%) were the most abundant in Ca. japonica (Figure 2a).
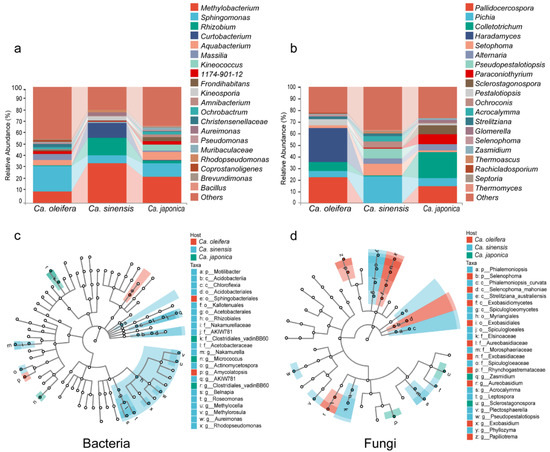
Figure 2.
The relative abundance of the top 20 bacterial (a) and fungal (b) genera. Marker species with significant differences between the Camellia samples were identified using LDA coupled with LEfSe for bacterial (c) and fungal (d) taxa.
The most abundant fungal phyla were Ascomycota and Basidiomycota. The fungal taxa in Ca. oleifera were mainly Haradamyces (29.62%), Pallidocercospora (22.15%), Colletotrichum (8.04%), and Pestalotiopsis (5.36%); Pichia (23.06%), Setophoma (9.82%), Pseudopestalotiopsis (8.59%), and Colletotrichum 0.96% in Ca. sinensis; Colletotrichum (22.85%), Pallidocercospora (14.56%), Paraconiothyrium (8.44%), and Sclerostagonospora (8.35%) in Ca. japonica (Figure 2b). At the phylum level, the three plant samples exhibited a similar core microbial composition and abundance with specific bacterial and fungal phyla. However, the composition and the abundance varied at the genus level, especially in the fungal community.
3.3. Differential Abundance of the Phyllosphere Bacterial and Fungal Taxa Among Three Camellia Plants
Linear discriminant analysis of the effect size of core ASVs was applied to investigate the differential abundance of bacterial and fungal taxa across plant samples. We found that different plant species had specific bacterial and fungal taxonomic community under infection. According to the analysis, 24 bacterial and 26 fungal OTUs were identified and thought to be responsible for the anthracnose of the three Camellia plants. For the bacterial community, o_Sphingobacteriales and g_Amycolatopsis were significantly enriched in Ca. oleifera; P_Motilibacter, g_Nakamurella, g_Actinomycetospora, g_Belnapia, g_Roseomonas, g_Methylocella, g_Aureimonas, and g_Rhdopseudomonas were enriched in Ca. sinensis, and g_Clostridiales vadinBB6 and g_Micrococcus were enriched in the Ca. japonica samples (Figure 2c).
For the fungal community, c_Selenophoma mahoniae, g_Aureobasidium, g_Exobasidium, and g_Papiliotrema were significantly enriched in Ca. oleifera; c_Phialemoniopsis curvata, c_Strelitziana australiensis, c_Spiculogloeomycetes, f_Elsinoaceae, g_Acrocalymma, g_Letospora, g_Plectosphaerella, and g_Pseudope were enriched in Ca. sinensis, and g_Zasmidium and g_Scelrostagonspora were enriched in the Ca. japonica samples (Figure 2d). The bacteria in the Actinomycetes phylum and the fungi in the Dothideomycetes class were enriched among the three plants under pathogen infection.
3.4. Phyllosphere Key Microbial Taxa and Their Associations with Three Camellia Plants Under Anthracnose
To investigate the key microbial taxa and potential interactions among the foliar microbiota of three Camellia plants upon Colletotrichum infection, the co-occurrence networks were built using microbial profiling data to characterize the comprehensive connectivity patterns (positive, neutral, or negative). The network analysis showed that the bacterial correlation networks of the Camellia samples included 62, 53, and 66 nodes, respectively. The predominant hubs exhibiting high degree and closeness centrality values within the bacterial co-occurrence networks were mainly Methylobacterium, Sphingomonas, Massilia, Allorhizobium, and Burkholderiaceae, which were shared by the three plant samples. The nodes of the network are divided into three modules. All three samples showed a higher proportion of positive edges. The fungal co-occurrence networks included 41, 52, and 53 nodes, respectively. Colletotrichum, Pallidocercospora, Pichia, Septophoma and Septoria were the top hub taxa shared by the three plant samples. The network nodes were also divided into three modules and showed more negative edges. The complexity of bacterial networks in the three samples generally exceeded that of fungal networks. Similar patterns were apparent in the three plant samples (Figure 3, Table 2).
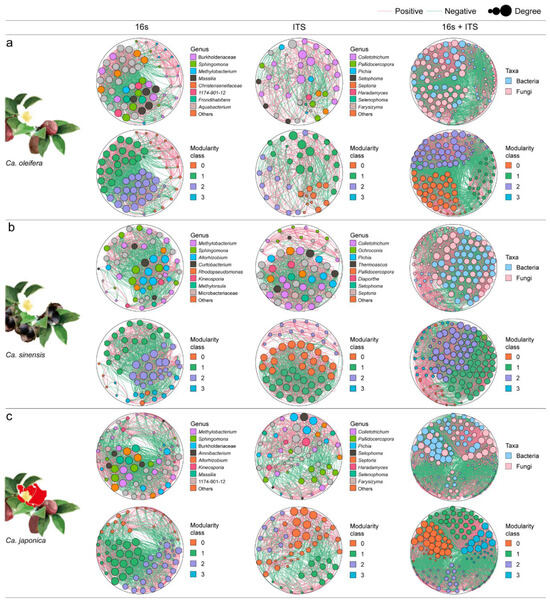
Figure 3.
Co-occurrence analysis of microbial networks. (a) bacteria–bacteria (16s), fungi–fungi (ITS) and bacteria–fungi (16s + ITS) network analysis in infected Ca. oleifera, (b) Ca. sinensis, and (c) Ca. japonica samples. Node colors indicate genus–level taxonomy. Green and pink links refer to negative and positive correlations. The size of the node is assigned in terms of node degree. If the Spearman’s correlation coefficient is between ρ > 0.05 and p < 0.6, the co-occurrence is considered robust.

Table 2.
Characteristics of microbial co-occurrence network.
The co-occurrence and heatmap analysis of the microbial taxa associated with the relative abundance of pathogens (Colletotrichum) in leaf lesions of the three samples were also conducted. The fungi with significantly negative correlations were mainly Diaporthe, Setophoma, and Neopestalotiopsis, and the fungi with positive correlations were Sclerostagonospora and Paraconintohyrium. For bacteria, Sphingomonas showed a positive correlation with the relative abundance of Colletotrichum, while Massilia and Bacillus exhibited significantly negative correlations (Figure 4 and Figure S1).
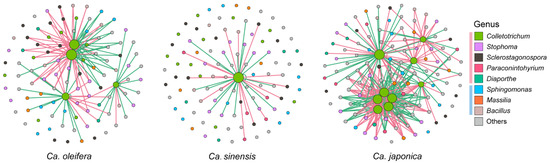
Figure 4.
Co-occurrence analysis of hub taxa with significance correlated with the abundance of Colletotrichum in infected Ca. oleifera, Ca. sinensis, and Ca. japonica samples. Node colors indicate genus–level taxonomy. Green and pink links refer to negative and positive correlations. The color boxes pink and blue refer to fungi and bacteria.
3.5. Microorganisms Associated with the Three Camellia Phyllosphere Anthracnose Lesion Microbiome
We selectively cultivated bacteria and fungi from the foliar anthracnose lesions of three Camellia plants (Figure S2). Among them, 40, 39, and 24 Colletotrichum isolates were obtained from the Ca. oleifera, Ca. sinensis, and Ca. japonica, respectively. A multi-locus phylogenetic analysis showed that these Colletotrichum isolates primarily clustered into nine known species within the C. gloeosporioides and C. acutatum complexes. C. fructicola was the dominant taxon in the three Camellia anthracnose lesions, while C. siamense, C. fioriniae, C. wuxiense, and C. fructicola were present in all three hosts, with the remaining five Colletotrichum species only isolated from one or two plant hosts (Figure 5a). The pathogenicity test of all Colletotrichum isolates showed that the diseased lesion lengths at 14 dpi varied among the isolates (Figure 5a). Furthermore, the difference analysis of representative strains revealed that the pathogenicity of Colletotrichum isolates varied between the three Camellia plants (p < 0.05) (Figure 5b).
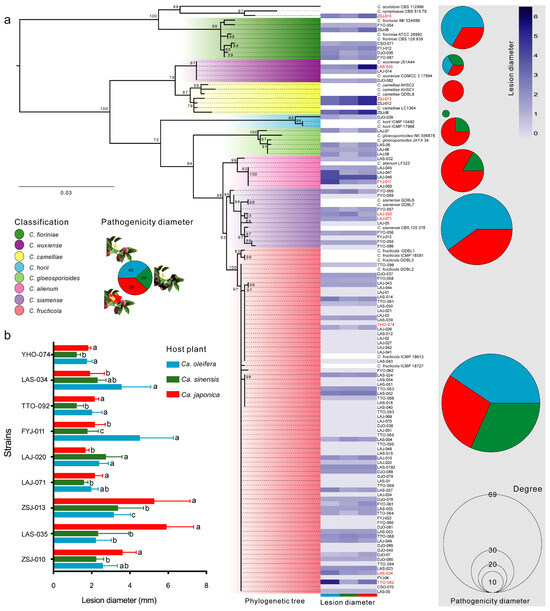
Figure 5.
Characterization and pathogenicity test of Colletotrichum pathogens isolated from infected Camellia samples. (a) Phylogenetic tree of 103 Colletotrichum isolates using the maximum livelihood method. Phylogenetic tree using combined ACT, TUB2, GAPDH and ITS sequences alignments, Monilochaetes infuscans (CBS 869.96) was selected as the outgroup. Bootstrap values ≥ 50%, in 1000 replication. Scale bar indicates an expected change of 0.05 per site. (b) The pathogenicity of different Colletotrichum species to three Camellia plants. Letters over the error bars indicate a significant difference at the p = 0.05 level.
3.6. Members of the Phyllosphere Microbiome Could Affect Colletotrichum Infection
To further verify the interactions between the hub taxa and pathogenic Colletotrichum, species of Sclerostagonospora sp., Paraconiothyrium sp., Neopestalotiopsis sp., Diaporthe sp., Setophoma sp., and Bacillus sp., which showed significant correlations with the abundance of Colletotrichum sp., were co-cultured with Colletotrichum fructicola and C. camelliae on the MEA plates for 7 days. The results are shown in Figure 6. The fungal strain Sclerostagonospora sp. had no obvious boundary when co-cultured, and Paraconiothyrium sp. could grow on the upper part of Colletotrichum hyphae. Other fungal strains such as Diaporthe sp., Setophoma sp., and Neopestalotiopsis sp. with significantly negative correlation had obvious boundary with the Colletotrichum sp. at the seventh day of growth. The Bacillus sp. with a significantly negative correlation showed an antagonistic effect against two Colletotrichum strains (Figure 6a,b).
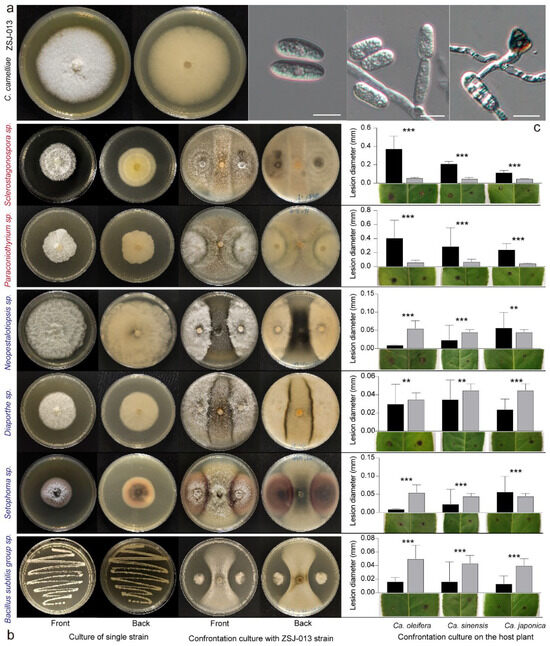
Figure 6.
Co-culture and combination tie-back of related fungal and bacterial isolates. (a) The morphological characteristics including views of 6 d-old MEA culture, conidia, conidiophores, and appressoria of C. camelliae ZSJ-013. (b) Co-culture results of Sclerostagonospora sp., Paraconiothyrium sp., Neopestalotiopsis sp., Diaporthe sp., Setophoma sp., Bacillus sp., and pathogenic C. camelliae ZSJ-013. (c) The significant difference analysis of combined tie-back pathogenicity. ** p < 0.01; *** p < 0.001.
The combined tie-back tests were conducted on three Camellia leaves. The results indicated a significantly higher diameter increment value compared to the CK group (inoculated with only Colletotrichum isolates) when Paraconiothyrium sp. (YNS-07) or Sclerostagonospora sp. (LAJ-029) were combined with C. camelliae (ZSJ-013) inoculation (Figure 6c). The bacterial strain Bacillus subtilis group sp. (LAJ-B33) and the fungal strains Neopestalotiopsis sp. (LAJ-84), Diaporthe sp. (LAJ-85), and Setophoma sp. (LAF-1) negatively correlated with one another; the average diameter of the lesions showed a significantly lower increment compared to the CK group (Figure 6c). The result indicated that the microbial groups significantly correlated with the relative abundance of Colletotrichum pathogen and may have promoting or inhibiting effects on the infection.
4. Discussion
Plant infection caused by pathogenic microbes is always related to interactions between microbe–microbe and microbe–plant within specific plant niches [33]. Recently, the composition of microbial communities in the leaves and roots of various plant species has been clarified. The knowledge regarding what and how interactions between foliar microbiota members with this plant disease has, however, been less studied [34,35]. Here, we investigated the diversity, composition, and co-occurrence patterns of the foliar microbiome between three Camellia plants infected by the anthracnose pathogen. Furthermore, we emphasized that the members of phyllosphere microbiome could assist or inhibit the infection of pathogenic fungi. This work advanced our understanding on hub taxa’s role in phyllosphere microbial networks upon pathogen infection. This is of great significance on Camellia anthracnose prevention and control.
Microbial community analysis results indicated an insignificant difference in composition at both the phylum and genus levels. However, the relative abundance of major groups varies among the three Camellia samples (Figure 2). The genera Sphingomonas and Methylobacterium were dominant in three samples (relative abundance > 5%), and they were also the hub taxa with high centrality in the bacterial networks (Figure 2a and Figure 4). Many Methylobacterium species are the dominant endophytic bacteria residing in the leaves of Triticum, Medicago, and Arabidopsis, etc. [36,37,38]. They have the function of fixing nitrogen, dissolving phosphorus and iron, producing plant hormones, and antagonizing plant pathogens. This makes it possible for them to be widely utilized as plant growth-promoting bacteria (PGPB), improving the production of wheat, corn, tomato, and other food crops [39]. Nevertheless, another study showed that the phyllosphere microbial community infected by the melanose pathogen can recruit Pantoea asv90 and Methylobacterium asv41 to antagonize Diaporthe citri by inhibiting spore germination and mycelial growth [35]. For species of Sphingomonas, they possess the capability to promote aromatic compound degradation and strengthen plant resistance against pests and diseases [40]. The genera Massilia and Bacillus exhibited a significant negative correlation with the relative abundance of Colletotrichum in foliar lesions of the three plants (Figure 5). The Massilia belongs to the phylum Proteobacteria and is widely found in water, soil, rhizosphere, leaf, and air. Studies showed that the Massilia species participate in the carbon and nitrogen cycle, secrete plant auxin, and improve plant resistance [41]. The Bacillus belongs to the Bacilli class of the phylum Firmicutes, of which the species Bacillus subtilis, B. amloliquefaciens, and B. velezensis, have antagonistic effects against pathogenic Colletotrichum species [42,43,44]. These findings disclosed that the abundant taxa and hub taxa, including the species significantly associated with the abundance of the pathogen microbes, were mostly beneficial bacteria, which can influence the progression of plant diseases through various mechanisms, including competitive exclusion, antibiotic secretion, and the induction of host immune responses [45].
The fungal community analysis result showed that Pallidocercospora, Colletotrichum and Pichia were the dominant taxa of the three Camellia plants (relative abundance > 5%) (Figure 2b). They were also the hub taxa in the fungal network, in addition to Pichia, Septophoma, and Septoria (Figure 4). Among them, both Setophoma and Septoria belong to the class Dothideomycetes of Ascomycetes. Most members of these two genera are pathogens that cause root and leaf diseases on various plants. For example, Setophoma terrestris can cause root powder rot in a range of plant hosts including Cucurbita, Benincasa, Brassica, and Allium [46,47,48,49]. The Septoria species can cause leaf spot disease on Populus and Solanum [50,51]. Furthermore, the fungal taxa that exhibited a significant correlation with the abundance of Colletotrichum in the leaf lesion microbial community of the three infected Camellia plants were determined. They were mainly Sclerostagonospora and Paraconiothyrium, which were positively correlated with the abundance of Colletotrichum and both affiliated with the class Dothideomycetes of Ascomycetes (Figure 5). The Paraconiothyrium species is characterized by its diverse host habitats, wide distribution, and potential pathogenicity in plants [52]. Paraconiothyrium sporulosum causes hawthorn branch canker [53] while P. variabile results in leaf spot disease on apple trees, pear trees, and other hosts [54]. The genus with a significantly negative correlation is Diaporthe, and its species are known to cause rot diseases on plants of Camellia, Citrus, Vitis, and Pyrus [55,56,57,58]. The fungal taxa that significantly negatively correlated with the abundance of Colletotrichum were mainly potential plant pathogens. They may have negative competition with pathogenic Colletotrichum in overlapping ecological niche.
We further cultured the bacteria and fungi from the foliar anthracnose lesions of the three Camellia plants and selected the above microbial isolates with positive or negative correlation with the Colletotrichum abundance for co-culture and combined tie-back test (Figure 6). The results demonstrated that positively/negatively correlated taxa had non-competitive and competitive/antagonistic growth modes during co-culture and promoted/inhibited the expansion of lesion diameter in the combined tie-back test, respectively. We verified that the relative abundance of microbial taxa with positive or negative correlations influenced pathogenicity by promoting or inhibiting the growth of pathogens. Considering the interesting findings, which include exploring binary interactions between hub taxa members and pathogenic Colletotrichum in the phyllosphere microbiome, this in-depth study on the relationship and mechanism of the higher-order interactions in mixed colonies and comparison with the uninfected leaves deserves further investigation, which will provide a more comprehensive understanding of the microbial community structure caused by Colletotrichum infection.
5. Conclusions
Camellia plants have significant ecological and economic values globally, especially in Southeast Asia. However, anthracnose caused by Colletotrichum species has become the major challenge. The highlights of the study were to explore the Camellia anthracnose microbial communities upon infection and their pathogenicity. Three important Camellia species with anthracnose symptoms were studied and the results impose the importance of phytopathological conditions for the interactions among microbial members of phyllosphere fungal and bacterial communities. The findings provide new understanding on the microbial community structure under pathogen infection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f15122080/s1, Figure S1: Correlation heatmap analysis of fungal and bacterial communities; Figure S2: Colony morphology of isolates obtained from the anthracnose lesions.
Author Contributions
X.P. performed the laboratory experiments, analyzed the data and prepared the manuscript draft. H.W. participated in conceptual design and manuscript preparation. X.Z. coordinated the study, performed conceptual design, and manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by “the Launching Funds for Talents of Zhejiang A & F University” Program (2022LFR0005), China.
Data Availability Statement
Accession codes: 16S rRNA and ITS sequences have been deposited into the Sequence Read Archive (SRA) database with the accession code PRJNA967982.
Acknowledgments
We thank Shouke Zhang for the technical support provided in the laboratory.
Conflicts of Interest
The authors declare no competing interests.
References
- de Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.T.; Whiteman, N.K. Insect herbivory reshapes a native leaf microbiome. Nat. Ecol. Evol. 2020, 4, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Zhang, X.Y.; Zhang, Z.C.; Chen, Y.; Tian, Q.; Zeng, D.D.; Xu, M.; Wang, Y.; Dong, S.M.; Ma, Z.H.; et al. Fusarium–produced vitamin B6 promotes the evasion of soybean resistance by Phytophthora sojae. J. Integr. Plant Biol. 2023, 65, 2204–2217. [Google Scholar] [CrossRef]
- Yin, C.; Casa Vargas, J.M.; Schlatter, D.C.; Hagerty, C.H.; Hulbert, S.H.; Paulitz, T.C. Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome 2021, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Ameye, M.; Landschoot, S.; De Zutter, N.; De Saeger, S.; De Boevre, M.; Audenaert, K. At the scene of the crime: New insights into the role of weakly pathogenic members of the fusarium head blight disease complex. Mol. Plant Pathol. 2020, 21, 1559–1572. [Google Scholar] [CrossRef]
- Espinoza, J.G.; Briceo, E.X.; Keith, L.M.; Latorre, B.A. Canker and twig dieback of blueberry caused by Pestalotiopsis spp. and a Truncatella sp. in Chile. Plant Dis. 2008, 92, 1375–1475. [Google Scholar] [CrossRef]
- Shi, Y.L. Studies on Molecular Mechanism of Tea Plant Against Anthracnose Infection and Assessment of Freezing Tolerance of Tea Cultivars. Ph.D. Dissertation, Zhejiang University, Hangzhou, China, 2020. [Google Scholar]
- Zhang, Z.; Luo, L.; Tan, X.; Kong, X.; Yang, J.; Wang, D.; Zhang, D.; Jin, D.; Liu, Y. Pumpkin powdery mildew disease severity influences the fungal diversity of the phyllosphere. Peerj 2018, 6, e4559. [Google Scholar] [CrossRef]
- Chaudhry, V.; Runge, P.; Sengupta, P.; Doehlemann, G.; Parker, J.E.; Kemen, E. Shaping the leaf microbiota: Plant–microbe–microbe interactions. J. Exp. Bot. 2021, 72, 36–56. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Gong, T.Y.; Xin, X.F. Phyllosphere microbiota: Community dynamics and its interaction with plant hosts. J. Integr. Plant Biol. 2021, 63, 297–304. [Google Scholar] [CrossRef]
- Fan, X.; Matsumoto, H.; Xu, H.; Fang, H.; Pan, Q.; Lv, T.; Zhan, C.; Feng, X.; Liu, X.; Su, D.; et al. Aspergillus cvjetkovicii protects against phytopathogens through interspecies chemical signalling in the phyllosphere. Nat. Microbiol. 2024, 9, 2862–2876. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.M.; Sousa, C. A Review on the Biological Activity of Camellia Species. Molecules 2021, 26, 2178. [Google Scholar] [CrossRef]
- Fu, M.; Crous, P.W.; Bai, Q.; Zhang, P.F.; Xiang, J.; Guo, Y.S.; Zhao, F.F.; Yang, M.M.; Hong, N.; Xu, W.X.; et al. Colletotrichum species associated with anthracnose of Pyrus spp. in China. Persoonia 2019, 42, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, V.; Groenewald, J.Z.; Polizzi, G.; Crous, P.W. High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia 2018, 39, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.R.; Xu, X.M.; Che, H.Y.; West, J.S.; Luo, D.Q. Characteristics and distribution of Colletotrichum species in coffee plantations in Hainan, China. Plant Pathol. 2019, 68, 1146–1156. [Google Scholar] [CrossRef]
- Peng, X.J.; Wang, Q.C.; Zhang, S.K.; Guo, K.; Zhou, X.D. Colletotrichum species associated with Camellia anthracnose in China. Mycosphere 2023, 14, 130–157. [Google Scholar] [CrossRef]
- Wang, Y.C.; Hao, X.Y.; Wang, L.; Xiao, B.; Wang, X.C.; Yang, Y.J. Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Sci. Rep. 2016, 6, 35287. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Cao, H.; Hao, X.; Zeng, J.; Yang, Y.; Wang, X. Transcriptome analysis of an anthracnose-resistant tea plant cultivar reveals genes associated with resistance to Colletotrichum camelliae. PLoS ONE 2017, 11, e0148535. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High–resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson–Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence–based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open–source, platform–independent, community–supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Chang, F.; He, S.; Dang, C. Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data. J. Vis. Exp. 2022, 11, 537–558. [Google Scholar] [CrossRef]
- Parente, E.; Cocolin, L.; De Filippis, F.; Zotta, T.; Ferrocino, I.; O’Sullivan, O.; Neviani, E.; De Angelis, M.; Cotter, P.D.; Ercolini, D. FoodMicrobionet: A database for the visualisation and exploration of food bacterial communities based on network analysis. Int. J. Food Microbiol. 2016, 219, 28–37. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, F.J.R.M.; White, T.; Lee, S.H.; Taylor, L.; Shawetaylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990, 38, 315–322. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNAITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Templeton, M.D.; Rikkerink, E.H.; Solon, S.L.; Crowhurst, R.N. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 1992, 122, 225. [Google Scholar] [CrossRef]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.K.M.; Wang, M.M.; Zhou, X.; Zhang, A.M.; Cai, L. Disease–induced changes in plant microbiome assembly and functional adaptation. Microbiome 2021, 9, 187. [Google Scholar] [CrossRef]
- Li, P.D.; Zhu, Z.R.; Zhang, Y.; Xu, J.; Wang, H.; Wang, Z.; Li, H. The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome 2022, 10, 56. [Google Scholar] [CrossRef]
- Aziz, M.Z.; Yaseen, M.; Abbas, T.; Naveed, M.; Mustafa, A.; Hamid, Y.; Saeed, Q.; Ming-Gang, X. Foliar application of micronutrients enhances crop stand, yield and the biofortification essential for human health of different wheat cultivars. J. Integr. Agric. 2019, 18, 1369–1378. [Google Scholar] [CrossRef]
- Sy, A.; Timmers, A.C.; Knief, C.; Vorholt, J.A. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 2005, 71, 7245–7252. [Google Scholar] [CrossRef]
- Knief, C.; Frances, L.; Vorholt, J.A. Competitiveness of Diverse Methylobacterium Strains in the Phyllosphere of Arabidopsis thaliana and Identification of Representative Models, Including M. extorquens PA1. Microb. Ecol. 2010, 60, 440–452. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, M.Y.; Khan, N.; Tan, L.L.; Yang, S. Potentials, Utilization, and Bioengineering of Plant Growth–Promoting Methylobacterium for Sustainable Agriculture. Sustainability 2021, 13, 3941. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Sedláček, I.; Holochová, P.; Busse, H.J.; Koublová, V.; Králová, S.; Švec, P.; Sobotka, R.; Staňková, E.; Pilný, J.; Šedo, O.; et al. Characterisation of Waterborne Psychrophilic Massilia Isolates with Violacein Production and Description of Massilia antarctica sp. nov. Microorganisms 2022, 10, 704. [Google Scholar] [CrossRef]
- Kanitkar, S.; Sawant, S.; Adsule, P.; Kulkarni, M.; Kadam, M.; Raut, V. Bio-Efficacy of Milastin–K (Bacillus subtilis KTSB 1015 1.5% A.S.) as a Potential Bio–Control Agent for Management of Bacterial Blight (Xanthomonas axonopodis) and Anthracnose (Colletotrichum gloeosporioides) Diseases in Pomegranate. Soc. Sci. Electron. Publ. 2020, 7, 2349–8889. [Google Scholar] [CrossRef]
- Wang, X.; Yuan, Z.; Shi, Y.; Cai, F.; Wang, Y. Bacillus amyloliquefaciens HG01 induces resistance in loquats against anthracnose rot caused by Colletotrichum acutatum. Postharvest Biol. Technol. 2020, 160, 111034. [Google Scholar] [CrossRef]
- Huang, L.; Li, Q.C.; Hou, Y.; Li, G.Q.; Ye, J.R. Bacillus velezensis strain HYEB5-6 as a potential biocontrol agent against anthracnose on Euonymus japonicus. Biocontrol. Sci. Technol. 2017, 27, 636–653. [Google Scholar] [CrossRef]
- Bashir, I.; War, A.F.; Rafiq, I.; Reshi, Z.A.; Rashid, I.; Shouche, Y.S. Phyllosphere microbiome: Diversity and functions. Microbiol. Res. 2022, 254, 126888. [Google Scholar] [CrossRef]
- López-López, A.M.; León-Félix, J.; Allende, R.; Lima, N.B.; García–Estrada, R.S. First Report of Setophoma terrestris Causing Corky and Pink Root of Tomato in Sinaloa, Mexico. Plant Dis. 2020, 104, 1553. [Google Scholar] [CrossRef]
- Zhang, F.B.; Zheng, H.L.; Cui, W.G.; Zhang, M.Q.; Yin, Y.S.; Cui, M.; Gao, M. First Report of Setophoma terrestris Causing Pink Root of Garlic in China. Plant Dis. 2019, 103, 584. [Google Scholar] [CrossRef]
- Ikeda, K.; Kuwabara, K.; Urushibara, T.; Soyai, P.; Miki, S.; Shibata, S. Pink root rot of squash caused by Setophoma terrestris in Japan. J. Gen. Plant Pathol. 2012, 78, 372–375. [Google Scholar] [CrossRef]
- Yoshida, N. Seasonal dynamics of the pink root fungus (Setophoma terrestris) in rhizosphere soil: Effect of crop species and rotation. Plant Pathol. 2022, 71, 361–372. [Google Scholar] [CrossRef]
- Feau, N.; Weiland, J.E.; Stanosz, G.R.; Bernier, L. Specific and sensitive PCR–based detection of Septoria musiva, S. populicola and S. populi, the causes of leaf spot and stem canker on poplars. Mycol. Res. 2005, 109, 1015–1028. [Google Scholar] [CrossRef]
- Satelis, F.J.; Boiteux, S.L.; Reis, A. Resistance to Septoria lycopersici in Solanum (section Lycopersicon) species and in progenies of S. lycopersicum × S. peruvianum. Sci. Agric. 2010, 67, 334–341. [Google Scholar] [CrossRef]
- Forootanfar, H.; Movahednia, M.M.; Yaghmaei, S.; Tabatabaei-Sameni, M.; Rastegar, H.; Sadighi, A.; Faramarzi, M.A. Removal of chlorophenolic derivatives by soil isolated ascomycete of Paraconiothyrium variabile and studying the role of its extracellular laccase. J. Hazard. Mater. 2012, 209, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Montecchio, L.; Causin, R.; Vettorazzo, M. A Twig Canker on English Hawthorn Caused by Coniothyrium sporulosum in Italy. Plant Dis. 2002, 86, 1403. [Google Scholar] [CrossRef]
- Cloete, M.; Fourie, P.H.; Damm, U.; Crous, P.W.; Mostert, L. Fungi associated with die-back symptoms of apple and pear trees, a possible inoculum source of grapevine trunk disease pathogens. Phytopathol. Mediterr. 2011, 50, 176–190. [Google Scholar]
- Gao, Y.; Liu, F.; Cai, L. Unravelling Diaporthe species associated with Camellia. Syst. Biodivers. 2016, 14, 102–117. [Google Scholar] [CrossRef]
- Gai, Y.; Xiong, T.; Xiao, X.; Li, P.; Zeng, Y.; Li, L.; Riely, B.K.; Li, H. The Genome Sequence of the Citrus Melanose Pathogen Diaporthe citri and Two Citrus–Related Diaporthe Species. Phytopathology 2021, 111, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.J.; Liu, M.; Zhang, W.; Chen, Z.; Udayanga, D.; Chukeatirote, E.; Li, X.; Yan, J.; Hyde, K.D. Morphological and molecular characterisation of Diaporthe species associated with grapevine trunk disease in China. Fungal Biol. 2015, 119, 283–294. [Google Scholar] [CrossRef]
- Guo, Y.S.; Crous, P.W.; Bai, Q.; Fu, M.; Yang, M.M.; Wang, X.H.; Du, Y.M.; Hong, N.; Xu, W.X.; Wang, G.P. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 2020, 45, 132–162. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).