Abstract
Macadamia is an economically significant crop, with its kernel oil being abundant in monounsaturated fatty acids (MUFA). Analyzing the expression of genes related to MUFA biosynthesis is essential for understanding the complex regulatory networks in Macadamia. However, there are few reports on the identification of suitable reference genes for use as internal controls in this species. Consequently, selecting a reliable reference gene for gene expression studies under various conditions is critical. In this study, we evaluated the expression stability of 11 traditional housekeeping genes: α-tubulin (TUBa), β-tubulin (TUBb), malate dehydrogenase (MDH), 18S ribosomal RNA (18S), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α-elongation factor 1 (EF1a), β-elongation factor 1 (EF1b), ubiquitin (UBQ), ubiquitin-conjugating enzyme (UBC), cyclophilin (CYP), and actin (ACT) under abiotic stresses, hormonal treatments and in variety of plant tissues using the online tool RefFinder, which integrates four commonly used software programs: ΔCt, geNorm (version 3.4), NormFinder (version 0953), and BestKeeper (version 1.0). A comprehensive expression stability ranking was established by integrating results from these four methods based on the geometric mean. The findings indicated that ACT was the most stable gene across all samples, including those subjected to cold stress, NaCl stress, PEG stress, ABA treatment, MeJA treatment, and both stem and leaf tissues. EF1b was identified as the most stable gene in GA treatment and heat stress samples, while UBC and CYP were ranked highest in ethrel treatment and root tissue samples, respectively. Finally, the reliability of these findings was further validated using the target gene SAD through qRT-PCR. In summary, this study evaluated and validated appropriate reference genes for qRT-PCR, which will facilitate future investigations into the molecular mechanisms in Macadamia.
1. Introduction
Macadamia is a genus within the Proteaceae family and is considered the most economically important crop. This long-lived, evergreen tree originates from subtropical and tropical regions, specifically native to Australia [1]. Macadamia produces edible dried fruits [2,3,4] and is also used as an ingredient in pastries, chocolate, oils, and the cosmetic and pharmaceutical industries [5]. The kernels of macadamia nuts are rich in lipids, proteins, minerals, and bioactive compounds [6,7]. Long-term consumption of macadamia nuts can reduce cholesterol levels and platelet viscosity in the blood, prevent arteriosclerosis, and lower the risk of heart disease, myocardial infarction, and other cardiovascular conditions [8,9,10,11]. Therefore, macadamia nuts can be beneficial for health maintenance and disease prevention.
Macadamia oil contains a higher concentration of monounsaturated fatty acids compared to other vegetable oils, making oil content a key indicator of macadamia kernel quality. The molecular mechanisms regulating fatty acid biosynthesis are the focus of our research. The completion of the macadamia genome sequencing has enabled large-scale identification of functional genes [12,13,14]. To investigate the gene expression of fatty acid biosynthesis-related genes in macadamia, quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) is one of the most commonly used technologies [15,16]. This method is widely utilized for analyzing gene expression levels due to its high sensitivity, specificity, reproducibility, and accuracy [17,18,19]. However, the accuracy of qRT-PCR results depends not only on gene-specific primers but also on the selection of appropriate internal references. Studies have shown that the expression of housekeeping genes can vary depending on experimental conditions and organ specificity. If changes in internal reference gene expression are not considered, the accuracy of qRT-PCR results may be compromised, potentially leading to contradictory or incorrect conclusions [20,21]. Therefore, it is essential to select suitable reference genes as internal controls under varying experimental conditions [22,23]. Furthermore, while several studies have investigated the functional genes related to fatty acid biosynthesis in macadamia [24,25,26,27], the stability of reference genes has yet to be evaluated. Additionally, the mechanism of oil accumulation in plant seeds is complex and closely related to seed development, hormonal responses, and environmental stress. To further explore the molecular mechanisms of fatty acid biosynthesis in macadamia, it is urgent to identify suitable reference genes for qRT-PCR analysis.
In this study, we selected 11 traditional candidate reference genes: α-tubulin (TUBa), β-tubulin (TUBb), malate dehydrogenase (MDH), 18S ribosomal RNA (18S), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α-elongation factor 1 (EF1a), β-elongation factor 1 (EF1b), ubiquitin (UBQ), ubiquitin-conjugating enzyme (UBC), cyclophilin (CYP), and actin (ACT). We tested their expression patterns and expression stability in abiotic stresses, hormonal treatments, and a variety of plant tissues using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). The expression stability of these candidate reference genes was validated and evaluated using statistical algorithms, including ΔCt [28], geNorm [29], NormFinder [30], and BestKeeper [31]. A recommended comprehensive stability ranking of these reference genes under each specific experimental condition was also performed in this test. Additionally, we used one target gene, Δ9-stearoyl-ACP desaturase (SAD), which introduces the first desaturation in the synthesis of aliphatic chains in plants, to validate the effectiveness of the selected reference genes. Finally, this work provides a foundation for further research into gene expression profiling in macadamia oil accumulation.
2. Materials and Methods
2.1. Plant Materials and Stress Treatments
NanYa No. 1 is a macadamia cultivar developed by the South Subtropical Crops Research Institute of the Chinese Academy of Tropical Agricultural Sciences (CATAS), Zhanjiang, Guangdong Province, China. Seeds from this cultivar were provided by the National Field Genebank for Tropical Fruits, located at the South Subtropical Crops Research Institute, the Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, China (110°27′ E; 21°17′ N). The macadamia seeds were sown and maintained in a glass bottle containing 1/2 SD solid medium (without hormones) at 20 °C ± 2 °C. When the stem diameters of the macadamia seedlings reached 2.1–3.3 mm, various treatments could be conducted in the glass bottle.
For the experiments, each treatment group was established in triplicate, with each group consisting of 7–8 sterile seedlings grown on a solid culture medium to ensure adequate sample collection. All treatments were conducted in sterile containers. The control group was treated with water. Organ-specific samples (root, stem, leaf) were collected from the seedlings. For drought treatment, a 20% PEG-6000 solution (w/v, polyethylene glycol, Sangon, Shanghai, China) was applied to incubate the plants for 0, 12, 24, 48, and 72 h. For cold and heat stress, the plants in their pots were placed in chambers at temperatures of 4 °C and 42 °C, respectively, for 0, 2, 6, 12, and 24 h. For salinity treatment, seedlings were transferred to 1/2 SD medium containing 150 mmol NaCl for 0, 12, 24, 48, and 72 h. For hormone treatment, plants were exposed to 100 μmol/L methyl jasmonate (MeJA, S30685, Shanghai Yuanye Biotech, Shanghai, China), abscisic acid (ABA, S18006, Shanghai Yuanye Biotech), gibberellins (GA, S18001, Shanghai Yuanye Biotech), or ethrel (ETH, S18030, Shanghai Yuanye Biotech) for 0, 2, 6, 12, and 24 h. Subsequently, the roots, leaves, and stems were sampled separately at different time points for expression analysis. Detailed information regarding the samples collected from various tissues and experimental conditions is provided in Supplementary Table S1. All samples were frozen in liquid nitrogen and stored at −80 °C prior to RNA isolation.
2.2. Total RNA Isolation and cDNA Synthesis
The frozen samples were ground into a fine powder in liquid nitrogen using a pestle and mortar. Total RNA was extracted from the samples using the Plant Total RNA Isolation Kit Plus (Chengdu Fuji Biotech, Chengdu, China), following the manufacturer’s instructions. RNA purity and concentration were assessed using a NanoDrop spectrophotometer (NanoDrop Technologies, ThermoScientific, Waltham, MA, USA), while RNA integrity was verified through agarose gel electrophoresis with ethidium bromide staining. First-strand cDNAs were synthesized from 1 µg of total RNA in a final volume of 20 µL using the RTIII all-in-one mix (Monad Biotech, Suzhou, China). The cDNA synthesis mixture was incubated at 37 °C for 2 min, followed by incubation at 55 °C for 15 min. The reaction was terminated by heating at 85 °C for 10 s. The cDNAs were diluted 1:50 with nuclease-free water prior to use in subsequent experiments.
2.3. Selection of Candidate Reference Genes and Primer Design
In this study, 11 traditional candidate reference gene sequences were collected from RNA-seq data (unpublished) and genomic sources [13]. The sequences of candidate reference genes were cloned in a previous study [32], and these genes are listed in Table 1. These primers were designed with melting temperatures ranging from 60 °C to 70 °C, primer lengths of 19–25 bases, GC content between 40% and 60%, and amplification lengths of 150–300 bp for the experiments. The result of the PCR and qRT-PCR experiment proved that the 11 candidate reference genes primer could amplify a single predicted band and a unique absorption peak through. The reference gene Actin has previously been used in studies of gene expression in Macadamia [33,34], while the stability of the other reference genes has not yet been evaluated. In this study, 11 classical housekeeping genes from different functional classes were selected to avoid potential co-regulation. The primers were designed based on the 3’ UTRs of these genes. Amplification primers for real-time PCR were created using Primer-BLAST from NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (accessed on 22 October 2021). The criteria for primer design included lengths of 20–24 bp, GC contents of 45%–55%, melting temperatures (Tm) ranging from 55 °C to 60 °C, and amplification lengths of 100–150 bp. The specificity of the designed primers was verified using NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) (accessed on 22 October 2021).

Table 1.
Primer sequences of the candidate reference genes.
2.4. Real-Time Quantitative Polymerase Chain Reaction
qRT-PCR was conducted in 96-well plates using the LightCycler 480 (Roche Molecular Biochemicals, Mannheim, Germany). The reaction mixture comprised 1 μL of diluted cDNA (~3 ng), 5 μL of Tap SYBR Green qPCR Premix (Yugong Biolabs, Lianyungang, China), 0.5 μL of specific primer pairs (0.5 μmol/L), and 3.5 μL of ddH2O, resulting in a final volume of 10 μL. Three biological replicates of all samples and three technical replicates for each biological replicate were performed. The qRT-PCR protocol was as follows: 95 °C for 3 min, followed by 45 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 10 s. A melting curve analysis was included to verify the specificity of each primer. The mean amplification efficiency of each primer pair was calculated using the equation: E = 10(−1/slope) − 1 [35].
2.5. Analysis of Real-Time PCR Data
The comparative ΔCt algorithm evaluates the expression stability of reference genes by calculating the mean standard deviation values from test samples [28]. The ΔCt value reflects the variability in transcription among the candidate genes.
The geNorm VBA applet determines the most suitable reference gene based on the geometric mean and pairwise variation in each gene relative to all other candidate reference genes in the total sample. Two parameters are utilized to assess the expression stability of reference genes: the average expression stability value (M value), which is derived from pairwise variation between a specific gene and all others, and the pairwise variation (Vn/n+1), which indicates the minimum number of genes needed for more accurate normalization [29]. A cut-off value of Vn/n+1 < 0.15 signifies that the inclusion of an additional reference gene does not significantly enhance normalization.
While geNorm employs a stepwise exclusion of the least stable genes, NormFinder utilizes a model-based approach that computes both inter- and intra-group variability to estimate gene expression stability. NormFinder identifies the optimal normalization genes from a panel of candidates based on their expression stability within a specific sample set or experimental design. This algorithm assesses not only the overall expression variation in candidate reference genes but also the variation among subgroups of samples. Additionally, NormFinder can analyze expression data from methods beyond real-time PCR, such as microarrays. It ranks stability based on the stability value (SV), where a lower stability value indicates higher gene expression stability, and vice versa [30].
BestKeeper assesses the stability of reference genes by calculating the coefficient of variance (CV) and the standard deviation (SD) of the average Ct values. A lower CV ± SD value indicates greater stability among candidate reference genes; conversely, genes with an SD > 1 are deemed unacceptable and should be excluded [31].
RefFinder is an accessible web-based tool developed for evaluating and screening reference genes from extensive experimental datasets. It integrates major computational programs (geNorm (version 3.4), NormFinder (version 0953), BestKeeper (version 1.0), and the comparative ΔCt method) to compare and rank the tested candidate reference genes according to the geometric mean of the rankings produced by the four algorithms [36]. We analyzed the qRT-PCR data using the RefFinder mirror site (http://blooge.cn/RefFinder/, accessed on 22 October 2021).
2.6. Evaluating Reference Genes Expression
acyl-ACP thioesterase (FAT), fatty acid desaturase (FAD), and stearoyl-acyl carrier protein 9 desaturase (SAD) are key enzymes involved in the fatty acid biosynthesis pathway in plants. The expression levels of the target gene SAD were analyzed using the most stable and least stable reference genes after normalization across all experimental sets. Primer pairs for these genes were synthesized by BGI. The sequences for the primers are as follows: SAD-F1: GAAGTCTACATGACAAGAACCCCAC; SAD-R1: CTTGGAGCTAGAACGAAGGGTGAT; FATA_qF1: GGTACAGTTATTGGGAGAGCC; FATA_qR1: TGGAAAAGCTAACCGTGGAG; FAD_qF1: GTCTGCTCCGGGTGTATAATG; FAD_qR1: TGCAACGCCACCCTAAT. The amplification efficiencies of the target genes were estimated using the LinRegPCR program. The average Ct values were calculated from three biological and technical replicates and utilized for relative expression analyses. Relative expression data were computed according to the qRT-PCR 2−ΔCt method and presented as fold changes [37].
3. Results
3.1. Verification of Primer Specificity and PCR Efficiency
The primer sequences of the 11 candidate reference genes are described in Table 1. The results of agarose gel electrophoresis revealed a single, specific band of the expected size. The melting curve analysis exhibited a single peak, indicating the specificity of the primers (Supplementary Figure S1). The amplification efficiency of the primers was assessed by qRT-PCR after 45 cycles across all samples at different gradient concentrations (Supplementary Figure S2), ranging from 91.9% to 102.5%, with a correlation coefficient (R2) close to 0.99 (Table 1). These results indicate that all primer pairs are specific and effective.
3.2. Expression Profile of the Candidate Reference Genes
The cycle threshold (Ct) value of quantitative reverse transcription polymerase chain reaction (qRT-PCR) represents the number of cycles required for the fluorescence signal intensity to reach a predetermined threshold level of detection. This value correlates with the transcript levels of genes in the test samples. The transcript levels of 11 reference genes exhibited significant variation across all experimental sets, as shown in Figure 1. The median Ct values ranged from 16.73 for 18S rRNA to 27.94 for TUBa, with most values falling between 20 and 23 across all samples. The 18S rRNA gene exhibited the highest expression level, indicated by the lowest mean Ct value (17.66 ± 2.62), followed by CYP (21.05 ± 2.39), MDH (21.97 ± 2.26), ACT (22.82 ± 2.20), UBQ (23.05 ± 3.08), UBC (23.25 ± 2.39), EF1a (23.79 ± 2.98), EF1b (23.80 ± 2.41), TUBb (25.40 ± 2.21), and GAPDH (27.69 ± 2.86). The candidate gene TUBa exhibited the lowest expression level among all test samples (27.94 ± 3.22). Genes with higher standard deviations (SD) of Ct values displayed more variable expression compared to those with lower SD. ACT showed the smallest variation in gene expression, with the lowest SD (22.82 ± 2.20), whereas TUBa (27.94 ± 3.22) exhibited the most variable expression levels (Figure 1).
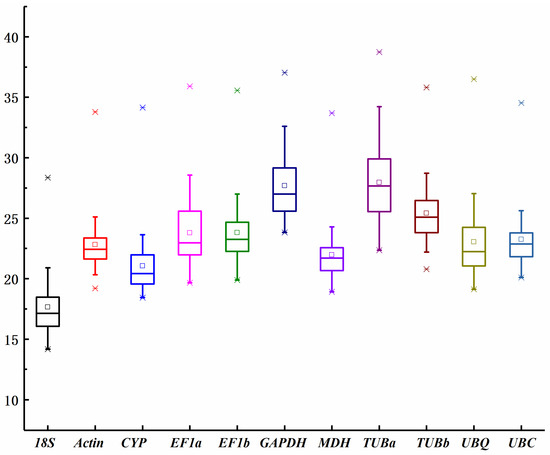
Figure 1.
Distribution of qRT-PCR Ct values of candidate reference genes across all samples (includes the control and treatment samples). X-axis represents reference genes. Y-axis represents the Ct value. The final Ct value of each sample was the mean of three biological and technical replicates. Box graph represents the interquartile range. The line across the box is the median value. The two bars represent the minimum and maximum values, respectively; the small square in the box shows the mean values. ӿ represents the non-outlier and outlier.
3.3. Expression Stability Analysis of Candidate Reference Genes
To identify the most suitable reference genes across different treatments and tissue types, the RefFinder online tool was employed to rank the expression stability values. This tool incorporates four widely used algorithms: comparative ΔCt, geNorm, NormFinder, and BestKeeper. The recommended comprehensive ranking was generated by averaging the results from the four programs across different experimental setups, as shown in Figure 2 and Table 2.
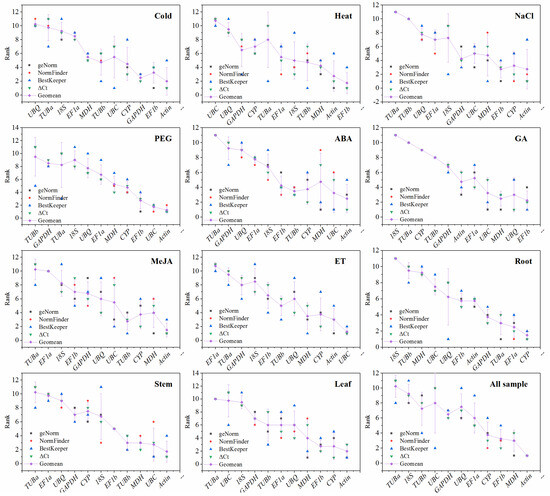
Figure 2.
Aggregation of four rankings. Each plot contains five types of ranking including (geNorm, NormFinder, BestKeeper, ΔCt and Geomean). The geometric ranking of candidate genes was calculated comprehensively by four types of rankings (geNorm, NormFinder, BestKeeper and ΔCt) under different treatments.

Table 2.
Expression stability ranking of the 11 candidate reference genes.
3.3.1. ΔCt Algorithm Analysis
The comparative ΔCt algorithm ranks potential candidate reference genes based on their expression stability, as indicated by standard deviation values (Supplementary Table S3) [28]. According to the ΔCt analysis, ACT was consistently identified as the most stable gene across all samples (1.05), as presented in Table 2. In macadamia, CYP emerged as the most stable reference gene in root (1.04) and leaf tissues (1.09), while ACT was the most stable reference gene in the stem (1.00) across various organ datasets. ACT remained the most stable gene in subsets subjected to cold stress (0.80), NaCl stress (0.70), PEG stress (0.34), ABA treatment (0.82), and MeJA treatment (0.82) among the 11 candidate reference genes, whereas TUBa was identified as the least stable gene under cold stress (1.54), NaCl stress (1.73), all samples (1.81), ABA treatment (2.04), MeJA treatment (1.49) and stem tissue (2.04). During heat stress, EF1b (1.38) surpassed ACT (1.40) to become the most stable reference gene. Under GA treatment, UBQ (0.64) was ranked as the stable reference gene, while 18S (1.30) was identified as the least stable reference gene. In the ethrel treatment, UBC (0.78) was the highest-ranked gene, whereas EF1a (1.35) exhibited the least stability (Figure 2; Table 2; Supplementary Table S3).
3.3.2. geNorm Analysis
The geNorm program utilized the M-value to assess the stability of reference gene expression. The M-value was calculated at each step during the stepwise exclusion of the least stable reference genes until the two most stable genes were identified [29]. Our finding revealed that all examined genes exhibited M values below 1.5, with the exceptions of TUBa (1.597) and UBC (1.87) under heat stress. Across the 12 tested conditions, ACT and MDH ranked as the two most stable genes, with M values of 0.527, 0.500, and 0.419 under MeJA treatment, leaf samples, and across all samples, respectively. Additionally, ACT and EF1b were among the two most stable genes under cold stress (0.245), heat stress (0.332), NaCl stress (0.183), and PEG stress (0.157). MDH and UBC were the most stable under GA treatment (0.231) and ABA treatment (0.209), while ACT and UBC exhibited the greatest stability in stem (0.403) and ethrel treatments (0.331). CYP/TUBa emerged as the most stable reference gene with an M value of 0.369. Furthermore, as shown in Figure 2, Table 2, and Supplementary Table S4, the least stable expression was observed for TUBa under cold stress, NaCl stress, ABA treatment, MeJA treatment, stem samples, and across all samples; UBC was the least stable under heat stress and leaf samples; TUBb was least stable under PEG stress; 18S under GA treatment and root samples; and EF1a under ethrel treatment.
In gene expression analysis, relying solely on a single reference gene for normalization may not fulfill experimental requirements. Consequently, the use of two or more reference genes is essential to reduce experimental errors and achieve more accurate results. The impact of introducing additional reference genes can be assessed by calculating the pairwise variation (V) values of the new normalization factor following the design of primers for these reference genes, and based on the Vn/Vn+1 ratio. The pairwise variation analysis of 11 candidate reference genes is shown in Table 3. For ABA treatment, V2/3 is 0.159, which is greater than the threshold of 0.15, while V3/4 is 0.103, falling below the threshold. This suggests that the optimal number of reference genes is three, namely MDH, UBC, and ACT. In other treatments, all V2/3 and V3/4 values are less than 0.15, indicating that the top two reference genes can be selected for normalization.

Table 3.
Pairwise variation analysis.
3.3.3. NormFinder Analysis
The NormFinder algorithm ranks candidate reference gene stability based on the stability value assigned to each reference gene, with lower stability values indicating more stable gene expression. Our findings indicate that the ranking orders produced by NormFinder are similar to those obtained through geNorm analysis. According to this analysis (Supplementary Figure S4; Table 2; Supplementary Table S5), the most stable gene for cold stress (0.269), ABA treatment (0.28), MeJA treatment (0.395), stem (0.461) and all samples (0.497) was ACT, while EF1b ranked highest for heat stress (0.621), CYP for leaf and NaCl stress (0.175), UBC for ethrel treatment (0.78) and PEG stress (0.076), UBQ for GA treatment (0.64) and EF1a for root samples (0.269) (Supplementary Figure S5). Conversely, TUBa emerged as the most unstable gene across all samples (1.602): NaCl stress (1.652), MeJA stress (1.337), stem (1.9) and ABA stress (1.943). UBC was identified as the least stable gene in the leaf sample (1.905) and heat treatment (2.872), while 18S exhibited instability under GA treatment (1.3) and in root samples (1.933), and EF1a under ethrel treatment (1.35). In general, the NormFinder results confirmed the geNorm analysis, but the results were not all the same.
3.3.4. BestKeeper Analysis
The BestKeeper algorithm assessed the stability of candidate reference genes based on the coefficient of variation (CV) ± standard deviation (SD) values [31]. The stability rankings generated by BestKeeper were significantly different from those determined by the ΔCt, geNorm, and NormFinder algorithms (Supplementary Figure S6). ACT was identified as the most stable gene across all samples (6.4 ± 1.44), PEG treatment (1.4 ± 0.31) and leaf tissue (5.8 ± 1.28). TUBb was recognized as the most stable gene under heat stress (8.57 ± 2.43), cold stress (9.28 ± 2.41), and MeJA treatment (3.86 ± 0.97); UBC exhibited the highest stability for ABA treatment (5.18±1.21) and stem tissue (6.17 ± 1.45); MDH demonstrated the most stability under NaCl stress (3.38 ± 0.71) and ethrel treatment (3.47 ± 0.79). EF1b was most stable under GA treatment (3.28 ± 0.72), while UBQ was the most stable for root tissue (5.32 ± 1.36). Additionally, 18S and TUBa were identified as the least stable reference genes across most samples according to the BestKeeper algorithm (Figure 2; Table 2; Supplementary Table S6).
3.3.5. Comprehensive Ranking
Through comprehensive analysis of the rankings generated by the four aforementioned algorithms, a recommended comprehensive ranking was established under abiotic stress, hormonal treatments, and a variety of plant tissues, as shown in Table 2, Figure 2, and Supplementary Tables S7 and S8. Among all samples—including conditions of cold stress, NaCl stress, PEG stress, ABA treatment, MeJA treatment, and both stem and leaf tissues—ACT was identified as the most stable gene. EF1b was recognized as the most stable gene for heat stress and GA treatment, while CYP exhibited the most stability for root tissue and UBC for ethrel treatment (Figure 2; Table 2). Conversely, TUBa was identified as the least stable reference gene in MeJA treatment, ABA treatment, NaCl stress, and across stem and leaf tissues. Additionally, 18S was the least stable gene in GA treatment and root tissues.
3.4. Validation of the Reference Genes with the SAD Gene
FATA, FAD, and SAD are key genes involved in the fatty acid biosynthesis pathway in plants. To further validate the selected reference genes, the target gene SAD in macadamia was evaluated under various experimental conditions using quantitative reverse transcription polymerase chain reaction (qRT-PCR). The expression levels were normalized using both the most stable reference genes and the two least stable reference genes as internal controls, applied individually and in combination, across various treatment subsets. The expression profiles of the SAD gene across 12 experimental treatments exhibited remarkable similarity when stable reference genes were employed as internal controls (Figure 3).
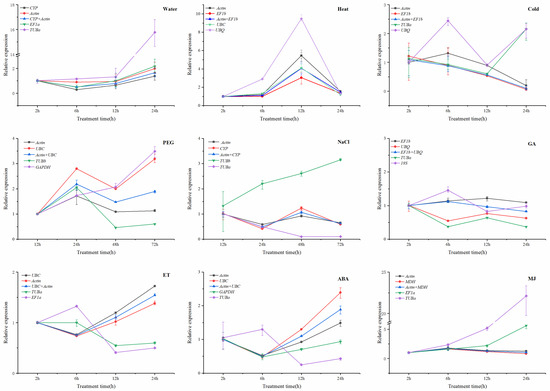
Figure 3.
Relative quantification of SAD expression using the validated reference gene(s). The broken line graph represents the relative expression trend when compared to non-treatment samples under different tissues and treatments. cDNA samples were taken from the same subset used for gene expression stability analysis. Roots, stems, and leaves were collected from Macadamia integrifolia seedlings subjected to various treatments.
Water treatment was established as a negative control for this study. When the most stable genes, ACT and CYP, were employed either separately or together to normalize the expression levels of the SAD gene, expression abundance was not significantly affected. Under cold, heat and salt stress, the expression level of the SAD gene remained consistent, with slight fluctuations when ACT, EF1b, and CYP were used as internal controls either singly or in combination. In treatments involving MeJA, GA, and ET hormones, a similar expression pattern was observed when either ACT, UBC, UBQ, EF1b or MDH was used alone or in combination for normalization. Conversely, when unstable genes were used as internal controls in the analysis of SAD gene expression under the 12 different treatments, a distinct pattern emerged. Specifically, the expression level of the SAD gene in plants treated with MeJ, water, PEG, cold stress, and heat stress was overestimated by up to 10 times as much compared to other treatments when the least stable reference gene TUBa, UBQ or GAPDH were employed. On the contrary, in ET treatment, NaCl, and ABA treatment, the expression level of the SAD gene was significantly underestimated when the least stable reference gene, TUBa, EF1a, or GAPDH, was used for normalization. In GA treatment, the expression of the SAD gene was consistent across both the most and least stable reference genes, indicating that most reference genes could be stably expressed.
Overall, the results indicated that ACT and EF1b were more suitable for most treatment subsets, while TUBa is ineffective in standardizing the expression data.
Additionally, to validate the selected reference genes, we compared four genes involved in fatty acid accumulation—FATA, FAD, Oleosin, and SAD—using qRT-PCR with expression patterns derived from RNA-seq results. In the qRT-PCR analysis, Actin served as the normalizer for quantifying expression levels. As shown in Figure 4, all selected genes exhibited similar expression profiles in both methods. Overall, these results provide reliable reference genes for Macadamia.
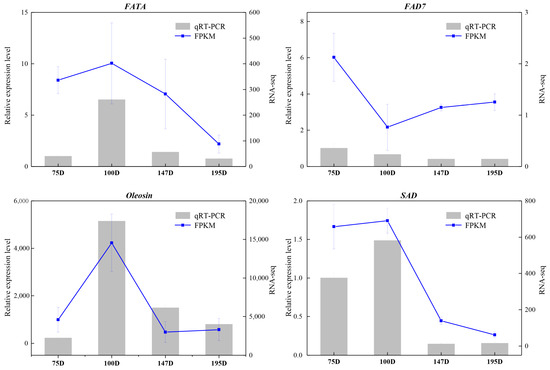
Figure 4.
Validation of qRT-PCR results through comparison with RNA-seq expression profiles. X-axis represents the days after the anthesis of macadamia kernel samples. Y-axis represents the gene relative expression level by qRT-PCR. The histograms show the qRT-PCR results involved fatty acid biosynthesis in kernel of Macadamia after anthesis; the line charts show the FPKM values of these fatty acid biosynthesis genes in kernel of Macadamia. qRT-PCR results represent the mean (±SD) of three biological replicates. 75D (75 days after anthesis), 100D (100 days after anthesis), 147D (147 days after anthesis), and 195D (195 day after anthesis).
4. Discussion
Gene expression quantification is a crucial method for characterizing gene function and has been widely accepted and applied in genetic research [16]. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) is commonly regarded as a practical method for the accurate analysis of gene expression profiling, demonstrating high sensitivity, specificity, and reproducibility, while requiring fewer samples in accordance with the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [38]. There are no universally suitable reference genes for every experimental condition; rather, the expression of reference genes is tissue-specific and varies based on the physiological status of the organs or experimental conditions [20,21]. Numerous studies have reported that while the expression of housekeeping genes may remain constant under certain experimental conditions, it can vary considerably in other instances [39,40]. Current protocols suggest that utilizing specific and stable reference genes for normalization is essential for ensuring accurate results [41]. Consequently, the selection and evaluation of appropriate reference genes are critically important for data normalization in gene expression analysis [42]. Recent studies in the field of plant biology have focused on suitable reference gene selection, including research on cucumber [43], Setaria viridis [44], sugarcane [45], wheat [46], soybean [47], rapeseed [48], yew [49], Gentiana macrophylla [50], and various other plants. To date, however, few studies have investigated reference gene selection in macadamia, which has impeded molecular functional studies of important genes related to stress conditions and specific organisms. In the present study, we systematically validated the stability of 11 traditional reference genes under specific environmental conditions and treatments, as well as in different tissues of Macadamia integrifolia seedlings. The selected traditional housekeeping genes, which are involved in cytoskeletal structure (ACT, TUBa, TUBb), protein synthesis (EF1a, EF1b, 18S), biological metabolic processes (GAPDH and UBQ), and multifunctional proteins (CYP and MDH), had their open reading frame sequences and 3′ non-coding regions cloned from Macadamia integrifolia.
It is recommended that the comprehensive RefFinder algorithm provides a reliable assessment of the expression stability of candidate reference genes under various experimental conditions for real-time quantitative reverse transcription PCR (qRT-PCR) analysis [51]. Four statistical methods—ΔCt, geNorm, NormFinder, and BestKeeper—have been developed to evaluate the expression stability of candidate reference genes for accurate normalization in gene expression studies. The ranking results generated by these algorithms largely coincide. In the heat treatment subset, ACT and EF1b were identified as the two most stable genes in the geNorm analysis; ACT was also deemed the most stable gene in both the ΔCt and NormFinder analyses, whereas it ranked fifth in the BestKeeper assessment. In the ABA treatment, NaCl stress, stem, and across all sample sets, TUBa consistently ranked as the least stable gene according to all four algorithms. However, significant differences in stability rankings were sometimes observed due to the complementary nature of the different statistical programs (Table 2). For instance, under NaCl stress, ACT was ranked first by both the ΔCt and geNorm methods, while it was ranked seventh by BestKeeper. In NormFinder analysis, CYP was ranked first, followed closely by ACT. The ΔCt method compares the relative expression of gene pairs within each sample to accurately identify the stability of reference genes [28]. GeNorm is one of the most commonly used methods for systematically validating reference gene stability [29]; it identifies two reference genes based on the similarity of expression profiles across samples and the lowest intra-group variation [52,53], but it is not suitable for distinguishing genes with similar expression patterns. NormFinder, developed by Andersen [30], accounts for both intra- and inter-group variations, combining them into a stability value to rank genes with minimal variability. Typically, the results from NormFinder align closely with those from geNorm. For example, in the heat stress samples, EF1b and ACT were ranked as the two most stable reference genes, while TUBa and UBQ were the least stable, with slight variations in the ranks assigned by geNorm and NormFinder. The BestKeeper software (version 1.0) not only assesses the stability of reference genes but also of target genes [31], determining stability rankings based on standard deviation (SD) and coefficient of variation (CV) values [54,55]. Compared to geNorm and NormFinder, the results from BestKeeper can differ significantly due to its distinct calculation strategy. For example, in the all-sample subset, ACT was consistently ranked first by all programs. Under cold stress, ACT was identified as the most stable gene by the ΔCt, geNorm, NormFinder, and comprehensive ranking methods, except for BestKeeper. Given that these programs frequently rank different most stable reference genes [56,57,58], we selected the most reliable reference gene using the comprehensive RefFinder tool, which ranks reference genes based on the geometric mean of the weights assigned to each gene by the various programs [36]. The results derived from the statistical analyses conducted using ΔCt, geNorm, NormFinder, and BestKeeper demonstrated considerable consistency in this study. Additionally, the choice of which reference gene and which screening tool to use should be made according to the specific requirements of the experimental materials, objectives, and design.
ACT is widely recognized as a reliable reference gene [59]. Although some studies have provided evidence that the expression of ACT varies considerably in Zea mays [60], our findings support the conclusion that ACT is the most stable reference gene for relative quantification in Macadamia, across various conditions including cold stress, salt stress, PEG stress, ABA treatment, and MeJA treatment. Elongation factors, which are involved in the translocation of aminoacyl-transfer RNA to the ribosome during protein synthesis, are typically used as internal control genes due to their high conservation in sequence and expression among eukaryotes. In this study, EF1b was identified as the most appropriate reference gene under GA and heat stress treatments, while EF1a was deemed the most stable gene during complex developmental processes in Populus [61]. In ethrel treatment, UBC ranked higher than ACT, which is consistent with results observed in Platycladus orientalis [54]. CYP (Cyclophilin), a specific cytosolic binding protein, was ranked as the most stable gene in the roots of Macadamia. Although TUBa was found to be unsatisfactory for qRT-PCR analysis in this study, it demonstrated sufficient stability in Heterosigma akashiwo (Raphidophyceae) [62] and has been selected as a reference gene in Ulva linza [63] and the diatom Pseudo-nitzschia multistriata [64].
Our previous studies have demonstrated that Δ9-Stearoyl-ACP desaturases (SAD) play a crucial role in the synthesis and accumulation of unsaturated fatty acids in macadamia oil. It was found that the MiSAD gene was highly expressed in the kernel and consistent with oleic acid accumulation in macadamia, suggesting that MiSAD significantly contributes to this process. Extensive research has established that SAD is a key acyl-ACP desaturase that introduces a double bond into the aliphatic chain during de novo fatty acid biosynthesis. This enzyme is responsible for the production of unsaturated fatty acids. In Arabidopsis thaliana, the FAB2 (SSI2) gene, encoding stearoyl-ACP desaturase (SAD), is associated with oleic acid accumulation and a reduction in stearic acid level [65]. The C. sativa actin gen (GenBank accession number: KJ670375) was used as an internal reference to normalize the relative amount of cDNAs for CsSAD, MtSAD and DuSAD [26]. Furthermore, the expression profile of SAD and the fatty acid levels were influenced in Arabidopsis crown galls under drought and hypoxia stress conditions [66]. In this study, we quantified SAD gene expression using the two most stable reference genes, both individually and in combination, along with the two least stable genes as internal controls in qRT-PCR analysis to validate the selected reference genes in macadamia. When the least stable gene TUBb was used for normalization, the expression pattern of SAD was significantly overestimated. In contrast, normalization with TUBa resulted in an underestimation of the SAD expression under NaCl treatment (Figure 3). The expression pattern of the SAD gene exhibited slight fluctuations when normalized with the stable reference gene. However, it was either overestimated or underestimated when normalized with the two least stable reference genes, TUBa and EF1a, following six hours of ET treatment. These findings emphasize the critical importance of selecting appropriate internal controls is critically important for normalization, as the use of unvalidated references can lead to misinterpretation of the results.
RNA-seq is a method for analyzing the transcriptome profiles of various species and has recently been employed to identify candidate reference genes. In this study, the expression profiles of the target genes were consistent in both qRT-PCR and RNA-seq results, thereby supporting each other. Therefore, our findings indicate that the results of this experiment are credible. The reference genes evaluated in this study will be valuable for future gene expression analyses related to the molecular mechanisms of fatty acid accumulation under different treatments in macadamia.
5. Conclusions
The reference genes selected in the current study will aid in the accurate normalization of qRT-PCR data for M. integrifolia. We identified the most stable reference genes across different tissue types of macadamia seedlings and under various treatments. Our results indicate that ACT demonstrated strong stability for most treatment subsets. UBC can be used for normalization in samples treated with ethrel. In the GA treatment and heat samples, EF1b emerged as the best reference gene, while CYP is suitable as a reference gene for root samples. The identification of suitable reference genes in this study will facilitate future gene expression studies in macadamia, enhancing our understanding of the molecular mechanisms of fatty acid biosynthesis and accumulation under different experimental conditions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f15111966/s1, Supplementary Figure S1: Melting curve analysis following qRT-PCR. Supplementary Figure S2: Amplification efficiency of primers under different gradient concentrations. Supplementary Figure S3: ΔCt ranking of 11 candidate reference gene. Supplementary Figure S4: geNorm ranking of 11 candidate reference gene. Supplementary Figure S5: NormFinder ranking of 11 candidate reference gene. Supplementary Figure S6: BestKeepr ranking of 11 candidate reference gene. Supplementary Figure S7: Agarose gel analysis of RNA in abiotic stresses, hormonal treatment and plant tiuues. Supplemental Table S1: The detailed information of samples collected from various tissues/experimental conditions. Supplemental Table S2: The raw CT values of all samples under multiple treatment. Supplemental Table S3: ΔCt algorithm analysis. Supplemental Table S4: geNorm analysis. Supplemental Table S5: NormFinder analysis. Supplemental Table S6: Bestkeeper Analysis. Supplemental Table S7: Comprehensive Ranking by RefFinder. Supplemental Table S8: Comprehensive stability ranking of 14 candidate reference genes based on the results of the four software programs.
Author Contributions
Q.Y. and Z.Y. contributed equally to this work. Q.Y. drafted the manuscript. Z.Y. revised this article, including pre- or post-stage. H.Z. oversaw this research activity planning and execution. M.Z. verified the replication and reproducibility of results. X.S. produced and collected all the metadata. J.W. analyzed all the data. Z.W. provided the experimental method in this research. J.C. performed the investigation of research status before this project. L.L. provided the seedlings for the experiment in this article. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Central Public-interest Scientific Institution Basal Research Fund for the Chinese Academy of Tropical Agricultural Sciences (No. 1630062021015, No. 1630062022002), the National Key Research and Development Program of China (No. 2023YFD2200701). The funding bodies played no role in the study design, the collection and analysis of the data, data interpretation, and writing the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Acknowledgments
We thank the National Tropical Plants Germplasm Resource Center and National Field GenBank for Tropical Fruits for providing macadamia seeds.
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
| SAD | Δ9-Stearoyl-ACP desaturases |
| 18S | 18S ribosome RNA |
| ACT | Actin |
| CYP | Cyclophilin |
| EF1a | Elongation factor 1-α |
| EF1b | Elongation factor 1-β |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| MDH | Malate dehydrogenease |
| UBC | Ubiquitin-conjugating enzyme |
| TUBa | Tubulin-α |
| TUBb | Tubulin-β |
| UBQ | Ubiquitin-A |
| FATA | fatty acid thioesterase |
| FAD | fatty acid desaturation |
| MeJA | jasmonate |
| ABA | abscisic acid |
| GA | gibberrellins |
| ETH | ethyrel |
References
- Hardner, C.M.; Peace, C.; Lowe, A.J.; Neal, J.; Pisanu, P.; Powell, M.; Schmidt, A.; Spain, C.; Williams, K. Genetic resources and domestication of macadamia. Hortic. Rev. 2009, 35, 1–125. [Google Scholar] [CrossRef]
- Toft, B.D.; Alam, M.; Topp, B. Estimating genetic parameters of architectural and reproductive traits in young macadamia cultivars. Tree Genet. Genomes 2018, 14, 50. [Google Scholar] [CrossRef]
- Wallace, H.M.; Walton, D.A. Macadamia (Macadamia integrifolia, Macadamia tetraphylla and hybrids). In Woodhead Publishing Series in Food Science, Technology and Nutrition, Postharvest Biology and Technology of Tropical and Subtropical Fruits; Yahia, E.M., Ed.; Woodhead Publishing: Sawston, UK, 2011; pp. 450–473. [Google Scholar] [CrossRef]
- Cavaletto, C.G. Macadamia nuts. In Handbook of Tropical Foods; Chan, H.T., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1983; pp. 361–397. [Google Scholar]
- Navarro, S.L.B.; Rodrigues, C.E.C. Macadamia oil extraction with alcoholic solvents: Yield and composition of macadamia oil and production of protein concentrates from defatted meal. Eur. J. Lipid Sci. Technol. 2018, 120, 1800092. [Google Scholar] [CrossRef]
- Maguire, L.S.; O’Sullivan, S.M.; Galvin, K.; O’Connor, T.P.; O’Brien, N.M. Fatty acid profile, tocopherol, squalene and phytosterol content of walnuts, almonds, peanuts, hazelnuts and the macadamia nut. Int. J. Food Sci. Nutr. 2004, 55, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Rengel, A.; Pérez, E.; Piombo, G.; Ricci, J.; Servent, A.; Tapia, M.S.; Gibert, O.; Montet, D. Lipid profile and antioxidant activity of macadamia nuts (Macadamia integrifolia) cultivated in Venezuela. Nat. Sci. 2015, 7, 535–547. [Google Scholar] [CrossRef]
- Vadivel, V.; Kunyanga, C.N.; Biesalski, H.K. Health benefits of nut consumption with special reference to body weight control. Nutrition 2012, 28, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Griel, A.E.; Cao, Y.; Bagshaw, D.D.; Cifelli, A.M.; Holub, B.; Kris-Etherton, P.M. A macadamia nut-rich diet reduces total and LDL-cholesterol in mildly hypercholesterolemic men and women. J. Nutr. 2008, 138, 761–767. [Google Scholar] [CrossRef]
- Kaijser, A.; Dutta, P.; Savage, G. Oxidative stability and lipid composition of macadamia nuts grown in New Zealand. Food Chem. 2000, 71, 67–70. [Google Scholar] [CrossRef]
- Wang, X.F.; Yang, R.N.; Xue, L.; Zhang, L.X.; Wang, X.P.; Zhang, Q.; Li, P.W. Determination of fatty acid composition of 28 kinds of functional vegetable oil. J. Food Saf. Qual. 2017, 8, 4336–4343. [Google Scholar] [CrossRef]
- Nock, C.J.; Baten, A.; King, G.J. Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae. BMC Genom. 2014, 15, S13. [Google Scholar] [CrossRef][Green Version]
- Lin, J.S.; Zhang, W.P.; Zhang, X.T.; Ma, X.K.; Zhang, S.C.; Chen, S.A.; Wang, Y.B.; Jia, H.F.; Liao, Z.Y.; Jing, L.; et al. Signatures of selection in recently domesticated macadamia. Nat. Commun. 2022, 13, 242. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.F.; Li, G.H.; Ni, S.B.; He, X.Y.; Zheng, C.; Liu, Z.Y.; Gong, L.D.; Kong, G.H.; Li, W.; Liu, J. The chromosome-scale reference genome of Macadamia tetraphylla provides insights into fatty acid biosynthesis. Front. Genet. 2022, 13, 835363. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR-a perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, H.; Liu, X.; Lin, Z.; Guo, Y.; Deng, H.; Wang, J.; Lin, L.; Deng, Q.; Lv, X.; et al. Identification of suitable reference genes for qRT-PCR normalization in Kiwifruit. Horticulturae 2022, 8, 170. [Google Scholar] [CrossRef]
- Lian, C.L.; Zhang, B.; Yang, J.F.; Lan, J.X.; Yang, H.; Guo, K.H.; Li, J.J.; Chen, S.Q. Validation of suitable reference genes by various algorithms for gene expression analysis in Isodon rubescens under different abiotic stresses. Sci. Rep. 2022, 12, 19599. [Google Scholar] [CrossRef]
- VanGuilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008, 44, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.X.; Tan, H.X.; Yu, J.; Chen, Y.; Guo, Z.Y.; Wang, G.Q.; Zhang, Q.L.; Chen, J.F.; Zhang, L.; Yong, D. Stable internal reference genes for normalizing real-time quantitative PCR in Baphicacanthus cusia under Hormonal Stimuli and UV Irradiation, and in different plant organs. Front. Plant Sci. 2017, 8, 668. [Google Scholar] [CrossRef]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and validation of appropriate reference genes for quantitative real-Time PCR analysis of gene expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalization; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Fei, P.; Lei, Z.; Cai, S.B.; Tang, X.N.; Mehmood, A.; Alnadari, F.; Tuersuntuoheti, T.; Zhou, N.; Ai, X. Prediction and evaluation of the 3D structure of Macadamia integrifolia antimicrobial protein 2 (MiAMP2) and its interaction with palmitoleic acid or oleic acid: An integrated computational approach. Food Chem. 2022, 367, 130677. [Google Scholar] [CrossRef]
- Gummeson, P.O.; Lenman, M.; Lee, M.; Singh, S.; Stymne, S. Characterisation of acyl-ACP desaturases from Macadamia integrifolia Maiden & Betche and Nerium oleander L. Plant Sci. 2000, 154, 53–60. [Google Scholar] [CrossRef]
- Rodríguez, M.F.; Sánchez-García, A.; Salas, J.J.; Garcés, R.; Martínez-Forc, E. Characterization of soluble acyl-ACP desaturases from Camelina sativa, Macadamia tetraphylla and Dolichandra unguis-cati. J Plant Physiol. 2015, 178, 35–42. [Google Scholar] [CrossRef]
- Moreno-Pérez, A.J.; Sánchez-García, A.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Acyl-ACP thioesterases from macadamia (Macadamia tetraphylla) nuts: Cloning, characterization and their impact on oil composition. Plant Physiol. Biochem. 2011, 49, 82–87. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, Z.P.; Zhou, Y.L.; Chen, D.Q.; Liu, H. Screening of stable reference genes for qRT-PCR analysis in Macadamia integrifolia. Chin. J. Trop. Crops 2020, 41, 1504–1512. [Google Scholar]
- Yang, W.H.; Guo, S.X.; Xu, T.; Xiao, Y.; Lei, W.J. Expression of genes related to photosynthetic energy metabolism during leaf yellowing of macadamia. Chin. J. Trop. Crops 2024, 45, 1110–1119. [Google Scholar]
- Yang, Q.; Yang, Z.P.; Zou, M.H.; Song, X.M.; Wan, J.F.; Chen, J.; Luo, L.F.; Zeng, H. Cloning and expressing analysis of stearoyl-acyl-carrier-protein desaturase (SAD) from Macadamia intergrifolia. Chin. J. Trop. Crops 2023, 44, 254–263. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; Van den Hoff, M.J.B.; Moorman, A. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Xiao, P.; Chen, D.L.; Xu, L.; Zhang, B.H. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Remans, T.; Smeets, K.; Opdenakker, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Normalizations of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 2008, 227, 1343–1349. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Brunner, A.M.; Yakovlev, I.A.; Strauss, S.H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004, 4, 14. [Google Scholar] [CrossRef]
- Migocka, M.; Papierniak, A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2011, 28, 343–357. [Google Scholar] [CrossRef]
- Martins, P.K.; Mafra, V.; Souza, W.R.; Ribeiro, A.P.; Vinecky, F.; Basso, M.F.; da Cunha, B.A.; Kobayashi, A.K.; Molinari, H.B. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci. Rep. 2016, 6, 28348. [Google Scholar] [CrossRef]
- Guo, J.L.; Ling, H.; Wu, Q.B.; Xu, L.P.; Que, Y.X. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 2014, 4, 7042. [Google Scholar] [CrossRef]
- Long, X.Y.; Wang, J.R.; Ouellet, T.; Rocheleau, H.; Wei, Y.M.; Pu, Z.E.; Jiang, Q.T.; Lan, X.J.; Zheng, Y.L. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 2010, 74, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.H.; Niu, H.W.; Liu, C.J.; Zhang, J.; Hou, C.Y.; Wang, D.M. Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS ONE 2013, 8, e75271. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Y.; Fang, H.D.; Shi, H.F.; Chen, K.P.; Zhang, Z.Y.; Tan, X.L. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genom. 2014, 289, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.K.; Fan, W.; Chen, D.F.; Jiang, L.Y.; Li, Y.F.; Yao, Z.W.; Yang, Y.F.; Qiu, D.Y. Selection and validation of reference genes for quantitative gene expression normalization in Taxus spp. Sci. Rep. 2020, 10, 22205. [Google Scholar] [CrossRef]
- He, Y.H.; Yan, H.L.; Hua, W.P.; Huang, Y.Y.; Wang, Z.Z. Selection and validation of reference genes for quantitative real-time PCR in Gentiana macrophylla. Front. Plant Sci. 2016, 7, 945. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, B.Y.; Tan, Z.Q.; Liu, J.; Yang, Z.M.; Li, Z.H.; Huang, B.R. Selection of reference genes for quantitative real-time PCR normalization in creeping bentgrass involved in four abiotic stresses. Plant Cell Rep. 2015, 34, 1825–1834. [Google Scholar] [CrossRef]
- Jian, B.; Liu, B.; Bi, Y.R.; Hou, W.S.; Wu, C.X.; Han, T.F. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Bio. 2008, 9, 59. [Google Scholar] [CrossRef]
- Cruz, F.; Kalaoun, S.; Nobile, P.; Colombo, C.; Almeida, J.; Barros, L.M.; Romano, E.; Grossi-de-Sá, M.F.; Vaslin, M.; Alves-Ferreira, M. Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol. Breed. 2009, 23, 607–616. [Google Scholar] [CrossRef]
- Chang, E.M.; Shi, S.Q.; Liu, J.F.; Cheng, T.L.; Xue, L.; Yang, X.Y.; Yang, W.J.; Lan, Q.; Jiang, Z.P. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS ONE 2012, 7, e33278. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.L.; Ma, J.B.; Wang, J.R.; Wu, X.M.; Li, P.B.; Yao, Y.A. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 2015, 5, 788. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Guertler, R.; Naim, S.; Nixdorf, S.; Fedier, A.; Hacker, N.F.; Heinzelmann-Schwarz, V. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS ONE 2013, 8, e59180. [Google Scholar] [CrossRef]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef]
- Rivera, L.; Lopez-Patino, M.; Milton, D.; Nieto, T.; Farto, R. Effective qPCR methodology to quantify the expression of virulence genes in Aeromonas salmonicida subsp. salmonicida. J. Appl. Microbiol. 2015, 118, 792–802. [Google Scholar] [CrossRef]
- Pu, Q.; Li, Z.; Nie, G.; Zhou, J.Q.; Liu, L.; Peng, Y. Selection and validation of reference genes for quantitative real-time PCR in white clover (Trifolium repens L.) involved in five abiotic stresses. Plants 2020, 9, 996. [Google Scholar] [CrossRef]
- Díaz-Camino, C.; Conde, R.; Ovsenek, N.; Villanueva, M.A. Actin expression is induced and three isoforms are differentially expressed during germination in Zea mays. J. Exp. Bot. 2005, 56, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef]
- Ji, N.J.; Li, L.; Lin, L.X.; Lin, S.J. Screening for suitable reference genes for quantitative real-Time PCR in Heterosigma akashiwo (Raphidophyceae). PLoS ONE 2015, 10, e0132183. [Google Scholar] [CrossRef]
- Dong, M.T.; Zhang, X.W.; Chi, X.Y.; Mou, S.L.; Xu, J.F.; Xu, D.; Wang, W.Q.; Ye, N.H. The validity of a reference gene is highly dependent on the experimental conditions in green alga Ulva linza. Curr. Geneti. 2012, 58, 13–20. [Google Scholar] [CrossRef]
- Adelfi, M.G.; Borra, M.; Sanges, R.; Montresor, M.; Fontana, A.; Ferrante, M.I. Selection and validation of reference genes for qPCR analysis in the pennate diatoms Pseudo- nitzschia multistriata and P. arenysensis. J. Exp. Mar. Biol. Ecol. 2014, 451, 74–81. [Google Scholar] [CrossRef]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 2007, 63, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Klinkenberg, J.; Faist, H.; Saupe, S.; Lambertz, S.; Krischke, M.; Stingl, N.; Fekete, A.; Mueller, M.J.; Feussner, I.; Hedrich, R.; et al. Two fatty acid desaturases, STEAROYL-ACYL CARRIER PROTEIN Ɗ9-DESATURASE6 and FATTY ACID DESATURASE3, are involved in drought and hypoxia stress signaling in Arabidopsis crown galls. Plant Physiol. 2014, 164, 570–583. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).