Molecular Cloning and Functional Characterization of a Cytochrome P450 Enzyme (SaCYP736A167) Promoter from Santalum album
Abstract
1. Introduction
2. Materials and Methods
2.1. Extraction and Analysis of the SaCYP736A167 Gene Promoter
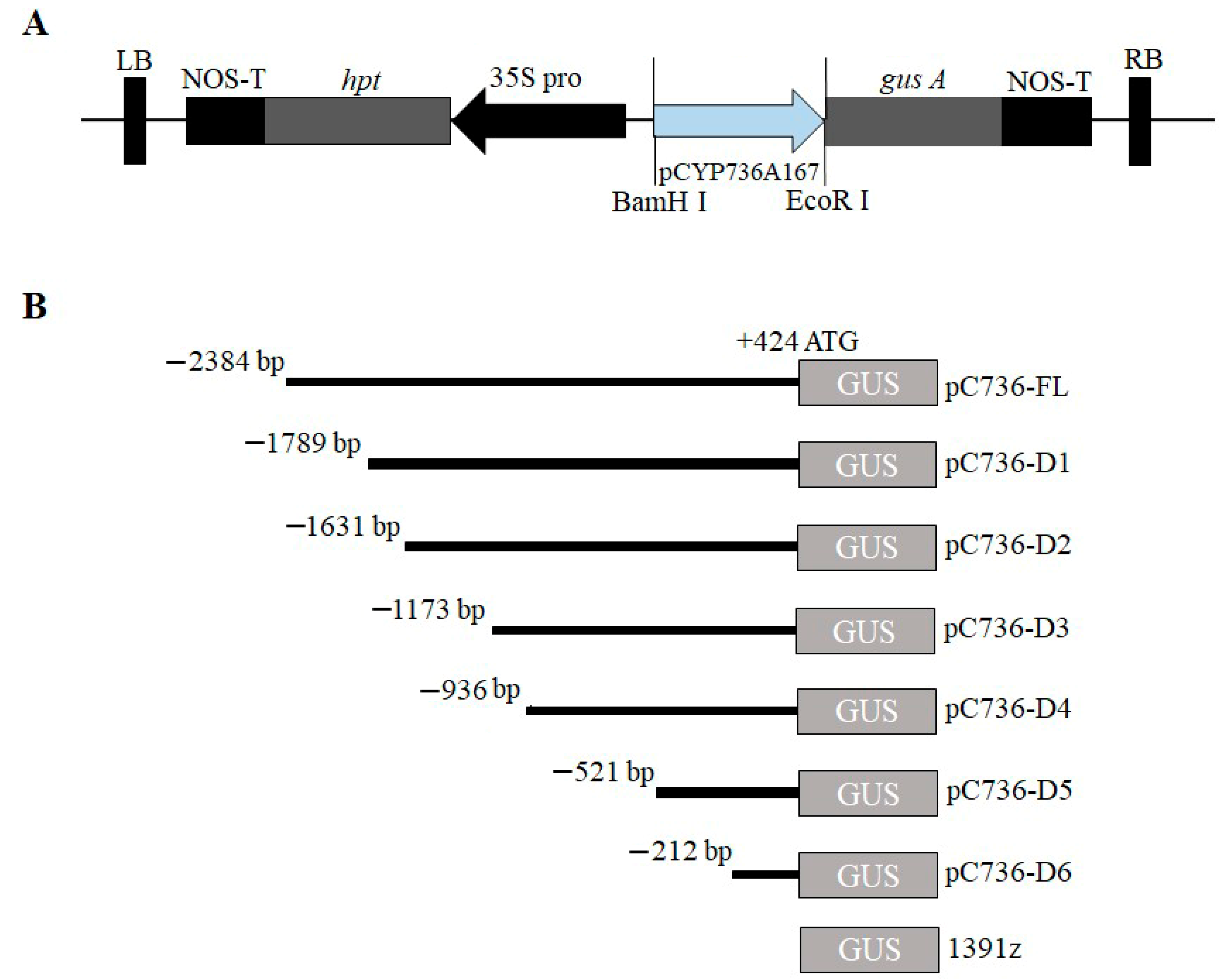
2.2. Fabrication of GUS-Containing Plasmids and Truncated Segments
2.3. Transformation and Identification of Successfully Modified Transgenic Lines of N. tabacum
2.4. Histochemical Analysis of β-Glucuronidase (GUS) Activity and Fluorometric Quantification of GUS Activity Assays
2.5. Statistical Analyses
3. Results
3.1. Characterization and Sequencing of the SaCYP736A167 Promoter Region
3.2. Tissue-Specific Expression Patterns of PSaCYP736A167
3.2.1. Screening of Positively Transgenic Tobacco Lines
3.2.2. The Tissue-Specific Expression Profiles of PSaCYP736A167
3.3. Transient Expression Verified the Activity of PSaCYP736A167
4. Deletion Analysis of PSaCYP736A167 in Transiently Expressed N. tabacum Leaves
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jones, C.G.; Keeling, C.I.; Ghisalberti, E.L.; Barbour, E.L.; Plummer, J.A.; Bohlmann, J. Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum album L. Arch. Biochem. Biophys. 2008, 477, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, M.; Xiong, Y.; Wu, J.; Teixeira da Silva, J.; Ma, G. Genome-Wide Characterization, Expression Profile Analysis of WRKY Family Genes in Santalum album and Functional Identification of Their Role in Abiotic Stress. Int. J. Mol. Sci. 2019, 20, 5676. [Google Scholar] [CrossRef] [PubMed]
- Baldovini, N.; Delasalle, C.; Joulain, D. Phytochemistry of the heartwood from fragrant Santalum species: A review. Flavour Fragr. J. 2010, 26, 7–26. [Google Scholar] [CrossRef]
- Srivastava, P.L.; Daramwar, P.P.; Krithika, R.; Pandreka, A.; Shankar, S.S.; Thulasiram, H.V. Functional Characterization of Novel Sesquiterpene Synthases from Indian Sandalwood, Santalum album. Sci. Rep. 2015, 5, 10095. [Google Scholar] [CrossRef]
- Nagegowda, D.A. Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010, 584, 2965–2973. [Google Scholar] [CrossRef]
- Jones, C.G.; Moniodis, J.; Zulak, K.G.; Scaffidi, A.; Plummer, J.A.; Ghisalberti, E.L.; Barbour, E.L.; Bohlmann, J. Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J. Biol. Chem. 2011, 286, 17445–17454. [Google Scholar] [CrossRef]
- Rani, A.; Ravikumar, P.; Reddy, M.D.; Kush, A. Molecular regulation of santalol biosynthesis in Santalum album L. Gene 2013, 527, 642–648. [Google Scholar] [CrossRef]
- Celedon, J.M.; Chiang, A.; Yuen, M.M.S.; Diaz-Chavez, M.L.; Madilao, L.L.; Finnegan, P.M.; Barbour, E.L.; Bohlmann, J. Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J. 2016, 86, 289–299. [Google Scholar] [CrossRef]
- Cirino, P.C.; Diaz-Chavez, M.L.; Moniodis, J.; Madilao, L.L.; Jancsik, S.; Keeling, C.I.; Barbour, E.L.; Ghisalberti, E.L.; Plummer, J.A.; Jones, C.G.; et al. biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS ONE 2013, 8, e75053. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Yasmeen, E.; Wang, J.; Riaz, M.; Zhang, L.; Zuo, K. Designing artificial synthetic promoters for accurate, smart, and versatile gene expression in plants. Plant Commun. 2023, 4, 100558. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Xiong, Y.; Teixeira da Silva, J.A.; Pang, J.; Zhang, T.; Yu, X.; Zhang, X.; Niu, M.; Ma, G. Molecular cloning and functional characterization of bisabolene synthetase (SaBS) promoter from Santalum album. Forests 2020, 11, 85. [Google Scholar] [CrossRef]
- Villao-Uzho, L.; Chávez-Navarrete, T.; Pacheco-Coello, R.; Sánchez-Timm, E.; Santos-Ordóñez, E. Plant promoters: Their identification, characterization, and role in gene regulation. Genes 2023, 14, 1226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-G.; Mitsukawa, N.; Oosumi, T.; Whittier, R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995, 8, 457–463. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, N. Efficient amplification of insert end sequences frombacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol. Biol. Rep. 1998, 16, 0735–9640. [Google Scholar] [CrossRef]
- Makhzoum, A.; Petit-Paly, G.; Pierre, B.S.; Bernards, M.A. Functional analysis of the DAT gene promoter using transient Catharanthus roseus and stable Nicotiana tabacum transformation systems. Plant Cell Rep. 2011, 30, 1173–1182. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Leonelli, L.; Erickson, E.; Lyska, D.; Niyogi, K.K. Transient expression in Nicotiana benthamiana for rapid functional analysis of genes involved in non-photochemical quenching and carotenoid biosynthesis. Plant J. 2016, 88, 375–386. [Google Scholar] [CrossRef]
- Jefferson, R.K.T.B.M. GUS fusion: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ljubej, V.; Karalija, E.; Salopek-Sondi, B.; Šamec, D. Effects of short-term exposure to low temperatures on proline, pigments, and phytochemicals level in Kale (Brassica oleracea var. acephala). Horticulturae 2021, 7, 341. [Google Scholar] [CrossRef]
- Muthusamy, M.; Lee, S.I. Abiotic stress-induced secondary metabolite production in Brassica: Opportunities and challenges. Front. Plant Sci. 2024, 14, 1323085. [Google Scholar] [CrossRef]
- Espinosa-Leal, C.A.; Mora-Vásquez, S.; Puente-Garza, C.A.; Alvarez-Sosa, D.S.; García-Lara, S. Recent advances on the use of abiotic stress (water, UV radiation, atmospheric gases, and temperature stress) for the enhanced production of secondary metabolites on in vitro plant tissue culture. Plant Growth Regul. 2022, 97, 1–20. [Google Scholar] [CrossRef]
- Abdollahi Mandoulakani, B.; Eyvazpour, E.; Ghadimzadeh, M. The effect of drought stress on the expression of key genes involved in the biosynthesis of phenylpropanoids and essential oil components in basil (Ocimum basilicum L.). Phytochemistry 2017, 139, 1–7. [Google Scholar] [CrossRef]
- Collins, J.M.; Wang, D. Cis-acting regulatory elements regulating CYP3A4 transcription in human liver. Pharm. Genom. 2020, 30, 107–116. [Google Scholar] [CrossRef]
- Cvetesic, N.; Lenhard, B. Core promoters across the genome. Nat. Biotechnol. 2017, 35, 123–124. [Google Scholar] [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.-J.; Xue, X.-Y.; Mao, Y.-B.; Wang, L.-J.; Chen, X.-Y. Arabidopsis MYC2 Interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Zhu, D.; Wu, Z.; Cao, G.; Li, J.; Wei, J.; Tsuge, T.; Gu, H.; Aoyama, T.; Qu, L.-J. TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol. Plant 2014, 7, 601–615. [Google Scholar] [CrossRef]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF Transcription Factor Family in Crop Improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Wei, S.; Zheng, X.; Tu, P.; Tao, F. APETALA2/ethylene-responsive factors in higher plant and their roles in regulation of plant stress response. Crit. Rev. Biotechnol. 2024, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Guo, H.; Zhang, Y.; Song, Y.; Pi, E.; Yu, C.; Zhang, L.; Dong, M.; Zheng, B.; Wang, H.; et al. Identification of potential genes that contributed to the variation in the taxoid contents between two Taxus species (Taxus media and Taxus mairei). Tree Physiol. 2017, 37, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Howat, S.; Park, B.; Oh, I.S.; Jin, Y.-W.; Lee, E.-K.; Loake, G.J. Paclitaxel: Biosynthesis, production and future prospects. New Biotechnol. 2014, 31, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Liang, Z.; Li, P.; Zhao, Z.; Chen, J. Tissue-specific chemical profiling and quantitative analysis of bioactive components of Cinnamomum cassia by combining laser-microdissection with UPLC-Q/TOF–MS. Chem. Cent. J. 2018, 12, 71. [Google Scholar] [CrossRef]
- Ye, P.; Su, J.; Lin, J.; Li, Y.; Wu, H. Identification of a cinnamoyl-CoA reductase from Cinnamomum cassia involved in trans-cinnamaldehyde biosynthesis. Planta 2024, 259, 138. [Google Scholar] [CrossRef]






| Cis-Acting Element | Signal Sequence | Start Position | Function |
|---|---|---|---|
| 3-AF1 binding site | AAGAGATATTT | −647 | light-responsive element |
| A-box | CCGTCC | −1918 | cis-acting regulatory element |
| ABRE | TACGTG | −1598 | cis-acting element involved in the abscisic acid responsiveness |
| ACGTGGC | −1572 | ||
| GCGACGTACA | −1155 | ||
| CACGTG | −526 | ||
| ACE | GCGACGTACA | −1155 | cis-acting element involved in light responsiveness |
| ACTACGTTGG | −197 | ||
| ARE | TGGTTT | −492 | cis-acting regulatory element essential for anaerobic induction |
| Box 4 | ATTAAT | −2211, −1863, −1164, −621 | part of a conserved DNA module involved in light responsiveness |
| Box I | TTTCAAA | −2452 | light responsive element |
| Box W1 | TTGACC | −1202 | fungal elicitor responsive element |
| CAT-box | GCCACT | −2159 | cis-acting regulatory element related to meristem expression |
| CCGTCC- box | CCGTCC | −1923 | cis-acting regulatory element related to meristem specific activation |
| G-box | CACGTA | −1603, −298 | cis-acting regulatory element involved in light responsiveness |
| TACGTG | −1598 | ||
| CACGTT | −1568, −312 | ||
| CACGTG | −526 | ||
| GAG-motif | AGAGATG | −648 | part of a light responsive element |
| GA-motif | AAGGAAGA | −2198 | part of a light responsive element |
| GATA-motif | GATAGGA | −1427 | part of a light responsive element |
| GCN4_motif | TGTGTCA | −2370 | cis-regulatory element involved in endosperm expression |
| GCC-like | GCCCGCC | −284 | - |
| HSE | CNNGAANNTTCNNG | −2277 | cis-acting element involved in heat stress responsiveness |
| AAAAGATTTC | −984 | ||
| AGAAAATTTG | −828 | ||
| AAAAAATTTA | −607 | ||
| AAAAAATTAC | −386 | ||
| I-box | CTCTTATGCT | −1711 | part of a light responsive element |
| LTR | CCGAAA | −471 | cis-acting element involved in low-temperature responsiveness |
| MBS | TAACTG | −1838, −1298 | MYB binding site involved in drought inducibility |
| Skn−1_motif | GTCAT | −67 | cis-acting regulatory element required for endosperm expression |
| Sp1 | CCCCCCACTA | −2133 | light responsive element |
| CCCACCTCCC | −363 | ||
| TCCCACCCACGT | −291 | ||
| TC-rich repeats | ATTTTCTTCC | −2187 | cis-acting element involved in defense and stress responsiveness |
| ATTCTCGAAC | −1186 | ||
| ATTTTCTCCC | −834 | ||
| as-2-box | GATAATGATG | −1912 | involved in shoot-specific expression and light responsiveness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Zhang, Y.; Wei, R.; Qiu, L.; Zhou, H.; Xiong, F.; Ma, G. Molecular Cloning and Functional Characterization of a Cytochrome P450 Enzyme (SaCYP736A167) Promoter from Santalum album. Forests 2024, 15, 1705. https://doi.org/10.3390/f15101705
Yan H, Zhang Y, Wei R, Qiu L, Zhou H, Xiong F, Ma G. Molecular Cloning and Functional Characterization of a Cytochrome P450 Enzyme (SaCYP736A167) Promoter from Santalum album. Forests. 2024; 15(10):1705. https://doi.org/10.3390/f15101705
Chicago/Turabian StyleYan, Haifeng, Yueya Zhang, Rongchang Wei, Lihang Qiu, Huiwen Zhou, Faqian Xiong, and Guohua Ma. 2024. "Molecular Cloning and Functional Characterization of a Cytochrome P450 Enzyme (SaCYP736A167) Promoter from Santalum album" Forests 15, no. 10: 1705. https://doi.org/10.3390/f15101705
APA StyleYan, H., Zhang, Y., Wei, R., Qiu, L., Zhou, H., Xiong, F., & Ma, G. (2024). Molecular Cloning and Functional Characterization of a Cytochrome P450 Enzyme (SaCYP736A167) Promoter from Santalum album. Forests, 15(10), 1705. https://doi.org/10.3390/f15101705






