Genome-Wide Identification, Expression Analysis, and Transcriptome Analysis of the NPF Gene Family under Various Nitrogen Conditions in Eucalyptus grandis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction, cDNA Synthesis, and Transcriptome Sequencing
2.3. Identification of NPF Genes and Analysis of Physicochemical Properties in E. grandis
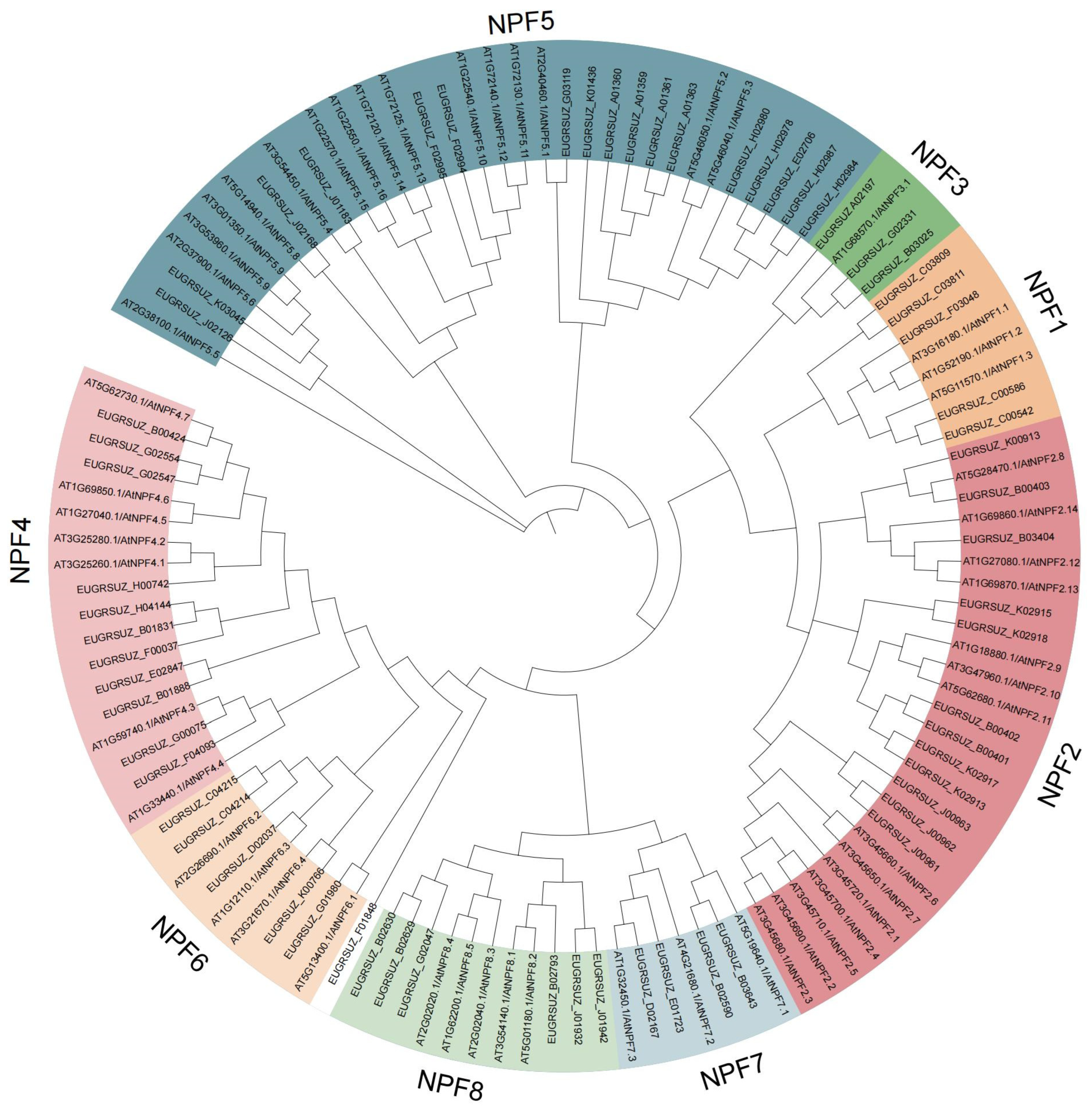
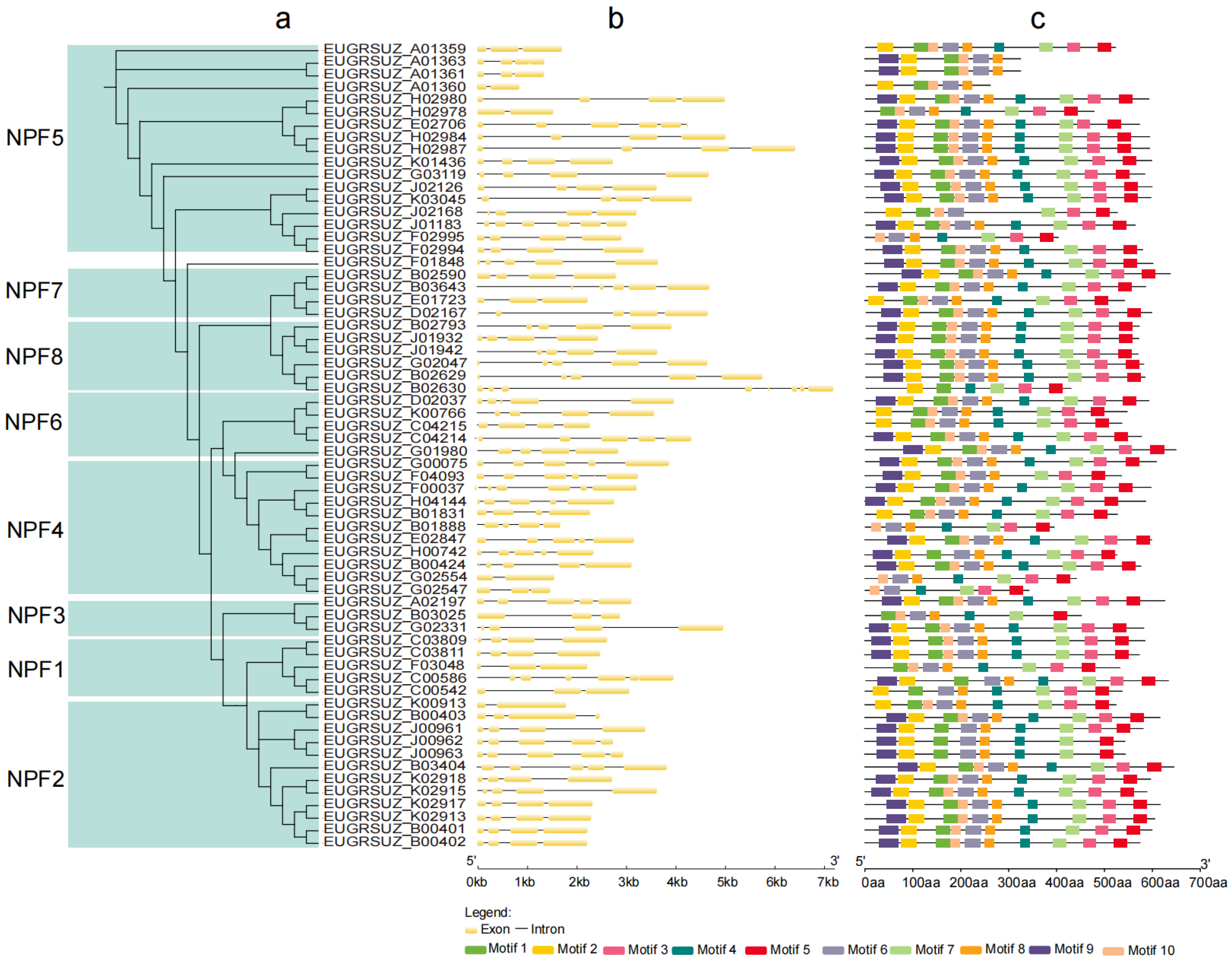
2.4. Phylogeny, Gene Structure, and Conserved Motif Analyses of EgNPF Genes
2.5. Analysis of Promoter of EgNPF Genes
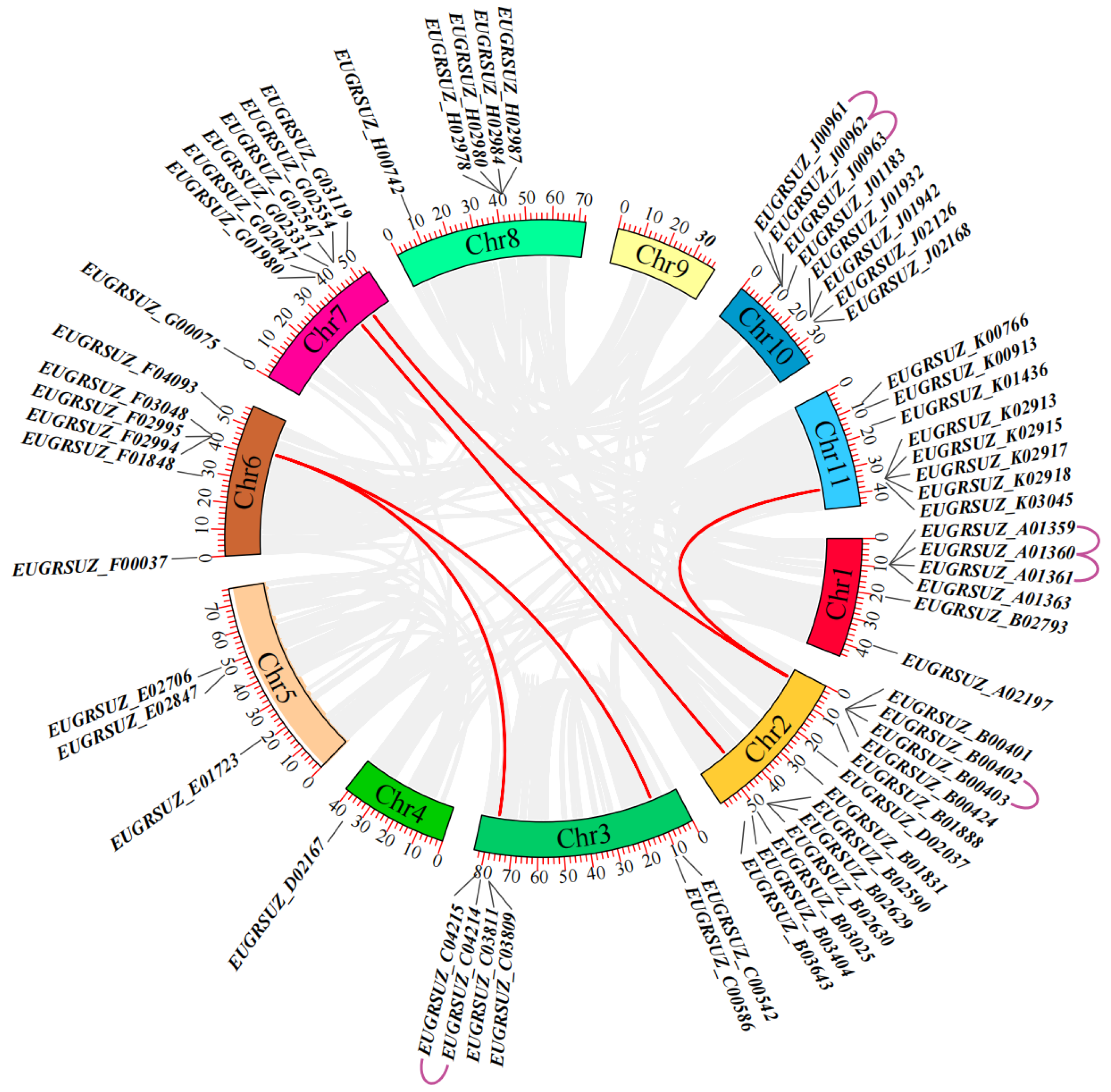
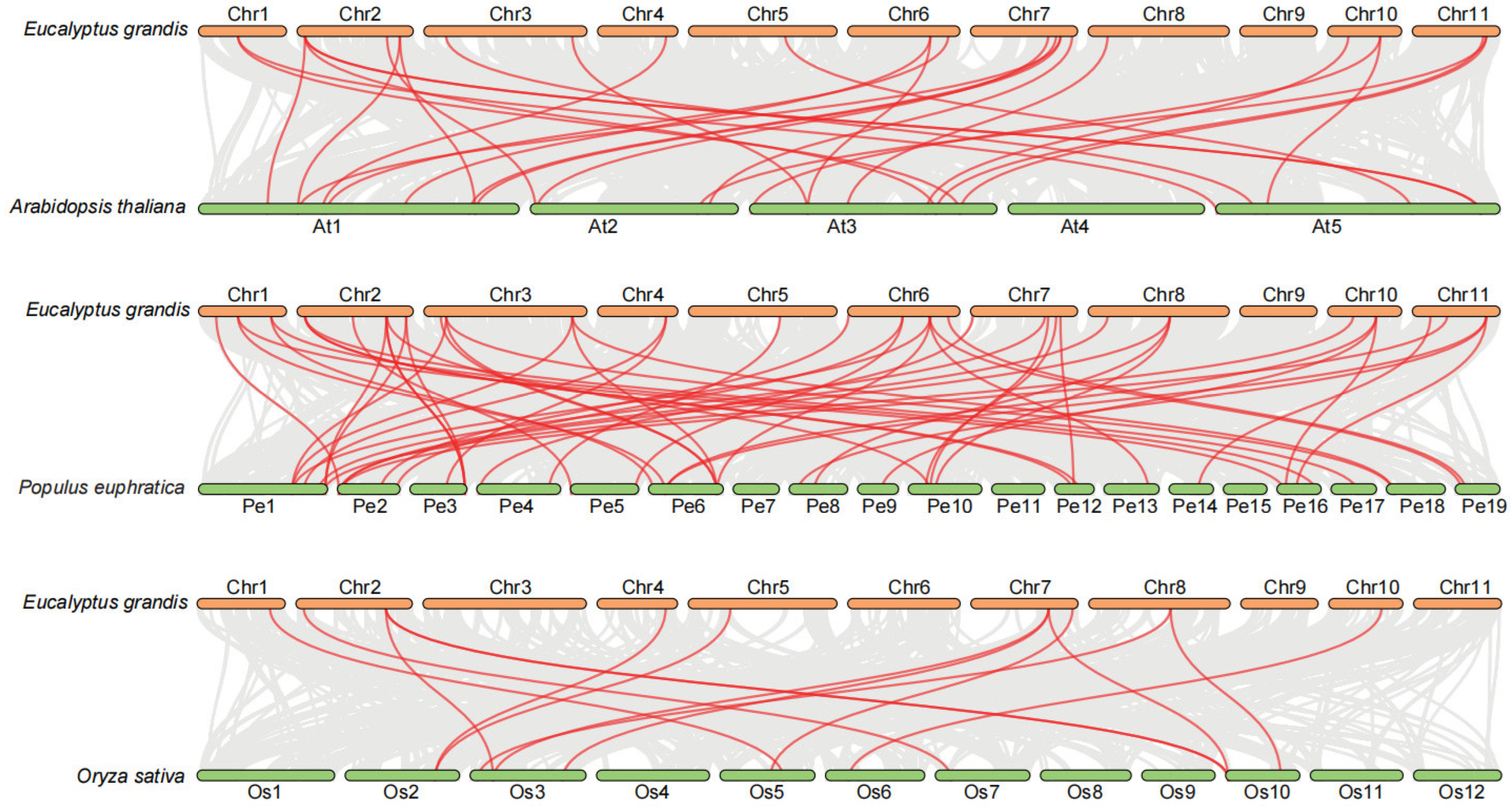
2.6. Chromosomal Localization and Collinearity Analysis of EgNPF Genes
2.7. Expression Analysis of Selected EgNPF Genes
2.8. Vector Construction and Subcellular Localization of EgNPF Gene
2.9. Statistical Analysis
3. Results
3.1. Genome-Wide Identification of the EgNPF Gene Family in E. grandis
3.2. Chromosome Location, Gene Duplication, and Synteny Analysis of EgNPF Genes
3.3. Phylogeny, Structural, and Conserved Motif Analysis of EgNPF Genes
3.4. Promoter Region Analysis of EgNPF Genes
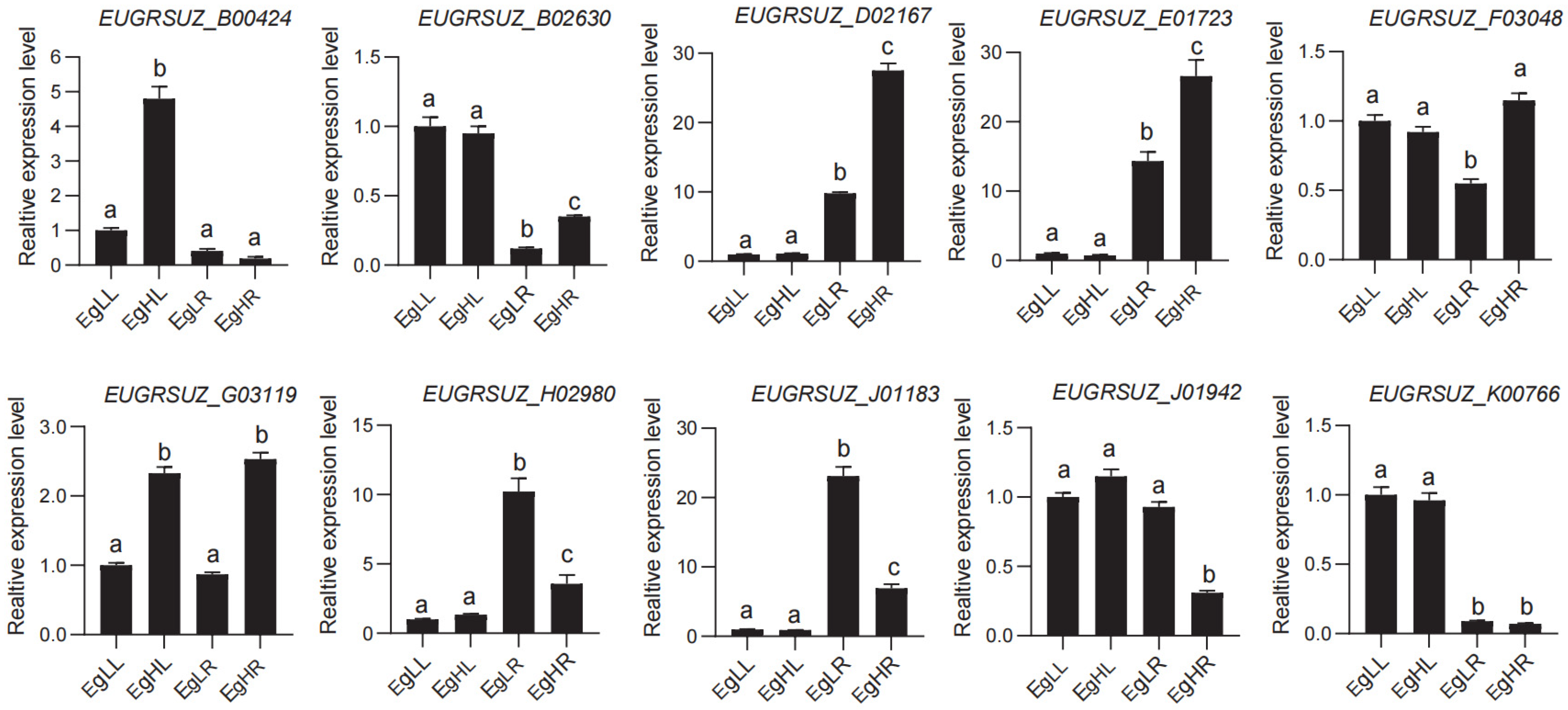
3.5. Expression Profiles of EgNPF Genes under Different N Supply Conditions
3.6. Subcellular Localization of EUGRSUZ_G03119
4. Discussion
4.1. Identification of NPF Genes in E. grandis
4.2. Chromosome Location, Duplication Events, and Synteny Relationships of EgNPF Genes
4.3. Phylogenetic Relationships, Structural Comparison, and Conserved Motifs of EgNPF Genes
4.4. Promoter Region Analysis of EgNPF Genes
4.5. Expression Profiles of EgNPF Genes under Different N Supply Conditions
4.6. Subcellular Localization of EUGRSUZ_G03119
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xuan, W.; Beeckman, T.; Xu, G. Plant nitrogen nutrition: Sensing and signaling. Curr. Opin. Plant Biol. 2017, 39, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty years from transport to signaling networks. Plant Cell 2020, 32, 2094–2119. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hu, B.; Chu, C. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef]
- Schulte-Uebbing, L.F.; Beusen, A.H.W.; Bouwman, A.F.; de Vries, W. From planetary to regional boundaries for agricultural nitrogen pollution. Nature 2022, 610, 507–512. [Google Scholar] [CrossRef]
- Kanter, D.R.; Chodos, O.; Nordland, O.; Rutigliano, M.; Winiwarter, W. Gaps and opportunities in nitrogen pollution policies around the world. Nat. Sustain. 2020, 3, 956–963. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Hsu, P.-K.; Tsay, Y.-F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Tsay, Y.-F.; Chiu, C.-C.; Tsai, C.-B.; Ho, C.-H.; Hsu, P.-K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Gojon, A.; Krouk, G.; Perrine-Walker, F.; Laugier, E. Nitrate transceptor(s) in plants. J. Exp. Bot. 2011, 62, 2299–2308. [Google Scholar] [CrossRef]
- Okamoto, M.; Kumar, A.; Li, W.; Wang, Y.; Siddiqi, M.Y.; Crawford, N.M.; Glass, A.D. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006, 140, 1036–1046. [Google Scholar] [CrossRef]
- Léran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Okamoto, M.; Vidmar, J.J.; Glass, A.D.M. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: Responses to nitrate provision. Plant Cell Physiol. 2003, 44, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Orsel, M.; Krapp, A.; Daniel-Vedele, F. Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 2002, 129, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1. 1B contributes to nitrate-use divergence between rice subspe-cies. Nat. Genet. 2015, 47, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Wang, G.; Iqbal, A.; Muhammad, N.; Wang, X.; Gui, H.; Zhang, H.; Kayoumu, M.; Li, X.; Zhang, X.; et al. Identification and expression analysis of the NPF genes in cotton. Int. J. Mol. Sci. 2022, 23, 14262. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, C.; Dong, Q.; Huang, D.; Li, C.; Li, P.; Ma, F. Genome-wide identification and analysis of apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) genes reveals MdNPF6.5 confers high capacity for nitrogen uptake under low-nitrogen conditions. Int. J. Mol. Sci. 2018, 19, 2761. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, K.; Ruan, L.; Bai, P.; Wu, L.; Wang, L.; Cheng, H. Systematic investigation and expression profiles of the nitrate transporter 1/peptide transporter family (NPF) in tea plant (Camellia sinensis). Int. J. Mol. Sci. 2022, 23, 6663. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q. Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 2020, 158, 297–307. [Google Scholar] [CrossRef]
- Marília, G.S.C.; Paulo, M.; Tiago, S.B. Insights into temperature modulation of the Eucalyptus globulus and Eucalyptus grandis antioxidant and lignification subproteomes. Phytochemistry 2017, 137, 15–23. [Google Scholar]
- Grassi, G.; Meir, P.; Cromer, R.; Tompkins, D.; Jarvis, P.G. Photosynthetic parameters in seedlings of Eucalyptus grandis as affected by rate of nitrogen supply. Plant Cell Environ. 2002, 25, 1677–1688. [Google Scholar] [CrossRef]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef]
- Qin, J.; Yue, X.L.; Fang, S.Z.; Qian, M.Y.; Zhou, S.T.; Shang, X.L.; Yang, W.X. Responses of nitrogen metabolism, photosynthetic parameter and growth to nitrogen fertilization in Cyclocarya paliurus. For. Ecol. Manag. 2021, 502, 119715. [Google Scholar] [CrossRef]
- Chen, J.; Wan, H.; Zhu, W.; Dai, X.; Yu, Y.; Zeng, C. Identification and expression analysis of the isopentenyl transferase (IPT) gene family under lack of nitrogen stress in oilseed (Brassica napus L.). Plants 2023, 12, 2166. [Google Scholar] [CrossRef]
- Lavigne, R.; Seto, D.; Mahadevan, P.; Ackermann, H.W.; Kropinski, A.M. Unifying classical and molecular taxonomic classi-fication: Analysis of the podoviridae using BLASTP-based tools. Res. Microbiol. 2008, 159, 406–414. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, J.; Qu, C.; Mo, X.; Zhang, X. Genome-wide identification of WRKY in Suaeda australis against salt stress. Forests 2024, 15, 1297. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinform. 2017, 60, 3–15. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLOS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef]
- Vega, A.; O’Brien, J.A.; Gutiérrez, R.A. Nitrate and hormonal signaling crosstalk for plant growth and development. Curr. Opin. Plant Biol. 2019, 52, 155–163. [Google Scholar] [CrossRef]
- Jiang, D.; Borg, M.; Lorković, Z.J.; Montgomery, S.A.; Osakabe, A.; Yelagandula, R.; Axelsson, E.; Berger, F. The evolution and functional divergence of the histone H2B family in plants. PLOS Genet. 2020, 16, e1008964. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, W.; Zheng, Z.; Zhang, Z.; Hua, B.; Liu, J.; Miao, M. Genome-wide identification and expression analysis of NPF genes in cucumber (Cucumis sativus L.). Plants 2023, 12, 1252. [Google Scholar] [CrossRef]
- Zhang, H.; Li, S.; Shi, M.; Wang, S.; Shi, L.; Xu, F.; Ding, G. Genome-wide systematic characterization of the NPF family genes and their transcriptional responses to multiple nutrient stresses in allotetraploid rapeseed. Int. J. Mol. Sci. 2020, 21, 5947. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Z.; Xu, G.; Bai, G.; Zhang, P.; Zhai, N.; Zheng, Q.; Chen, Q.; Liu, P.; Jin, L.; et al. Genome-wide identification and characterization of NPF family reveals NtNPF6.13 involving in salt stress in Nicotiana tabacum. Front. Plant Sci. 2022, 13, 999403. [Google Scholar] [CrossRef]
- Cheng, J.; Tan, H.; Shan, M.; Duan, M.; Ye, L.; Yang, Y.; He, L.; Shen, H.; Yang, Z.; Wang, X. Genome-wide identification and characterization of the NPF genes provide new insight into low nitrogen tolerance in Setaria. Front. Plant Sci. 2022, 13, 1043832. [Google Scholar] [CrossRef]
- Hou, X.; Wang, D.; Cheng, Z.; Wang, Y.; Jiao, Y. A near-complete assembly of an Arabidopsis thaliana genome. Mol. Plant 2022, 15, 1247–1250. [Google Scholar] [CrossRef]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Bai, H.; Euring, D.; Volmer, K.; Janz, D.; Polle, A. The nitrate transporter (NPF2.7) optimizes nitrogen use efficiency in plants under low-nitrogen availability. Plant Physiol. 2013, 162, 1246–1254. [Google Scholar]
- Xu, Q.; Wang, Y.; Sun, W.; Li, Y.; Xu, Y.; Cheng, B.; Li, X. Genome-wide identification of nitrate transporter 1/peptide transporter family (NPF) induced by arbuscular mycorrhiza in the maize genome. Physiol. Mol. Biol. Plants 2024, 30, 757–774. [Google Scholar] [CrossRef]
- He, Y.-N.; Peng, J.-S.; Cai, Y.; Liu, D.-F.; Guan, Y.; Yi, H.-Y.; Gong, J.-M. Tonoplast-localized nitrate uptake transporters involved in vacuolar nitrate efflux and reallocation in Arabidopsis. Sci. Rep. 2017, 7, 6417. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Yang, D.; Hu, Y.; Xu, J.; Li, J.; Lu, Z. Genome-Wide Identification, Expression Analysis, and Transcriptome Analysis of the NPF Gene Family under Various Nitrogen Conditions in Eucalyptus grandis. Forests 2024, 15, 1697. https://doi.org/10.3390/f15101697
Li G, Yang D, Hu Y, Xu J, Li J, Lu Z. Genome-Wide Identification, Expression Analysis, and Transcriptome Analysis of the NPF Gene Family under Various Nitrogen Conditions in Eucalyptus grandis. Forests. 2024; 15(10):1697. https://doi.org/10.3390/f15101697
Chicago/Turabian StyleLi, Guangyou, Deming Yang, Yang Hu, Jianmin Xu, Juan Li, and Zhaohua Lu. 2024. "Genome-Wide Identification, Expression Analysis, and Transcriptome Analysis of the NPF Gene Family under Various Nitrogen Conditions in Eucalyptus grandis" Forests 15, no. 10: 1697. https://doi.org/10.3390/f15101697
APA StyleLi, G., Yang, D., Hu, Y., Xu, J., Li, J., & Lu, Z. (2024). Genome-Wide Identification, Expression Analysis, and Transcriptome Analysis of the NPF Gene Family under Various Nitrogen Conditions in Eucalyptus grandis. Forests, 15(10), 1697. https://doi.org/10.3390/f15101697




