A Compatible Estimation Method for Biomass Factors Based on Allometric Relationship: A Case Study on Pinus densata Natural Forest in Yunnan Province of Southwest China
Abstract
:1. Introduction
- (1)
- To validate the compatibility between the total biomass and each component’s biomass;
- (2)
- To explore a more convenient and accurate method to calculate forest biomass via conversion coefficients.
2. Materials and Methods
2.1. Study Site
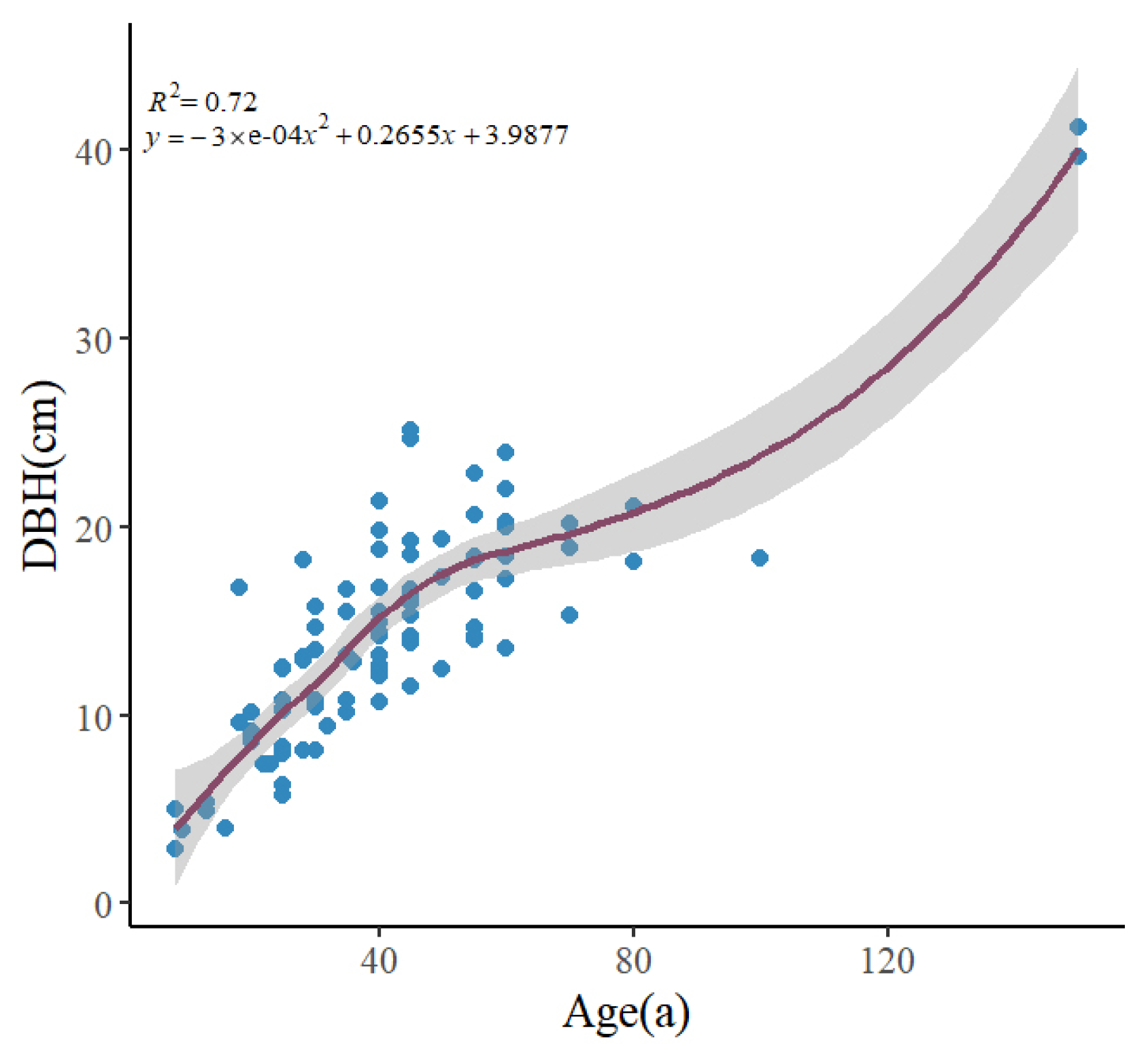
2.2. Date Investigation
2.3. Biomass and Biomass Factors
2.4. Biomass and Biomass Factor Compatibility Model
2.5. Model Evaluation and Validation
3. Results
3.1. Biomass Models for Pinus Densata Forest
3.2. BEF Models for Pinus Densata Forest
3.3. BCEF Models for Pinus Densata Forest
3.4. The Models of Rra, Rcs, Rbw, Rfb, and ρ
3.5. Comparison of Independent Models and Compatibility Models
3.6. Comparison of the BEF of the Compatibility Models and Independent Models
3.7. Comparison of the BCEF of the Independent Models and Compatibility Models
4. Discussion
4.1. The Performance of Compatibility Models
4.2. The Model Performance of the BEF and BCEF
4.3. Applicability and Limitations
5. Conclusions
- (1)
- The compatible biomass models were constructed to explore whether they could handle the compatibility problem between the total biomass and the biomass of different components. The results showed that the summation of each component’s biomass was equal to the total biomass.
- (2)
- The estimation accuracy of the compatible models was similar to that of the independent biomass models. Excepting the Bf model, which had a relatively lower accuracy, the accuracy for all of the component models was greater than 0.7. The accuracy was slightly lower than in other studies because the root biomass was added into the total biomass, which decreased the accuracy of the total biomass estimation.
- (3)
- The total error of BEF and BCEF models for most components was less than 0.1 and the estimation accuracy was higher than 0.87, indicating that reliable biomass estimations can be calculated using the corresponding conversion coefficient for the tree species in a specific region. The AGB and UGB of the forest can be accurately estimated by applying these compatible models or biomass factors (BEF, BCEF).
- (4)
- Although all ratios (Rra, Rcs, Rbw, Rfb, and ρ) can be used to calculate the BEF and BCEF, the Rcs was the most stable and easy to measure, and it was a unique ratio that could be used to infer all the other models. At the same time, the other ratios need to be composed to deduce all models.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- De Wit, H.A.; Palosuo, T.; Hylen, G.; Liski, J. A carbon budget of forest biomass and soils in southeast Norway calculated using a widely applicable method. For. Ecol. Manag. 2006, 225, 15–26. [Google Scholar] [CrossRef]
- Kurniawan, K.; Supriatna, J.; Sapoheluwakan, J.; Soesilo, T.E.B.; Mariati, S.; Gunarso, G. The analysis of forest and land fire and carbon and greenhouse gas emissions on the climate change in Indonesia. Agbioforum 2022, 24, 1–11. [Google Scholar]
- Sileshi, G.W. A critical review of forest biomass estimation models, common mistakes and corrective measures. For. Ecol. Manag. 2014, 329, 237–254. [Google Scholar] [CrossRef]
- Wu, H.; Xu, H. A review of sampling and modeling techniques for forest biomass inventory. Agric. Rural. Stud. 2023, 1, ar01010002. [Google Scholar] [CrossRef]
- Lehtonen, A.; Mäkipää, R.; Heikkinen, J.; Sievänen, R.; Liski, J. Biomass expansion factors (BEFs) for Scots pine, Norway spruce and birch according to stand age for boreal forests. For. Ecol. Manag. 2004, 188, 211–224. [Google Scholar] [CrossRef]
- Ameztegui, A.; Rodrigues, M.; Granda, V. Uncertainty of biomass stocks in Spanish forests: A comprehensive comparison of allometric equations. Eur. J. For. Res. 2022, 141, 395–407. [Google Scholar] [CrossRef]
- Ali, H.; Mohammadi, J.; Shataee Jouibary, S. Allometric Models and Biomass Conversion and Expansion Factors to Predict Total Tree-level Aboveground Biomass for Three Conifers Species in Iran. For. Sci. 2023, 69, fxad013. [Google Scholar] [CrossRef]
- Teobaldelli, M.; Somogyi, Z.; Migliavacca, M.; Usoltsev, V.A. Generalized functions of biomass expansion factors for conifers and broadleaved by stand age, growing stock and site index. For. Ecol. Manag. 2009, 257, 1004–1013. [Google Scholar] [CrossRef]
- Ochał, W.; Wertz, B.; Orzeł, S. Above-ground biomass allocation and potential carbon sink of black pine—A case study from southern Poland. Ann. For. Res. 2022, 65, 71–90. [Google Scholar] [CrossRef]
- Eggleston, H.; Buendia, L.; Miwa, K.; Ngara, T.; Tanabe, K. 2006 IPCC Guidelines for National Greenhouse Gas Inventories; Institute for Global Environmental Strategies: Hayama, Japan, 2006. [Google Scholar]
- Güner, Ş.T.; Diamantopoulou, M.J.; Poudel, K.P.; Çömez, A.; Özçelik, R. Employing artificial neural network for effective biomass prediction: An alternative approach. Comput. Electron. Agric. 2022, 192, 106596. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, H.; Xu, H. Study on establish and estimate method of compatible biomass model. Sci. Silvae Sin. 2000, 18, 19–27. [Google Scholar] [CrossRef]
- Xie, L.; Wang, T.; Miao, Z.; Hao, Y.; Dong, L.; Li, F. Considering random effects and sampling strategies improves individual compatible biomass models for mixed plantations of Larix olgensis and Fraxinus mandshurica in northeastern China. For. Ecol. Manag. 2023, 537, 120934. [Google Scholar] [CrossRef]
- Parresol, B.R. Assessing tree and stand biomass: A review with examples and critical comparisons. For. Sci. 1999, 45, 573–593. [Google Scholar]
- Parresol, B.R. Additivity of nonlinear biomass equations. Can. J. For. Res. 2001, 31, 865–878. [Google Scholar] [CrossRef]
- Affleck, D.L.; Diéguez-Aranda, U. Additive nonlinear biomass equations: A likelihood-based approach. For. Sci. 2016, 62, 129–140. [Google Scholar] [CrossRef]
- Zhang, C.; Peng, D.-L.; Huang, G.-S.; Zeng, W.-S. Developing aboveground biomass equations both compatible with tree volume equations and additive systems for single-trees in poplar plantations in Jiangsu Province, China. Forests 2016, 7, 32. [Google Scholar] [CrossRef]
- Zhao, D.; Westfall, J.; Coulston, J.W.; Lynch, T.B.; Bullock, B.P.; Montes, C.R. Additive biomass equations for slash pine trees: Comparing three modeling approaches. Can. J. For. Res. 2019, 49, 27–40. [Google Scholar] [CrossRef]
- Robinson, D. Implications of a large global root biomass for carbon sink estimates and for soil carbon dynamics. Proc. Royal Soc. B 2007, 274, 2753–2759. [Google Scholar] [CrossRef]
- Luo, W.; Lan, R.; Chen, D.; Zhang, B.; Xi, N.; Li, Y.; Fang, S.; Valverde-Barrantes, O.J.; Eissenstat, D.M.; Chu, C. Limiting similarity shapes the functional and phylogenetic structure of root neighborhoods in a subtropical forest. New Phytol. 2021, 229, 1078–1090. [Google Scholar] [CrossRef]
- Yu, X.; Ge, H.; Lu, D.; Zhang, M.; Lai, Z.; Yao, R. Comparative study on variable selection approaches in establishment of remote sensing model for forest biomass estimation. Remote Sens. 2019, 11, 1437. [Google Scholar] [CrossRef]
- Clough, B.; Dixon, P.; Dalhaus, O. Allometric relationships for estimating biomass in multi-stemmed mangrove trees. Aust. J. Bot. 1997, 45, 1023–1031. [Google Scholar] [CrossRef]
- Wang, C. Biomass allometric equations for 10 co-occurring tree species in Chinese temperate forests. For. Ecol. Manag. 2006, 222, 9–16. [Google Scholar] [CrossRef]
- Vorster, A.G.; Evangelista, P.H.; Stovall, A.E.; Ex, S. Variability and uncertainty in forest biomass estimates from the tree to landscape scale: The role of allometric equations. Carbon Balance Manag. 2020, 15, 1–20. [Google Scholar] [CrossRef]
- Puletti, N.; Grotti, M.; Ferrara, C.; Chianucci, F. Lidar-based estimates of aboveground biomass through ground, aerial, and satellite observation: A case study in a Mediterranean forest. J. Appl. Remote Sens. 2020, 14, 044501. [Google Scholar] [CrossRef]
- Chianucci, F.; Puletti, N.; Grotti, M.; Ferrara, C.; Giorcelli, A.; Coaloa, D.; Tattoni, C. Nondestructive Tree Stem and Crown Volume Allometry in Hybrid Poplar Plantations Derived from Terrestrial Laser Scanning. For. Sci. 2020, 66, 737–746. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, C.; Xu, H.; Wang, G. Estimating aboveground biomass of Pinus densata-dominated forests using Landsat time series and permanent sample plot data. J. For. Res. 2019, 30, 1689–1706. [Google Scholar] [CrossRef]
- Li, C.; Chen, D.; Wu, D.; Su, X. Design of an EIoT system for nature reserves: A case study in Shangri-La County, Yunnan Province, China. Int. J. Sustain. Dev. World Ecol. 2015, 22, 184–188. [Google Scholar] [CrossRef]
- Ma, F.; Zhao, C.; Milne, R.; Ji, M.; Chen, L.; Liu, J. Enhanced drought-tolerance in the homoploid hybrid species Pinus densata: Implication for its habitat divergence from two progenitors. New Phytol. 2010, 185, 204–216. [Google Scholar] [CrossRef]
- Wang, B.; Mao, J.; Gao, J.; ZHao, W.; Wang, X. Colonization of the Tibetan Plateau by the homoploid hybrid pine Pinus densata. Mol. Ecol. 2011, 20, 3796–3811. [Google Scholar] [CrossRef]
- Ou, G.; Li, C.; Lv, Y.; Wei, A.; Xiong, H.; Xu, H.; Wang, G. Improving aboveground biomass estimation of Pinus densata forests in Yunnan using Landsat 8 imagery by incorporating age dummy variable and method comparison. Remote Sens. 2019, 11, 738. [Google Scholar] [CrossRef]
- Zeng, W.; Luo, Q.; He, D. Research on weighting regression and modelling. Sci. Silvae Sin. 1999, 35, 5–11. [Google Scholar]
- Stovall, A.E.L.; Vorster, A.; Anderson, R.; Evangelista, P. Developing nondestructive species-specific tree allometry with terrestrial laser scanning. Methods Ecol. Evol. 2023, 14, 280–290. [Google Scholar] [CrossRef]
- Xin, S.; Mahardika, S.B.; Jiang, L. Stand-level biomass estimation for Korean pine plantations based on four additive methods in Heilongjiang province, northeast China. Cerne 2022, 28, e103008. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Fu, Y.; Lei, Y.; Zeng, W. Uncertainty Assessment in Regional-Scale Above Ground Biomass Estimation of Chinese Fir. Sci. Silvae Sin. 2014, 50, 79–86. [Google Scholar]
- Schepaschenko, D.; Moltchanova, E.; Shvidenko, A.; Blyshchyk, V.; Dmitriev, E.; Martynenko, O.; See, L.; Kraxner, F. Improved Estimates of Biomass Expansion Factors for Russian Forests. Forests 2018, 9, 312. [Google Scholar] [CrossRef]
- Langner, A.; Achard, F.; Grassi, G. Can recent pan-tropical biomass maps be used to derive alternative Tier 1 values for reporting REDD+ activities under UNFCCC? Environ. Res. Lett. 2014, 9, 124008. [Google Scholar] [CrossRef]
- Fang, J.-y.; Wang, G.G.; Liu, G.-h.; Xu, S.-l. Forest biomass of China: An estimate based on the biomass–volume relationship. Ecol. Appl. 1998, 8, 1084–1091. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, X.; Wang, X.; Ren, Y. Dissecting variation in biomass conversion factors across China’s forests: Implications for biomass and carbon accounting. PLoS ONE 2014, 9, e94777. [Google Scholar] [CrossRef]
- Jagodziński, A.M.; Dyderski, M.K.; Gęsikiewicz, K.; Horodecki, P.; Cysewska, A.; Wierczyńska, S.; Maciejczyk, K. How do tree stand parameters affect young Scots pine biomass?—Allometric equations and biomass conversion and expansion factors. For. Ecol. Manag. 2018, 409, 74–83. [Google Scholar] [CrossRef]
- Fang, J.; Chen, A.; Peng, C.; Zhao, S.; Ci, L. Changes in forest biomass carbon storage in China between 1949 and 1998. Science 2001, 292, 2320–2322. [Google Scholar] [CrossRef] [PubMed]
- Joosten, R.; Schumacher, J.; Wirth, C.; Schulte, A. Evaluating tree carbon predictions for beech (Fagus sylvatica L.) in western Germany. For. Ecol. Manag. 2004, 189, 87–96. [Google Scholar] [CrossRef]
- Haripriya, G. Estimates of biomass in Indian forests. Biomass Bioenergy 2000, 19, 245–258. [Google Scholar] [CrossRef]
- Jalkanen, A.; Mäkipää, R.; Ståhl, G.; Lehtonen, A.; Petersson, H. Estimation of the biomass stock of trees in Sweden: Comparison of biomass equations and age-dependent biomass expansion factors. Ann. For. Sci. 2005, 62, 845–851. [Google Scholar] [CrossRef]
- Sullivan, M.J.P.; Talbot, J.; Lewis, S.L.; Phillips, O.L.; Qie, L.; Begne, S.K.; Chave, J.; Cuni-Sanchez, A.; Hubau, W.; Lopez-Gonzalez, G.; et al. Diversity and carbon storage across the tropical forest biome. Sci. Rep. 2017, 7, 39102. [Google Scholar] [CrossRef]
- Zeng, W.; Zhang, L.; Chen, X.; Cheng, Z.; Ma, K.; Li, Z. Construction of compatible and additive individual-tree biomass models for Pinus tabulaeformis in China. Can. J. For. Res. 2017, 47, 467–475. [Google Scholar] [CrossRef]

| Components | Model Forms | Fitting | Test | ||||
|---|---|---|---|---|---|---|---|
| n | R2 | RMSE | n | MRE | MAE | ||
| Wood | Bw = 0.030 × DBH1.746 × H1.021 | 75 | 0.990 | 28.921 | 25 | −2.701 | 12.067 |
| Bark | Bb = 0.0034 × DBH1.222 × H1.640 | 75 | 0.898 | 11.175 | 25 | −9.796 | 31.204 |
| Foliage | Bf = 0.045 × DBH2.498 × H−1.143 | 75 | 0.674 | 4.389 | 25 | 13.115 | 41.071 |
| Branches | Bbr = 0.170 × DBH2.007 × H−0.386 | 75 | 0.831 | 18.989 | 25 | −18.635 | 42.069 |
| Roots | Br = 0.025 × DBH2.221 × H0.082 | 75 | 0.999 | 2.495 | 25 | −2.357 | 6.368 |
| Components | Fitting (n = 79) | Test (n = 19) | ||||||
|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | SD | Min | Max | Mean | SD | |
| Dg (cm) | 2.854 | 41.272 | 14.486 | 6.405 | 3.990 | 24.722 | 14.581 | 5.611 |
| Hm (m) | 2.200 | 24.296 | 10.610 | 4.453 | 2.821 | 15.515 | 10.178 | 3.490 |
| Vw (m3 ha−1) | 1.058 | 719.049 | 259.727 | 168.406 | 19.031 | 701.766 | 251.792 | 171.119 |
| Components | Number of Samples | Model Parameter Estimates | R2 | MSE | ||
|---|---|---|---|---|---|---|
| a | b | c | ||||
| Bw | 79 | 6.1035 | −0.9211 | 2.1432 | 0.7476 | 827.7531 |
| Bb | 79 | 0.8630 | 0.2662 | 0.8188 | 0.6106 | 22.2806 |
| Bs | 79 | 6.9594 | −0.7781 | 1.9851 | 0.7372 | 1072.9630 |
| Bbr | 79 | 15.4272 | −0.4155 | 0.8331 | 0.3382 | 137.6777 |
| Bf | 79 | 2.8332 | −0.1242 | 0.5168 | 0.1603 | 9.2828 |
| Bc | 79 | 18.2257 | −0.3665 | 0.7808 | 0.3207 | 199.0442 |
| Ba | 79 | 18.0495 | −0.6584 | 1.6106 | 0.6649 | 2067.4490 |
| Br | 79 | 2.1598 | −0.2416 | 1.0116 | 0.4684 | 23.3764 |
| Bt | 79 | 20.2150 | −0.6259 | 1.5621 | 0.6540 | 2482.2650 |
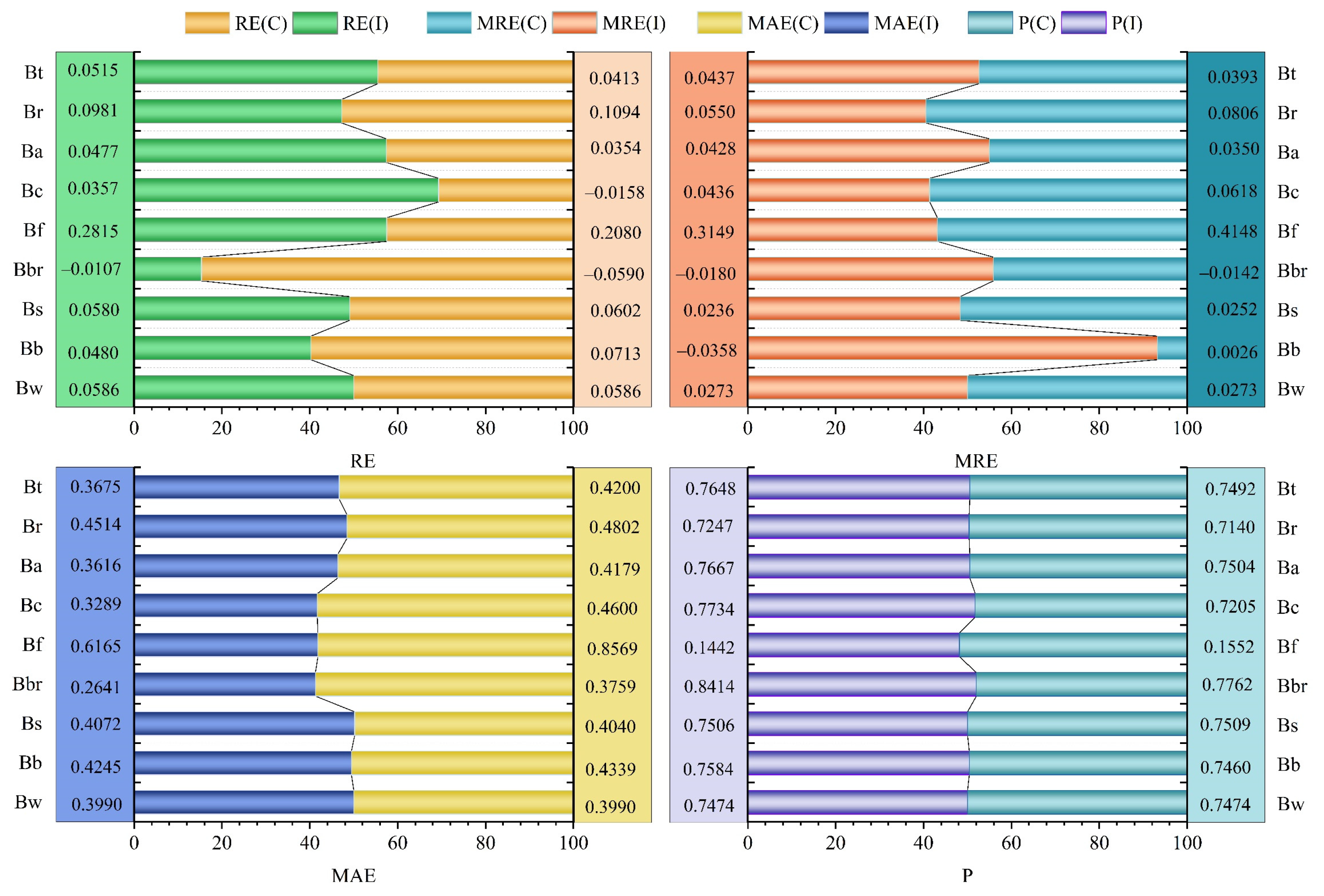
| Components | RE | MRE | MAE | p |
|---|---|---|---|---|
| Bw | 0.0586 | 0.0273 | 0.3990 | 0.7474 |
| Bb | 0.0713 | 0.0026 | 0.4339 | 0.7460 |
| Bs | 0.0602 | 0.0252 | 0.4040 | 0.7509 |
| Bbr | −0.0590 | −0.0142 | 0.3759 | 0.7762 |
| Bf | 0.2080 | 0.4148 | 0.8569 | 0.1552 |
| Bc | −0.0158 | 0.0618 | 0.4600 | 0.7205 |
| Ba | 0.0354 | 0.0350 | 0.4179 | 0.7504 |
| Br | 0.1094 | 0.0806 | 0.4802 | 0.7140 |
| Bt | 0.0413 | 0.0393 | 0.4200 | 0.7492 |
| Components | Number of Samples | Model Parameter Estimates | R2 | MSE | ||
|---|---|---|---|---|---|---|
| a | b | c | ||||
| BEFw | 79 | 0.9173 | −0.1492 | 0.1474 | 0.6538 | 0.0003 |
| BEFb | 79 | 0.0927 | 0.9490 | −0.9456 | 0.6412 | 0.0003 |
| BEFbr | 79 | 7.6920 | −0.1331 | −1.1075 | 0.9944 | 0.0012 |
| BEFf | 79 | 2.2482 | −0.0423 | −1.4085 | 0.9711 | 0.0004 |
| BEFc | 79 | 9.8850 | −0.1190 | −1.1590 | 0.9928 | 0.0024 |
| BEFa | 79 | 6.8847 | −0.1497 | −0.4803 | 0.9382 | 0.0202 |
| BEFr | 79 | 0.2837 | 0.5112 | −0.9008 | 0.7026 | 0.0006 |
| BEFt | 79 | 7.0382 | −0.0947 | −0.5152 | 0.9423 | 0.0211 |
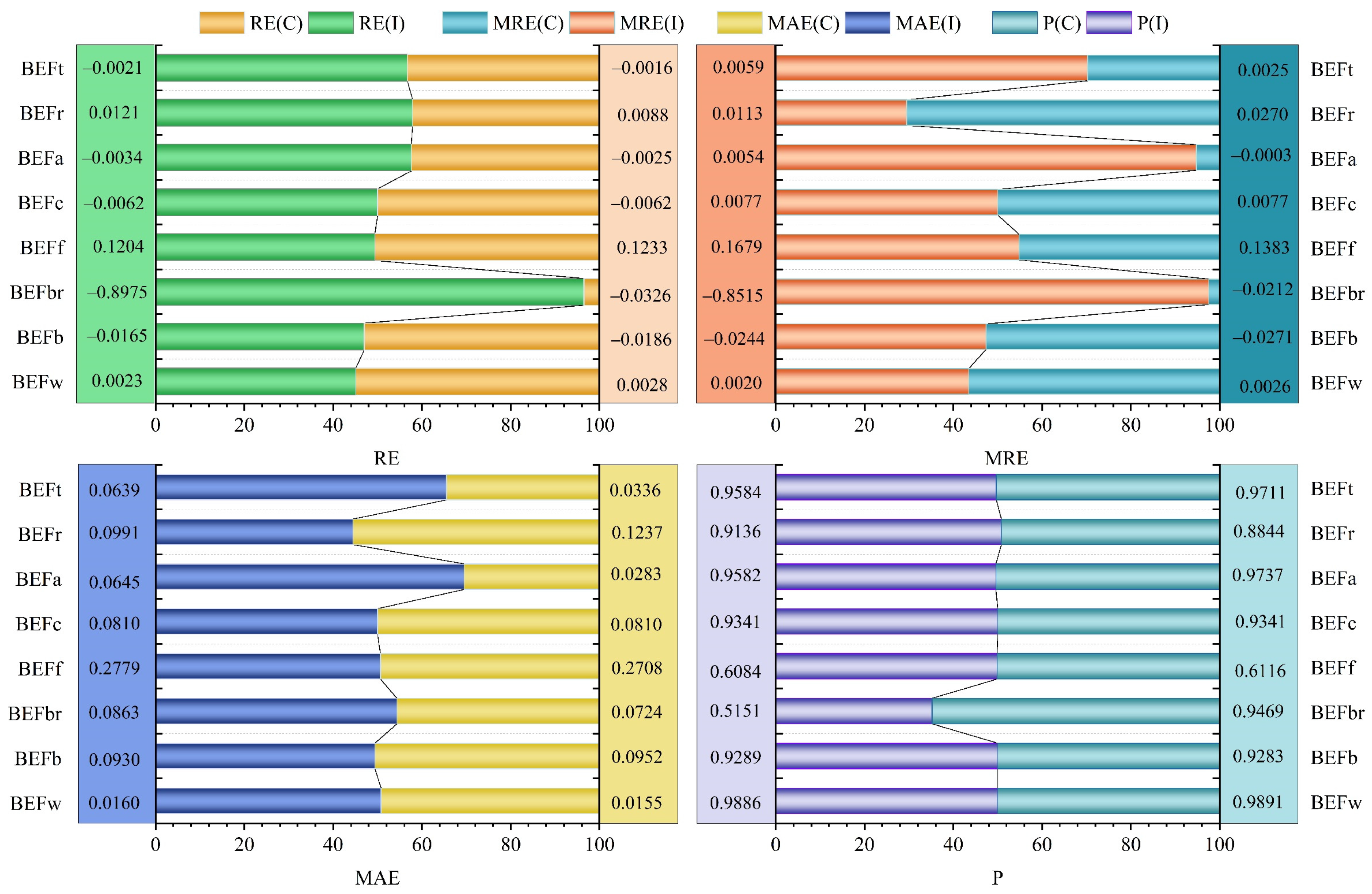
| Components | RE | MRE | MAE | p |
|---|---|---|---|---|
| BEFw | 0.0023 | 0.0020 | 0.0160 | 0.9886 |
| BEFb | −0.0165 | −0.0244 | 0.0930 | 0.9289 |
| BEFbr | −0.8975 | −0.8515 | 0.0863 | 0.5151 |
| BEFf | 0.1204 | 0.1679 | 0.2779 | 0.6084 |
| BEFc | −0.0062 | 0.0077 | 0.0810 | 0.9341 |
| BEFa | −0.0034 | 0.0054 | 0.0645 | 0.9582 |
| BEFr | 0.0121 | 0.0113 | 0.0991 | 0.9136 |
| BEFt | −0.0021 | 0.0059 | 0.0639 | 0.9584 |
| Components | Number of Samples | Model Parameter Estimates | R2 | MSE | ||
|---|---|---|---|---|---|---|
| a | b | c | ||||
| BCEFw | 79 | 0.4462 | −0.3730 | 0.2952 | 0.8700 | 0.0001 |
| BCEFb | 79 | 0.0446 | 0.7400 | −0.8104 | 0.5511 | 0.0000 |
| BCEFs | 79 | 0.4862 | −0.2233 | 0.1474 | 0.8298 | 0.0001 |
| BCEFbr | 79 | 3.8870 | −0.3726 | −0.9618 | 0.9945 | 0.0002 |
| BCEFf | 79 | 1.1394 | −0.2769 | −1.2718 | 0.9785 | 0.0001 |
| BCEFc | 79 | 4.9988 | −0.3576 | −1.0146 | 0.9936 | 0.0004 |
| BCEFa | 79 | 3.5430 | −0.3987 | −0.3317 | 0.9435 | 0.0036 |
| BCEFr | 79 | 0.1380 | 0.2986 | −0.7665 | 0.7501 | 0.0001 |
| BCEFt | 79 | 3.6139 | −0.3423 | −0.3668 | 0.9468 | 0.0038 |
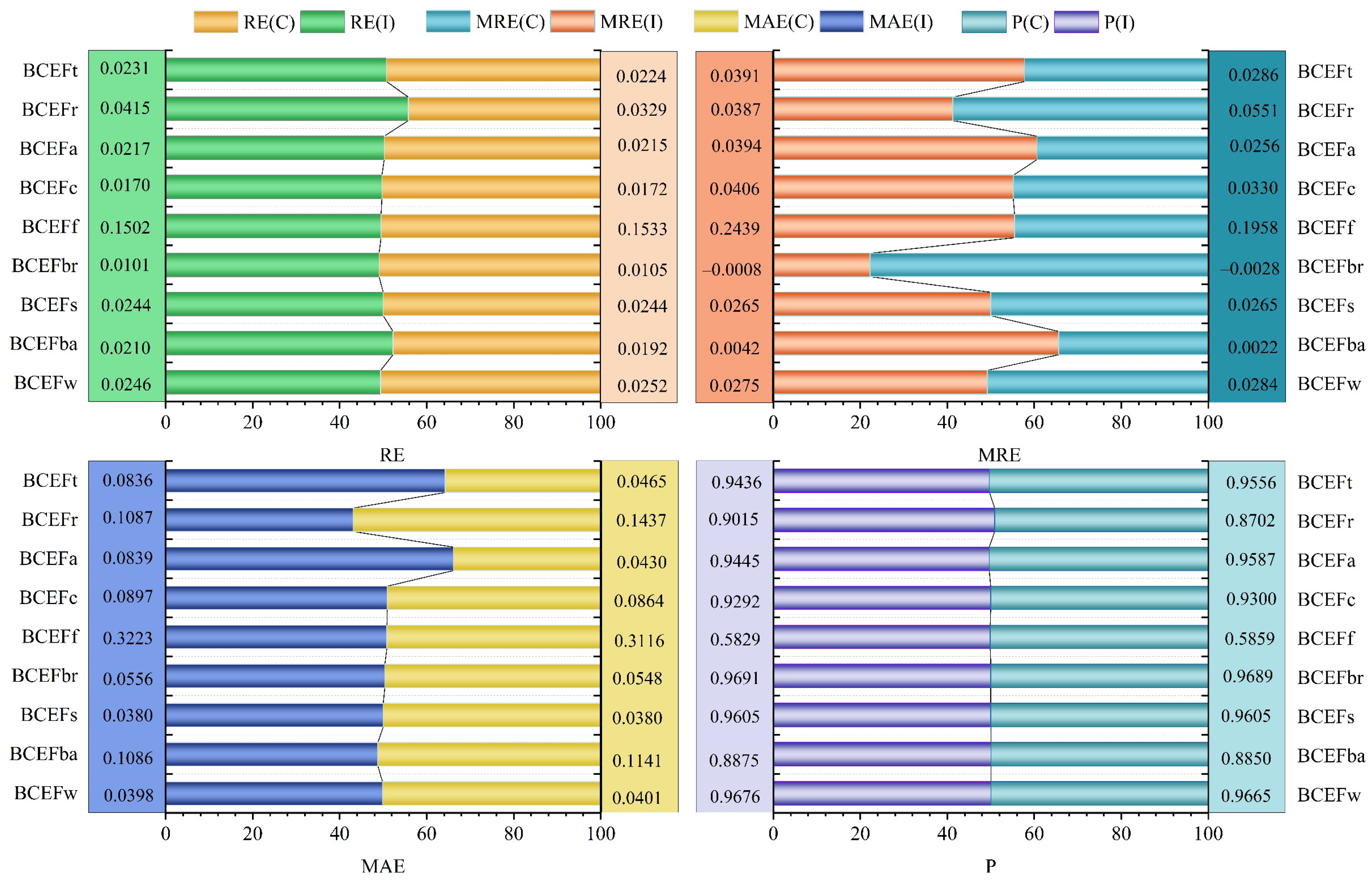
| Components | RE | MRE | MAE | p |
|---|---|---|---|---|
| BCEFw | 0.0246 | 0.0275 | 0.0398 | 0.9676 |
| BCEFb | 0.0210 | 0.0042 | 0.1086 | 0.8875 |
| BCEFs | 0.0244 | 0.0265 | 0.0380 | 0.9605 |
| BCEFbr | −0.0101 | −0.0008 | 0.0556 | 0.9691 |
| BCEFf | 0.1502 | 0.2439 | 0.3223 | 0.5829 |
| BCEFc | 0.0170 | 0.0406 | 0.0897 | 0.9292 |
| BCEFa | 0.0217 | 0.0394 | 0.0839 | 0.9445 |
| BCEFr | 0.0415 | 0.0387 | 0.1087 | 0.9015 |
| BCEFt | 0.0231 | 0.0391 | 0.0836 | 0.9436 |
| Components | Number of Samples | Model Parameter Estimates | R2 | MSE | ||
|---|---|---|---|---|---|---|
| a | b | c | ||||
| Rra | 79 | 0.0654 | 0.4656 | −0.4078 | 0.1417 | 0.0003 |
| Rcs | 79 | 9.8850 | −0.1190 | −1.1590 | 0.9928 | 0.0024 |
| Rfb | 79 | 0.2494 | 0.1625 | −0.3014 | 0.1122 | 0.0021 |
| Rbw | 79 | 0.1013 | 1.0878 | −1.0806 | 0.6125 | 0.0006 |
| ρ | 79 | 0.4862 | −0.2233 | 0.1474 | 0.8298 | 0.0001 |
| Components | RE | MRE | MAE | p |
|---|---|---|---|---|
| Rra | 0.0345 | 0.0269 | 0.1166 | 0.8930 |
| Rcs | −0.0062 | 0.0077 | 0.0810 | 0.9341 |
| Rfb | 0.2559 | 0.2273 | 0.3426 | 0.2964 |
| Rbw | −0.0144 | −0.0266 | 0.1116 | 0.9098 |
| ρ | 0.0244 | 0.0265 | 0.0380 | 0.9605 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Xu, H.; Wu, Y.; Zhang, X.; Liu, C.; Lu, C.; Yu, Z.; Ou, G. A Compatible Estimation Method for Biomass Factors Based on Allometric Relationship: A Case Study on Pinus densata Natural Forest in Yunnan Province of Southwest China. Forests 2024, 15, 26. https://doi.org/10.3390/f15010026
Li W, Xu H, Wu Y, Zhang X, Liu C, Lu C, Yu Z, Ou G. A Compatible Estimation Method for Biomass Factors Based on Allometric Relationship: A Case Study on Pinus densata Natural Forest in Yunnan Province of Southwest China. Forests. 2024; 15(1):26. https://doi.org/10.3390/f15010026
Chicago/Turabian StyleLi, Wenfang, Hui Xu, Yong Wu, Xiaoli Zhang, Chunxiao Liu, Chi Lu, Zhibo Yu, and Guanglong Ou. 2024. "A Compatible Estimation Method for Biomass Factors Based on Allometric Relationship: A Case Study on Pinus densata Natural Forest in Yunnan Province of Southwest China" Forests 15, no. 1: 26. https://doi.org/10.3390/f15010026
APA StyleLi, W., Xu, H., Wu, Y., Zhang, X., Liu, C., Lu, C., Yu, Z., & Ou, G. (2024). A Compatible Estimation Method for Biomass Factors Based on Allometric Relationship: A Case Study on Pinus densata Natural Forest in Yunnan Province of Southwest China. Forests, 15(1), 26. https://doi.org/10.3390/f15010026






