Abstract
As remote sensing transforms forest and urban tree management, automating tree species classification is now a major challenge to harness these advances for forestry and urban management. This study investigated the use of structural bark features from terrestrial laser scanner point cloud data for tree species identification. It presents a novel mathematical approach for describing bark characteristics, which have traditionally been used by experts for the visual identification of tree species. These features were used to train four machine learning algorithms (decision trees, random forests, XGBoost, and support vector machines). These methods achieved high classification accuracies between 83% (decision tree) and 96% (XGBoost) with a data set of 85 trees of four species collected near Krakow, Poland. The results suggest that bark features from point cloud data could significantly aid species identification, potentially reducing the amount of training data required by leveraging centuries of botanical knowledge. This computationally efficient approach might allow for real-time species classification.
1. Introduction
The identification of tree species is a fundamental aspect of measuring and monitoring forests and urban trees, providing critical insights into ecosystem dynamics, species-specific ecosystem services, and economic considerations such as timber values. Accurately identifying tree species is crucial for making informed decisions, ecological conservation, and sustainable resource management in urban and rural environments. With the growing adoption of remote sensing technologies in the fields of forestry and urban forestry, the task of automatic species classification has gained paramount importance.
Traditionally, species identification for inventories has relied on labor-intensive field surveys and manual observations by trained foresters or botanists. However, the emergence of remote sensing technologies has revolutionized this process by enabling large-scale non-invasive data collection. In this context, the automatic classification of tree species has become a critical challenge and a focus of extensive research efforts.
To date, several methods have been successfully employed for tree species classification, with many of them primarily relying on RGB images as the primary data source. While these methods have yielded promising results, they often overlook valuable information contained within the point cloud structure of a tree’s stem. Point clouds, generated through LiDAR (Light Detection and Ranging) or photogrammetry, offer a three-dimensional representation of the forest environment, including the stems of individual trees, when using a terrestrial platform [1].
Laser scanning is used in forestry and urban forestry for various applications, including forest inventory, tree mapping, and monitoring of individual tree characteristics. In forestry, laser scanning techniques, such as Airborne Laser Scanning (ALS), Terrestrial Laser Scanning (TLS), including mobile LiDAR systems such as handheld systems, and Mobile Laser Scanning (MLS), have been widely investigated for applications in forest inventory [2,3]. These techniques provide efficient means for acquiring detailed three-dimensional (3D) data from vegetation, enabling the extraction of tree and forest parameters such as tree height, crown dimensions, and biomass [4,5,6,7,8,9,10,11,12,13,14,15,16].
In urban forestry, laser scanning is used for mapping and monitoring single tree characteristics, providing a convenient tool for measuring tree attributes in cities and urban forests [12,17,18,19,20]. ALS can be used to generate high-resolution spatially explicit maps of urban forest structure, including the detection, mapping, and characterization of individual trees [21]. TLS has also been applied in urban forestry for capturing detailed 3D tree structures and monitoring tree growth and health [22,23].
Tree species classification using LiDAR data is typically based on extracting specific features from the point cloud data, such as geometric, radiometric, and full-waveform features [24]. These features can be used to differentiate between tree species based on their unique structural and reflectance properties. Researchers have developed various methods for tree species classification using LiDAR data, including deep learning models [25,26,27,28,29], individual tree segmentation and shape fitting [30], and also combined LiDAR with data from other sensor types like hyper- or multispectral data [31].
For example, a study using a 3D deep learning approach achieved an overall accuracy of 92.5% in tree species classification directly using ALS point clouds to derive the structural features of trees [27]. Another study proposed a method based on the crown shape of segmented individual trees extracted from ALS point clouds to identify tree species [30]. These and other studies, e.g., [24,25,26,28,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58], demonstrate the potential of LiDAR technology in providing accurate and efficient species classification in forestry and ecological applications.
However, individual tree segmentation and shape fitting methods using LiDAR data for species classification can face challenges in dense forests [32], be sensitive to data quality [59], have limitations in capturing species-specific features [47], and require significant computational resources [60]. Integrating additional data sources and developing algorithms that are more advanced can help address these drawbacks and improve the accuracy and efficiency of tree species classification using LiDAR data.
While textural features of tree organs like leaves or flowers vary notably because of seasonal change, the morphology of tree bark remains constant across seasons. Most previous work has been based on RGB images of tree stems. While this approach has been very successful, reaching accuracies well above 90% [61,62], the quality of such images can vary with contrasting or insufficient lighting conditions of the trunk, and such images can only be acquired during daylight hours. Although LiDAR point clouds should not be affected by lighting as much as RGB images are and can be collected even at night, the potential of utilizing structural bark characteristics derived from LiDAR point cloud data for tree species identification remains largely untapped.
While [44,63] have already used structural bark features, this paper tries to address this critical gap by exploring the utility of structural features traditionally used by experts to identify tree species based on their bark and stem characteristics. Apart from bark color, botanists use structural features like ridges, fissures, peeling, or scales, which have been described, amongst others, in [64]. To our knowledge, this is the first attempt to describe these structures mathematically and to derive features for machine learning from this description.
We test whether machine learning models can differentiate between various common Central European tree species (Acer platanoides L., Fraxinus excelsior L., Robinia pseudoacacia L., Larix decidua Mill., Fagus sylvatica L.). Our study goes beyond the conventional reliance on RGB images and demonstrates the potential of adding LiDAR point cloud information to enhance the automatic classification of tree species.
If successful, this approach has the potential to make a significant contribution to the field of remote sensing applications in forestry and urban forestry. Providing an additional tool for accurate tree species identification will have implications for improved forest management, conservation efforts, and sustainable urban planning.
2. Materials and Methods
2.1. Trees and Scanning
LiDAR point clouds of 85 trees (Table 1) were acquired from mature trees in forests around Krakow by the company ProGea 4D, Poland, using a Faro laser scanner.

Table 1.
Number of individuals sampled per species.
TLS point clouds were not very noisy. There were no moving objects in the scans (e.g., people or animals), and the scanning was performed on a windless day. The only noise that was created was probably in the tree canopy, at the edges of the leaves. Filter tools available in the Faro Scene 2023 software were completely sufficient to avoid processing the TLS cloud in external software (e.g., SOR filter in Cloud Compare 2.12.4).
All trees had fully developed mature bark features.
2.2. Feature Creation
The bark analysis method for tree species identification that is presented here is based on set theory and algebraic mathematical methods. The bark is characterized by several parameters. These parameters form a model of the bark. More precisely, it is a vector space that is provided with a distance measure, a metric space [65,66,67].
Each tree and its associated bark structure are represented as a vector in a vector space. The elements of a vector are the parameters listed in Table 2.

Table 2.
Features derived from point clouds.
Further information exists for each point, which is determined using LiDAR scanning. This includes, for example, time stamps and color values. However, these are of secondary importance for the bark analysis. The tree species therefore form a real subset of . Each tree species has a number of comparable elements per vector. The metric of the vector space is required to enable the tree species sets to be separated by a distance value.
To analyze tree bark surfaces using digital point clouds, they must be described and quantified through standardized methods. This project aims to propose a method for standardization that evaluates the parameters of rib structure, spacing, spatial orientation, and appearance.
Definition 1.
Ribs are elevations that differ from a “smooth” bark surface by an additive positive value ε as the difference value.
The clusters represent open subsets. This is the case because a cluster does not contain all accumulation points. The general concept of a vector space can therefore be made more precise. It is a topological space M in which the separation axiom [68], the Hausdorff separation axiom, applies. This means that no cluster exists that is identical to either its predecessor or its successor. Mathematically speaking, a point cloud in which only disjoint clusters exist is a Hausdorff space.
These clusters must be adjacent, i.e., they must be spatially close to each other and parallel to the trunk diameter, the abscissa axis. A point, as defined in this model, is a 15-dimensional vector. An important element is the cluster size . This specification makes it possible to form clusters with similar properties that satisfy the following conditions:
with x ∈ trunk diameter, y ∈ bark depth, z ∈ trunk length, and
The direction of x, y, and z coordinates is illustrated in Figure 1. By using clusters with similar sizes and properties, it is possible to trace structures along z. All clusters with similar structures represent elements of a meta-cluster, which in turn represents the bark rib. The clustering of the meta-cluster and the analysis of the bark structure is carried out using the AI software Dylogos 2.0. With the help of the software, it is possible to analyze the individual data of a point cloud.
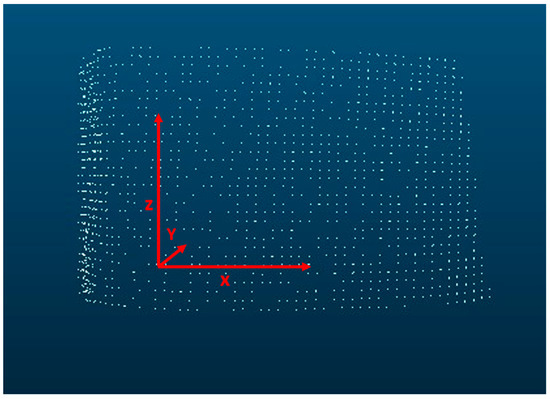
Figure 1.
Illustration of the orientation of x, y, and z coordinates.
For the study of the similarity of ribs, the following properties are considered:
- Rib width;
- Depth between two adjacent ribs;
- Distance between the ribs;
- Shape of the ribs, fissures, roughness, etc.
In the model described here, 11 evaluation criteria are defined which serve to classify the species (see Table 2).
These 11 evaluation criteria plus the three spatial axes and the cluster size form the 15 elements of the vector and thus of the cluster.
The following cluster axioms constitute the model:
- Axiom 1: Each model has at least one cluster.
- Axiom 2: Every model has no zero cluster.
- Axiom 3: More than one successor cluster can exist.
- Axiom 4: Each cluster contains information about its predecessors.
- Axiom 5: If no subsequent cluster exists, the number of predecessors defines the length of the ridge. This forms the 15th element of the cluster vector.
- Axiom 6: All clusters with similar properties and a spatial proximity form elements of a bark rib.
Figure 2 (Figure 2 and Figure 3 are visualizations of the clusters identified by Dylogos) of Robinia bark will explain this in more detail. It shows the bark structure as a cluster cloud. Based on this, axioms 1–6 will be shown in the following. Axioms 1 and 2 are fulfilled. The first cluster in the red circle has several successors (axiom 3 and 4).
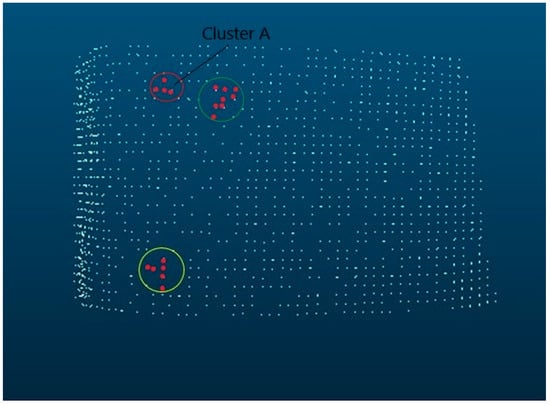
Figure 2.
Robinia pseudoacacia bark as cluster representation.
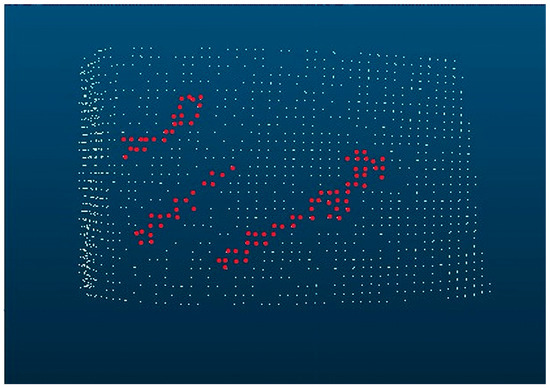
Figure 3.
Robinia pseudoacacia bark with diagonal, vertical, and branching alignments.
The cluster A is the last cluster of the row (axiom 5) and is the end element of the rib (axiom 6). The elements of the green circle form a meta-cluster structure. By this structure, we can also recognize the spatial orientation. In this case, it is diagonal. The clusters in the yellow circle are examples of horizontal as well as vertical structures. The middle cluster in the second row of the red circle is an example of branching. Structures that are more complex may have a combination of features. Each bark is individual, like a fingerprint, but shows characteristic relationships per species. These are shown in the figure below as an example for a Robinia pseudoacacia. The data shown therein are an excerpt from the training data.
In Figure 3, which is based on Figure 2, the diagonal and vertical alignments as well as the branching have been highlighted.
Areas without cluster points are areas that lie below the average bark surface. These points are suppressed in the display but are taken into account for the evaluation of roughness. The average bark surface is determined using average values from the decision grid and forms a meta-level for the entire trunk.
Figure 3 shows all clusters that have a positive distance from the average bark surface, the ϵ value. Figure 4 shows a section of Figure 2, in which the meta-plane, the average bark surface, and an example of ε are plotted.
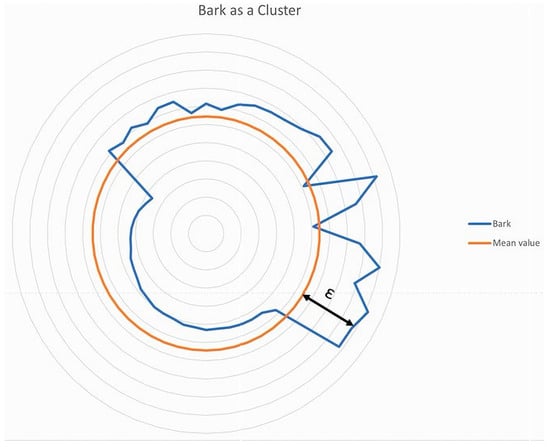
Figure 4.
Robinia pseudoacacia bark with the mean value (ochre color).
Figure 4 can be generated from Figure 3, using the mapping rule with = mean value and The mean value represents the average roughness of the meta-cluster, i.e., the reference bark. The circle is the graphical representation of the mean value of the meta-cluster. The blue bark curve, which is shown as a deviation from the mean value of the meta-cluster, is not formed by individual points but by clusters. The clusters represent a large number of points. All cluster elements that lie above the meta level are considered for the analysis; these are the elements of the set . All cluster elements are determining elements of the evaluation and recognition of the bark using this method. The only value that has no reference to the meta-level is that of the gradient. This value is an evaluation of the surface of each individual ridge. The evaluation consists of looking at each rib cluster and its orientation in space in comparison to its predecessor and successor. The number of successor clusters that show positive, negative, or no change along the Y-axis is decisive for the evaluation.
The LiDAR data were available as LAS files. For the bark analysis, a trunk section of 2 m was used, which was divided into 40 × 40 cm grids. These grids were used to determine an average bark grid and thus an average bark pattern. All parameter values were determined from this grid. If the bark is evenly comparable over large parts or the entire trunk, all grids are combined into one grid.
The evaluation was carried out using the AI software Dylogos 2.0. The Dylogos software transforms the LAS data into XYZ data. These are then clustered into the two groups:
- Geometric description of the bark;
- Rib characteristics.
The features created in this way were further analyzed with R.
2.3. Machine Learning
For the quality of the decision making, the clustering of the training data is of particular importance. In the following, the four methods applied in this study and their structural differences will be applied to the bark model. These are as follows:
- Decision trees;
- Random forests;
- XGBoost;
- Support vector machines.
Their results can be characterized by two parameters. These are the accuracy and the predictive power of the trained system. The way of calculating the accuracy by means of the confusion matrix is identical for all four methods. A confusion matrix is a 2 × 2 matrix scheme. The elements of the matrix are as follows:
- True Positive (TP);
- False Positive (FP);
- True Negative (RN);
- False Negative (FN).
All four elements are taken into account in the machine learning process.
Here, the evaluation of a condition and its future development is judged. The assessment can be true or false, and the respective expression can turn out to be positive or negative for the assessment model in the future. The rows are filled with the actual condition and the columns with the predicted condition.
Each data set is now classified into one of the four elements (classes) of the matrix according to the model generated using the training data. The accuracy value is determined as follows:
The accuracy value is between 0 and 1. A value > 0.9 is a good value. A value > 0.7 is good, and 0.7 is a fair result.
The four methods differ in the prediction condition. The decision tree and random forests methods use the roc auc value (Compute Area Under the Receiver Operating Characteristic Curve) to parameterize the prediction. The roc auc value is between 0 and 1. A value of 0.5 represents a random estimate.
The XGBoost and the support vector machine methods use κ (kappa) for the quality of the prediction. κ or Cohen’s kappa is a measure of interrater reliability and thus a parameter that reflects the agreement or disagreement between two observers on a decision. The authors of [69] suggest that κ < 0 = “poor agreement”, 0 < κ < 0.2 = “slight agreement”, 0.21 < κ < 0.40 = “fair agreement”, 0.41 < κ < 0.60 = “moderate agreement”, 0.61 < κ < 0.80 = “substantial agreement”, and κ > 0.81 = “almost perfect agreement”.
Table 2 shows the parameters used. This structure corresponds to the generated evaluation database. They are divided into two blocks:
- Geometric description of the bark;
- Rib characteristics.
These give the geometry of the bark, a description of the bark’s appearance, and the description of the individual ribs.
The machine learning methods we investigated differ not only in their functionalities, which will be discussed in more detail, but also in the different weighting and therefore relevance of the various parameters. The type and number of parameters taken into account vary from method to method.
2.3.1. Decision Trees
The decision tree method is based on the assumption that all information important for a decision is available in the training data sets. The trees have a uniform structure in the form that the leaves of the tree describe classes and the branches form conjunctions of features that then lead to a class.
Decisions are thus better structured. The path of a decision is not a linear path but has nodes with branches. The choice of which branch to select is made by means of a decision function that is derived from the training data. In most cases, the decision function separates within a cluster whether values are larger or smaller than a target value.
2.3.2. Random Forests
In the random forest method, the samples used to determine the tree structure are randomly selected from the training data. After a new node is created, the samples are added back to the training set. At each node, a randomly selected subset of criteria from the entire set of criteria is used to make a decision (branch). The selection of features is performed to minimize the impurity of the overall model. Due to the randomness, multiple decision trees are created per training set, forming a decision forest. The predictions of the individual trees are then aggregated to produce an overall prediction.
2.3.3. XGBoost
In the XGBoost model, the fitting of a tree structure is performed using a loss function. The tree structure is generated, starting from a starting point, by means of the Newton method. Each new node is considered as a new model and optimized by a loss function.
2.3.4. Support Vector Machines
A support vector machine (SVM) is a discriminative machine learning model that uses a hyperplane to separate training data into two classes. Unlike the DBSCAN clustering method, which searches for the elements with the smallest distance to the hyperplane, SVMs search for the elements with the widest distance to the hyperplane. This results in data clusters with sharp boundaries.
The analyses were implemented in R [70]. Models were fitted with 10-fold cross-validation on a training data set of 75% of the samples and tested on the remaining 25%. Features with near-zero variation and closely correlated features were removed prior to analysis. When necessary, features were Yeo-Johnson transformed. All features were normalized.
Finally, Figure 5 illustrates the entire process as a workflow for greater clarity.
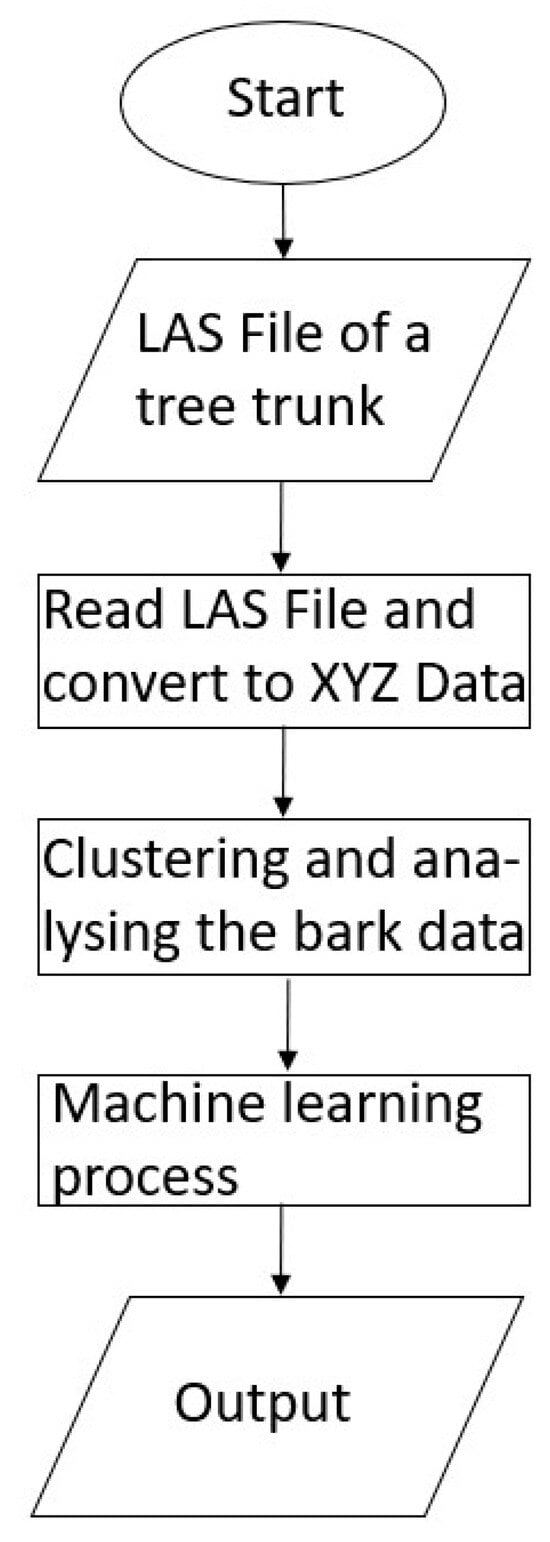
Figure 5.
Scheme of the workflow used in this study.
3. Results
3.1. Features
The calculation of the features was computationally very efficient and was conducted on a recent desktop PC (DELL Latitude, CPU Intel Core i5-6300, 8,00 GB Ram).
Some of the proposed features were correlated, e.g., roughness and smoothness (r = 0.81), behavior and smoothness (r = 0.91), or behavior and CL-LR (r = 0.85). Only one of each pair was used for further analyses. Before transformation, the distributions of most features were highly skewed.
Fagus sylvatica had the most distinctive set of bark features. It was the species with the smoothest bark (Figure 6a) and the lowest spacing between ribs (Figure 6b).
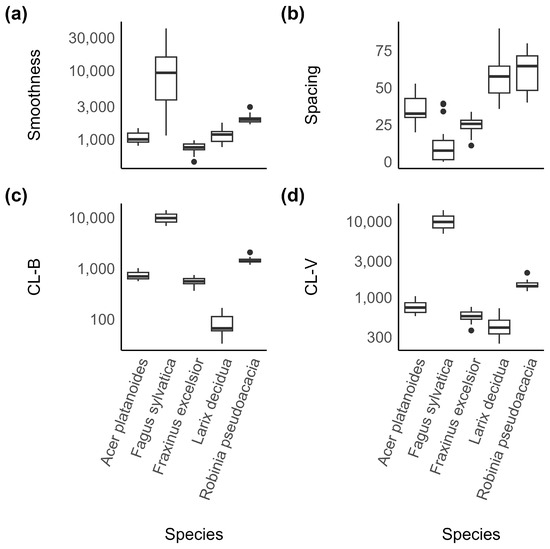
Figure 6.
Distribution of four selected features ((a): smoothness, (b): spacing, (c): branching, and (d): vertical alignment of clusters) of all species in the sample. They illustrate how the features differentiate between species.
3.2. Machine Learning
The decision tree (Figure 7) performed less well than the other approaches and reached an accuracy of 83% and a roc auc score of 94%. R. pseudoacacia with its very distinctive bark was the first species that was split from the others in this model.
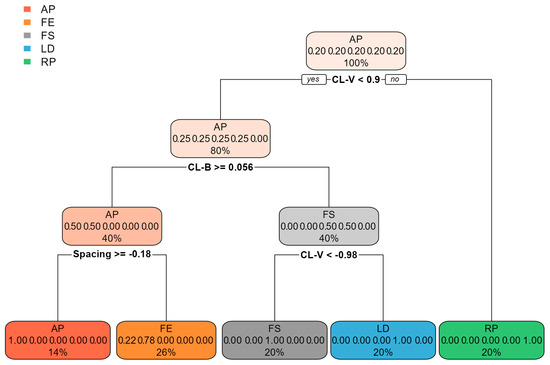
Figure 7.
Result of the classification based on a decision tree, illustrating the subset of features used by this model. AP: Acer pseudoplatanus, FE: Fraxinus excelsior, FS: Fagus sylvatica, LD: Larix decidua, RP: Robinia pseudoacacia.
The remaining three methods performed equally well, with accuracies between 92% and 96% even for this small data set of 85 trees (Table 3 and Figure 8).

Table 3.
Performance of the machine learning methods used in this study.
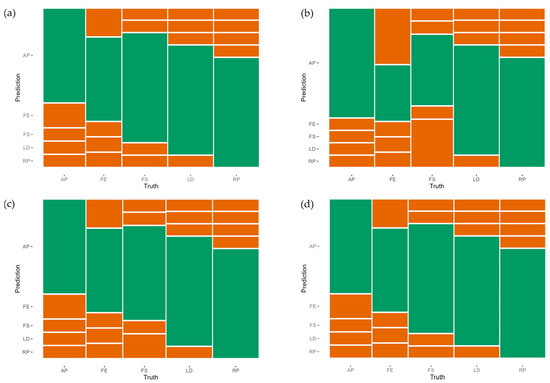
Figure 8.
Confusion matrices of (a) decision tree, (b) random forest, (c) SVM, and (d) XGBoost. AP: Acer pseudoplatanus, FE: Fraxinus excelsior, FS: Fagus sylvatica, LD: Larix decidua, RP: Robinia pseudoacacia. The green area represents the proportion of members of the test sample set (not used in the training of the models) classified correctly, while red represents false classifications.
The other more complex methods achieved 96% accuracy in predicting the species of the test population. However, the predictions of the random forest model were comparatively poor for F. excelsior (Figure 8b). Overall, the XGBoost model had the best results (Table 3 and Figure 8d). The difference between the accuracy value and κ, the value of the prediction, is the smallest of all the presented models.
The ranking of feature importance was not similar for all models. The most important feature for the XGBoost model was “Cluster cluster left/right” (CL-RL), followed by “clusters vertical” (CL-V) and bark roughness (Figure 9). The decision tree, on the other hand, used only the features “CL-V”, “CL-B”, and “Spacing” to classify species (Figure 7).
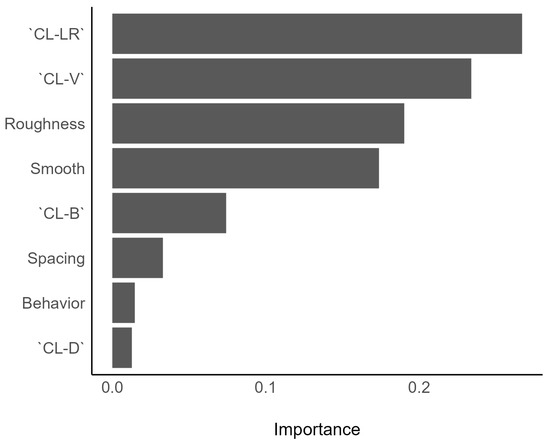
Figure 9.
Feature importance of the XGBoost model. Note that compared to the decision tree (Figure 7) a different subset of features was selected by this algorithm.
4. Discussion
Trees provide a wide range of ecosystem services in urban areas, including air and water purification, noise reduction, and temperature regulation [71]. Up-to-date tree inventories are essential for effective tree management and monitoring of their ecosystem services [72]. Terrestrial Laser scanning (TLS) can make tree inventory data collection much more efficient than traditional methods [73], but species identification from TLS point clouds is still challenging.
In this study, we hypothesized that bark features traditionally used by botanists, like ridges, crevices, and smoothness, could be described mathematically and applied to species identification from TLS point clouds. The approach we presented differs from the very few other recently published approaches, such as that of [44], in the kind and number of bark features considered.
Since different machine learning methods are structurally different, we tested the performance of several approaches. The accuracy of all methods was high, despite the rather small data set used. Our results suggest that the mathematical description of bark features used by botanists could be used to complement, or even provide advantages over, the black-box approaches used so far.
Correct species identification is essential for tree inventories, as it is the basis for, amongst other things, ecosystem services calculations, tree maintenance, and tree risk management. Our method will complement other approaches to identify tree species based on remote sensing data, will help to increase overall accuracy, and thus, will support the more efficient creation of tree inventories. This assumption is based on a timesaving potential as well as the future possibility of the partial automation of this process.
However, our study has some limitations. First, the number of trees and tree species in our data set was relatively small. Secondly, we only used trees with fully developed mature bark characteristics. This means that the trees examined had diameters corresponding to the age class. Future research should focus on increasing the size and diversity of the training data set to improve the accuracy and robustness of the machine learning models. Additionally, the models should be tested on younger trees because the bark structure can change significantly during the lifetime of a tree [74]. Although present on some trees, we did not study the effects of epiphytes growing on the bark on the accuracy of species identification. Furthermore, the models should be integrated into existing tree inventory workflows to assess their feasibility for practical use.
5. Conclusions
This study provides promising evidence that explainable bark features can be used to identify species from TLS point clouds. A model of bark features based on expert knowledge could potentially reduce the number of required samples in comparison to black-box approaches. This could result in more efficient and accurate collection of tree inventory data, which is crucial for the effective management and monitoring of their ecosystem services in urban areas. This computationally efficient approach might allow for real-time species classification.
Author Contributions
Conceptualization, S.R.; methodology, S.R. and B.S.; software, B.S.; validation, S.R.; formal analysis, S.R.; data curation, B.S.; writing—original draft preparation, S.R. and B.S.; writing—review and editing, S.R. and B.S.; visualization, S.R. and B.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are unavailable due to privacy restrictions.
Acknowledgments
We thank ProGea 4D, Krakow, Poland, for providing the point cloud data.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kushwaha, S.K.P.; Singh, A.; Jain, K.; Cabo, C.; Mokros, M. Integrating Airborne and Terrestrial Laser Scanning for Complete 3D Model Generation in Dense Forest. In Proceedings of the IGARSS 2023—2023 IEEE International Geoscience and Remote Sensing Symposium, Pasadena, CA, USA, 16–21 July 2023; pp. 3137–3140. [Google Scholar]
- Chen, S.; Liu, H.; Feng, Z.; Shen, C.; Chen, P. Applicability of Personal Laser Scanning in Forestry Inventory. PLoS ONE 2019, 14, e0211392. [Google Scholar] [CrossRef]
- Hyyppä, J.; Yu, X.; Kaartinen, H.; Kukko, A.; Jaakkola, A.; Liang, X.; Wang, Y.; Holopainen, M.; Vastaranta, M.; Hyyppa, H. Forest Inventory Using Laser Scanning; Shan, J., Toth, C.K., Eds.; CRC Press-Taylor & Francis Group: Boca Raton, FL, USA, 2018; pp. 379–412. ISBN 978-1-4987-7228-0. [Google Scholar]
- Brede, B.; Terryn, L.; Barbier, N.; Bartholomeus, H.M.; Bartolo, R.; Calders, K.; Derroire, G.; Krishna Moorthy, S.M.; Lau, A.; Levick, S.R.; et al. Non-Destructive Estimation of Individual Tree Biomass: Allometric Models, Terrestrial and UAV Laser Scanning. Remote Sens. Environ. 2022, 280, 113180. [Google Scholar] [CrossRef]
- Arseniou, G.; MacFarlane, D.W.; Calders, K.; Baker, M. Accuracy Differences in Aboveground Woody Biomass Estimation with Terrestrial Laser Scanning for Trees in Urban and Rural Forests and Different Leaf Conditions. Trees 2023, 37, 761–779. [Google Scholar] [CrossRef]
- Wang, F.; Sun, Y.; Jia, W.; Li, D.; Zhang, X.; Tang, Y.; Guo, H. A Novel Approach to Characterizing Crown Vertical Profile Shapes Using Terrestrial Laser Scanning (TLS). Remote Sens. 2023, 15, 3272. [Google Scholar] [CrossRef]
- Demol, M.; Verbeeck, H.; Gielen, B.; Armston, J.; Burt, A.; Disney, M.; Duncanson, L.; Hackenberg, J.; Kükenbrink, D.; Lau, A.; et al. Estimating Forest Above-ground Biomass with Terrestrial Laser Scanning: Current Status and Future Directions. Methods Ecol. Evol. 2022, 13, 1628–1639. [Google Scholar] [CrossRef]
- Vazirabad, Y.F.; Karslioglu, M.O. Lidar for Biomass Estimation; Matovic, D., Ed.; Intech Europe: Rijeka, Croatia, 2011; pp. 3–26. ISBN 978-953-307-492-4. [Google Scholar]
- Calders, K.; Verbeeck, H.; Burt, A.; Origo, N.; Nightingale, J.; Malhi, Y.; Wilkes, P.; Raumonen, P.; Bunce, R.G.H.; Disney, M. Laser Scanning Reveals Potential Underestimation of Biomass Carbon in Temperate Forest. Ecol. Solut. Evid. 2022, 3, e12197. [Google Scholar] [CrossRef]
- Dassot, M.; Barbacci, A.; Colin, A.; Fournier, M.; Constant, T. Tree Architecture and Biomass Assessment from Terrestrial LiDAR Measurements: A Case Study for Some Beech Trees (Fagus sylvatica). Silvilaser Full Proc. 2010, 206–215. [Google Scholar]
- Sun, C.; Huang, C.; Zhang, H.; Chen, B.; An, F.; Wang, L.; Yun, T. Individual Tree Crown Segmentation and Crown Width Extraction from a Heightmap Derived From Aerial Laser Scanning Data Using a Deep Learning Framework. Front. Plant Sci. 2022, 13, 914974. [Google Scholar] [CrossRef]
- Wu, L.; Shi, Y.; Zhang, F.; Zhou, Y.; Ding, Z.; Lv, S.; Xu, L. Estimating Carbon Stocks and Biomass Expansion Factors of Urban Greening Trees Using Terrestrial Laser Scanning. Forests 2022, 13, 1389. [Google Scholar] [CrossRef]
- Imai, Y.; Setojima, M.; Yamagishi, Y.; Fujiwara, N. Tree-Height Measuring Characteristics of Urban Forests by Lidar Data Different in Resolution. In Proceedings of the Geo-Imagery Bridging Continents, Istanbul, Turkey, 12–23 July 2004; pp. 2–5. [Google Scholar]
- Guo, Y.; Luka, A.; Wei, Y. Modeling Urban Tree Growth for Digital Twins: Transformation of Point Clouds into Parametric Crown Models. J. Digit. Landsc. Archit. 2022, 213–223. [Google Scholar] [CrossRef]
- Uzquiano, S.; Barbeito, I.; San Martín, R.; Ehbrecht, M.; Seidel, D.; Bravo, F. Quantifying Crown Morphology of Mixed Pine-Oak Forests Using Terrestrial Laser Scanning. Remote Sens. 2021, 13, 4955. [Google Scholar] [CrossRef]
- Ma, Q.; Lin, J.; Ju, Y.; Li, W.; Liang, L.; Guo, Q. Individual Structure Mapping over Six Million Trees for New York City USA. Sci. Data 2023, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Münzinger, M.; Prechtel, N.; Behnisch, M. Mapping the Urban Forest in Detail: From LiDAR Point Clouds to 3D Tree Models. Urban For. Urban Green. 2022, 74, 127637. [Google Scholar] [CrossRef]
- Zieba-Kulawik, K.; Skoczylas, K.; Wezyk, P.; Teller, J.; Mustafa, A.; Omrani, H. Monitoring of Urban Forests Using 3D Spatial Indices Based on LiDAR Point Clouds and Voxel Approach. Urban For. Urban Green. 2021, 65, 127324. [Google Scholar] [CrossRef]
- Zieba-Kulawik, K.; Wezyk, P. Monitoring 3D Changes in Urban Forests Using Landscape Metrics Analyses Based on Multi-Temporal Remote Sensing Data. Land 2022, 11, 883. [Google Scholar] [CrossRef]
- César de Lima Araújo, H.; Silva Martins, F.; Tucunduva Philippi Cortese, T.; Locosselli, G.M. Artificial Intelligence in Urban Forestry—A Systematic Review. Urban For. Urban Green. 2021, 66, 127410. [Google Scholar] [CrossRef]
- Matasci, G.; Coops, N.C.; Williams, D.A.R.; Page, N. Mapping Tree Canopies in Urban Environments Using Airborne Laser Scanning (ALS): A Vancouver Case Study. For. Ecosyst. 2018, 5, 31. [Google Scholar] [CrossRef]
- Plowright, A.A.; Coops, N.C.; Eskelson, B.N.I.; Sheppard, S.R.J.; Aven, N.W. Assessing Urban Tree Condition Using Airborne Light Detection and Ranging. Urban For. Urban Green. 2016, 19, 140–150. [Google Scholar] [CrossRef]
- Degerickx, J.; Roberts, D.A.; McFadden, J.P.; Hermy, M.; Somers, B. Urban Tree Health Assessment Using Airborne Hyperspectral and LiDAR Imagery. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 26–38. [Google Scholar] [CrossRef]
- Michałowska, M.; Rapiński, J. A Review of Tree Species Classification Based on Airborne LiDAR Data and Applied Classifiers. Remote Sens. 2021, 13, 353. [Google Scholar] [CrossRef]
- Sakharova, E.K.; Nurlyeva, D.D.; Fedorova, A.A.; Yakubov, A.R.; Kanev, A.I. Issues of Tree Species Classification from LiDAR Data Using Deep Learning Model. In Advances in Neural Computation, Machine Learning, and Cognitive Research V; Kryzhanovsky, B., Dunin-Barkowski, W., Redko, V., Tiumentsev, Y., Klimov, V.V., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 319–324. [Google Scholar]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban Tree Species Classification Using a WorldView-2/3 and LiDAR Data Fusion Approach and Deep Learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Han, Z.; Chen, Y.; Liu, Z.; Han, Y. Tree Species Classification of LiDAR Data Based on 3D Deep Learning. Measurement 2021, 177, 109301. [Google Scholar] [CrossRef]
- Xi, Z.; Hopkinson, C.; Rood, S.B.; Peddle, D.R. See the Forest and the Trees: Effective Machine and Deep Learning Algorithms for Wood Filtering and Tree Species Classification from Terrestrial Laser Scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 1–16. [Google Scholar] [CrossRef]
- Seidel, D.; Annighöfer, P.; Thielman, A.; Seifert, Q.E.; Thauer, J.-H.; Glatthorn, J.; Ehbrecht, M.; Kneib, T.; Ammer, C. Predicting Tree Species From 3D Laser Scanning Point Clouds Using Deep Learning. Front. Plant Sci. 2021, 12, 635440. [Google Scholar] [CrossRef]
- Qian, C.; Yao, C.; Ma, H.; Xu, J.; Wang, J. Tree Species Classification Using Airborne LiDAR Data Based on Individual Tree Segmentation and Shape Fitting. Remote Sens. 2023, 15, 406. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of Studies on Tree Species Classification from Remotely Sensed Data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Qiao, Y.; Zheng, G.; Du, Z.; Ma, X.; Li, J.; Moskal, L. Tree-Species Classification and Individual-Tree-Biomass Model Construction Based on Hyperspectral and LiDAR Data. Remote Sens. 2023, 15, 1341. [Google Scholar] [CrossRef]
- Hakula, A.; Ruoppa, L.; Lehtomäki, M.; Yu, X.; Kukko, A.; Kaartinen, H.; Taher, J.; Matikainen, L.; Hyyppä, E.; Luoma, V.; et al. Individual Tree Segmentation and Species Classification Using High-Density Close-Range Multispectral Laser Scanning Data. ISPRS Open J. Photogramm. Remote Sens. 2023, 9, 100039. [Google Scholar] [CrossRef]
- Ahlswede, S.; Schulz, C.; Gava, C.; Helber, P.; Bischke, B.; Förster, M.; Arias, F.; Hees, J.; Demir, B.; Kleinschmit, B. TreeSatAI Benchmark Archive: A Multi-Sensor, Multi-Label Dataset for Tree Species Classification in Remote Sensing. Earth Syst. Sci. Data 2023, 15, 681–695. [Google Scholar] [CrossRef]
- Liu, B.; Huang, H.; Su, Y.; Chen, S.; Li, Z.; Chen, E.; Tian, X. Tree Species Classification Using Ground-Based LiDAR Data by Various Point Cloud Deep Learning Methods. Remote Sens. 2022, 14, 5733. [Google Scholar] [CrossRef]
- Liu, B.; Chen, S.; Huang, H.; Tian, X. Tree Species Classification of Backpack Laser Scanning Data Using the PointNet++ Point Cloud Deep Learning Method. Remote Sens. 2022, 14, 3809. [Google Scholar] [CrossRef]
- Hell, M.; Brandmeier, M.; Briechle, S.; Krzystek, P. Classification of Tree Species and Standing Dead Trees with Lidar Point Clouds Using Two Deep Neural Networks: PointCNN and 3DmFV-Net. PFG 2022, 90, 103–121. [Google Scholar] [CrossRef]
- Cetin, Z.; Yastikli, N. The Use of Machine Learning Algorithms in Urban Tree Species Classification. ISPRS Int. J. Geo-Inf. 2022, 11, 226. [Google Scholar] [CrossRef]
- Faizal, S. Automated Identification of Tree Species by Bark Texture Classification Using Convolutional Neural Networks. Int. J. Res. Appl. Sci. Eng. Technol. 2022, 10, 1384–1392. [Google Scholar] [CrossRef]
- Misra, D.; Crispim-Junior, C.; Tougne, L. Patch-Based CNN Evaluation for Bark Classification. In Computer Vision—ECCV 2020 Workshops; Bartoli, A., Fusiello, A., Eds.; Springer: Cham, Switzerland, 2020; pp. 197–212. ISBN 978-3-030-65413-9. [Google Scholar]
- Abdollahnejad, A.; Panagiotidis, D. Tree Species Classification and Health Status Assessment for a Mixed Broadleaf-Conifer Forest with UAS Multispectral Imaging. Remote Sens. 2020, 12, 3722. [Google Scholar] [CrossRef]
- Terryn, L.; Calders, K.; Disney, M.; Origo, N.; Malhi, Y.; Newnham, G.; Raumonen, P.; Åkerblom, M.; Verbeeck, H. Tree Species Classification Using Structural Features Derived from Terrestrial Laser Scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 170–181. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, X.; Cao, L.; Coops, N.C.; Goodbody, T.R.H.; Zhong, T.; Zhao, W.; Sun, Q.; Ba, S.; Zhang, Z.; et al. Tree Species Classification Using UAS-Based Digital Aerial Photogrammetry Point Clouds and Multispectral Imageries in Subtropical Natural Forests. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102173. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Ishii, A.; Nakamura, H. Individual Tree Species Classification Based on Terrestrial Laser Scanning Using Curvature Estimation and Convolutional Neural Network. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2-W13, 1077–1082. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Demir, B.; Dalponte, M. A Weighted SVM-Based Approach to Tree Species Classification at Individual Tree Crown Level Using LiDAR Data. Remote Sens. 2019, 11, 2948. [Google Scholar] [CrossRef]
- Marrs, J.; Ni-Meister, W. Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data. Remote Sens. 2019, 11, 819. [Google Scholar] [CrossRef]
- Dalponte, M.; Frizzera, L.; Gianelle, D. Individual Tree Crown Delineation and Tree Species Classification with Hyperspectral and LiDAR Data. PeerJ 2019, 6, e6227. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, T.; Liu, X. A Review: Individual Tree Species Classification Using Integrated Airborne LiDAR and Optical Imagery with a Focus on the Urban Environment. Forests 2018, 10, 1. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Ishii, A.; Nakamura, H.; Inoue, T.; Takamatsu, H. Lidar-Based Individual Tree Species Classification Using Convolutional Neural Network. In Proceedings of the Videometrics, Range Imaging, and Applications XIV; Remondino, F., Shortis, M.R., Eds.; SPIE: Bellingham, WA, USA, 2017; Volume 10332, p. UNSP103320O. [Google Scholar]
- Shen, X.; Cao, L. Tree-Species Classification in Subtropical Forests Using Airborne Hyperspectral and LiDAR Data. Remote Sens. 2017, 9, 1180. [Google Scholar] [CrossRef]
- Matsuki, T.; Yokoya, N.; Iwasaki, A. Hyperspectral Tree Species Classification of Japanese Complex Mixed Forest with the Aid of LiDAR Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2177–2187. [Google Scholar] [CrossRef]
- Othmani, A.; Voon, L.F.C.L.Y.; Stolz, C.; Piboule, A. Single Tree Species Classification from Terrestrial Laser Scanning Data for Forest Inventory. Pattern Recognit. Lett. 2013, 34, 2144–2150. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree Species Classification and Estimation of Stem Volume and DBH Based on Single Tree Extraction by Exploiting Airborne Full-Waveform LiDAR Data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree Species Classification in the Southern Alps Based on the Fusion of Very High Geometrical Resolution Multispectral/Hyperspectral Images and LiDAR Data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Heinzel, J.; Koch, B. Exploring Full-Waveform LiDAR Parameters for Tree Species Classification. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 152–160. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree Species Classification Using Airborne LiDAR—Effects of Stand and Tree Parameters, Downsizing of Training Set, Intensity Normalization, and Sensor Type. Silva Fenn. 2010, 44, 156. [Google Scholar] [CrossRef]
- Weiss, U.; Biber, P.; Laible, S.; Bohlmann, K.; Zell, A. Plant Species Classification Using a 3D LIDAR Sensor and Machine Learning. In Proceedings of the 2010 Ninth International Conference on Machine Learning and Applications, Washington, DC, USA, 12–14 December 2010; pp. 339–345. [Google Scholar]
- Voss, M.; Sugumaran, R. Seasonal Effect on Tree Species Classification in an Urban Environment Using Hyperspectral Data, LiDAR, and an Object-Oriented Approach. Sensors 2008, 8, 3020–3036. [Google Scholar] [CrossRef]
- Luo, H.; Khoshelham, K.; Chen, C.; He, H. Individual Tree Extraction from Urban Mobile Laser Scanning Point Clouds Using Deep Pointwise Direction Embedding. ISPRS J. Photogramm. Remote Sens. 2021, 175, 326–339. [Google Scholar] [CrossRef]
- Hamraz, H.; Contreras, M.A.; Zhang, J. Forest Understory Trees Can Be Segmented Accurately within Sufficiently Dense Airborne Laser Scanning Point Clouds. Sci. Rep. 2017, 7, 6770. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Li, X.; Li, T.; Li, M. Improvement and Assessment of Convolutional Neural Network for Tree Species Identification Based on Bark Characteristics. Forests 2023, 14, 1292. [Google Scholar] [CrossRef]
- Kim, T.K.; Hong, J.; Ryu, D.; Kim, S.; Byeon, S.Y.; Huh, W.; Kim, K.; Baek, G.H.; Kim, H.S. Identifying and Extracting Bark Key Features of 42 Tree Species Using Convolutional Neural Networks and Class Activation Mapping. Sci. Rep. 2022, 12, 4772. [Google Scholar] [CrossRef]
- Othmani, A.A.; Jiang, C.; Lomenie, N.; Favreau, J.-M.; Piboule, A.; Voon, L.F.C.L.Y. A Novel Computer-Aided Tree Species Identification Method Based on Burst Wind Segmentation of 3D Bark Textures. Mach. Vis. Appl. 2016, 27, 751–766. [Google Scholar] [CrossRef]
- Whitmore, T.C. Studies in Systematic Bark Morphology. I. Bark Morphology in Dipterocarpaceae. New Phytol. 1962, 61, 191–207. [Google Scholar] [CrossRef]
- Zeidler, E. Springer-Handbuch Der Mathematik I–IV; Springer Fachmedien: Wiesbaden, Germany, 2013. [Google Scholar]
- Brieskorn, E. Lineare Algebra Und Analytische Geometrie III; Springer Spektrum: Wiesbaden, Germany, 2019; Volume III. [Google Scholar]
- Bronstein, I.N.; Semendjajew, K.A.; Zeideler, E.; Herausgeber, W. Teubner-Taschenbuch Der Mathematik; B. G. Teubner Stuttgart: Leibzig, Germany, 1996. [Google Scholar]
- Barner, M.; Flor, F. Analysis II; de Gruyter: Berlin, Germany; New York, NY, USA, 1989. [Google Scholar]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021.
- Salbitano, F.; Borelli, S.; Conigliaro, M.; Chen, Y. Guidelines on Urban and Peri-Urban Forestry; FAO Forestry Paper; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016; ISBN 978-92-5-109442-6. [Google Scholar]
- Edgar, C.B.; Nowak, D.J.; Majewsky, M.A.; Lister, T.W.; Westfall, J.A.; Sonti, N.F. Strategic National Urban Forest Inventory for the United States. J. For. 2020, 119, 86–95. [Google Scholar] [CrossRef]
- Morgenroth, J.; Östberg, J. Measuring and Monitoring Urban Trees and Urban Forests. In Routledge Handbook of Urban Forestry; Routledge: London, UK, 2017; pp. 33–48. [Google Scholar]
- Whitmore, T.C. Studies in Systematic Bark Morphology: IV. The Bark of Beech, Oak and Sweet Chestnut. New Phytol. 1963, 62, 161–169. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).