Abstract
Botrytis cinerea, a pathogenic fungus that causes necrosis in plants, is one of the most destructive pathogens of hazelnuts. This fungus is responsible for causing Husk Brown Rot, a significant threat to hazelnut production. The plant’s defense mechanism against this pathogen, as well as other pathogens, is a complex biological process that involves changes at molecular, biochemical, and physiological levels. To better understand the molecular responses of hazelnut plants to B. cinerea infection, we conducted a comparative transcriptome profiling study between a B. cinerea-resistant Ping’ou hybrid hazelnut variety (Dawei; DW) and a susceptible variety (Qiuxiang; QX). Our study focused on the transcriptome profiles of DW and QX plants after three days of B. cinerea infection. The results of our study showed moderate changes in the defense strategies of both DW and QX plants in response to B. cinerea infection. Specifically, we observed that the expression of 14 disease-resistant genes was significantly different between DW and QX. Our comparative analysis revealed that DW had a higher number and expression of immunity-related differentially expressed genes compared to QX, which indicates that these genes play a crucial role in inducing innate resistance in DW plants against B. cinerea infection. This study highlights that plant resistance to pathogens like B. cinerea is a complex process that is controlled by multiple genes and biological pathways, each playing a specific role. Our findings provide new insights into the development of hazelnut varieties that are resistant to B. cinerea infection. By using the candidate genes identified in this study, it may be possible to enhance the resistance of hazelnut plants to B. cinerea and reduce the impact of Husk Brown Rot on hazelnut production.
1. Introduction
Botrytis cinerea, a pathogenic necrotrophic fungus causing Hazelnut Husk Brown Rot is responsible for yield losses and reduced economical values of Hazelnut. It causes annual loss of 5 to 50% due to reduced yield [1]. The initiation of infection takes place from the cell surface level and gradually spreads all over the plant causing brown rot symptoms [2]. Different environmental factors affect plant immunity against pathogenic infections [3]. Despite the great level of research and development of resistant varieties by manipulating genes and proteins and their expression levels, complete genetic tolerance to disease and other stresses is yet not been achieved in plants [4]. The plant resistance mechanism to various pathogens including B. cinerea is a complex process that involves various biological changes at physiological, biochemical, molecular, and hormonal levels [5]. The main factors that affect the capability of the host to resist the disease include the plant’s ability to maintain cell wall integrity, production of reactive oxygen species, activation of antioxidant enzymes production, expression of immune-related genes and hormonal regulation of metabolites and phytohormones including jasmonic acid (JA), salicylic acid (SA), ethylene (ET), abscisic acid (ABA) [3]. Once the plant detects the pathogen, effective plant immune response activates signaling mechanism to initiate defense pathways [6]. Previous studies reported many genes that are involved in causing plant resistance in response to B. cinerea [3]. Plant hormones, including JA, SA, ABA, and ET are the primary regulators of plant resistance against pathogens and pests. Several reports have revealed the involvement of SA, JA, and ET pathways regulating genes against necrotrophic and biotrophic pathogens [7,8].
Plants have a variety of defense mechanisms against the pathogenic fungus B. cinerea. These mechanisms are triggered by the recognition of pathogenic microbial and damage-associated molecular patterns (MAMPs/DAMPs) by the plant’s innate pattern recognition proteins (PRRPs). Upon detection, the plant immune system is activated to defend against the pathogen [9,10]. A previous study has shown that overexpressing the DAMP recognition receptor Wall associated kinase 1 (WAK1) can enhance resistance in Arabidopsis plants against B. cinerea infection [10]. Additionally, the Arabidopsis Botrytis induced kinase 1 (BIK1) gene, which encodes a receptor-like cytoplasmic kinase (RLCK), plays a role in mediating MAMP-triggered immunity during pathogenic infections [11]. Mutations in the BIK1 gene can lead to increased susceptibility to fungal infections in plants. Upon initial recognition of a fungal infection, the plant activates a series of signals that includes the activation of mitogen-activated protein kinases (MAPKs). These MAPKs are responsible for conveying signals from the receptors to specific effectors and for regulating gene expression to induce resistance against the infection [11]. After a B. cinerea infection, MAPKs undergo a series of phosphorylation events that result in numerous transcriptional changes, including the activation of genes encoding transcription factors (TFs) [12]. The hypothesis of this study is that the defense mechanisms in plants against B. cinerea are regulated by a complex network of genes and signaling pathways, including PRRPs, MAPKs, and TFs, each playing a critical role in inducing resistance against the pathogen.
Many resistant varieties were developed to overcome the economic losses but the studies to understand the differences in molecular mechanisms between susceptible and resistant varieties are limited. The development of efficient tools in omics technologies has made it very easy to better understand the concept and transcriptional differences between resistant and susceptible varieties against plant pathogens. High-throughput experiments can generate large-scale genomic and transcriptomic data to directly understand the transcript and expression differences of immunity related genes in plants [13]. Several stresses induce pathogenesis related (PR) proteins encoding and other genes, such as chitinase, Defensin, Oxalate oxidase, Expensin like proteins, NAC, WRKY TFs, etc. that play major roles in inducing plant resistance [14,15,16,17].
In the current study, we used a comparative transcriptome profiling to elucidate the resistance mechanisms and gene expression behavior between a resistant variety Dawei (DW) and a susceptible one Qiuxiang (QX). We described the transcriptome profiles of DW and QX after 3 days of B. cinerea infection and revealed relatively moderate changes in their defense strategies, providing new insights into resistance mechanisms of Hazelnut to B. cinerea.
2. Material and Methods
2.1. Experiment Location, Hazelnut Varieties, and Treatments
The experiment was performed at Songmudao Base (121°75′ E, 39°40′ N) located in Dalian City, Liaoning Province, China from July 2020 to July 2021. Two Ping’ou hybrid hazelnut varieties (Corylus heterophylla Fisch × Corylus avellana L.) were used: Dawei (DW), a disease-resistant variety and Qiuxiang (QX), a disease susceptible variety. The treatments applied were non-inoculated control of Dawei (A-CK), Inoculated Dawei (A), Non-inoculated control of Qiuxiang (B-CK) and Inoculated Qiuxiang (B). Each treatment was replicated three times.
2.2. Botrytis Cinerea Z9 Cultivation and Inoculation
The pathogenic strain, Botrytis cinerea Z9 was cultured on a potato dextrose agar plate and incubated at 27 °C for seven days in the dark. Ten bract clusters of 5- to 6-year-old Ping’ou hybrid hazelnut were selected for Dawei (A) and Qiuxiang (B) varieties for inoculation. The wound was caused by pricking the fruit bud, and the freshly grown fungal plug with a diameter of 0.5 mm was punched in it with the help of a hole puncher to cover the wound surface on the hazelnut bud. The wound was then bagged for 36 h (Supplementary Figure S1).
2.3. Quantification of Disease Resistance
The disease index (DI) was calculated at 3rd day post inoculation (DPI) on wounded fruit buds to evaluate disease resistance level in Dawei and Qiuxiang. The symptoms on the fruit buds are divided into four levels, 1, 2, 3, and 4 depending on the severity of disease [18]. DI was calculated by using the following formula:
2.4. Transcriptome Analysis
2.4.1. Plant Sampling
Sampling for transcriptome sequencing of Dawei and Qiuxiang was done at 3 DPI. The fruit bracts were cut into 1–2 cm tissue blocks (weighing 2–3 g) and were immediately flash frozen in liquid nitrogen. Samples were collected in 3 replicates for each inoculated and non-inoculated treatment. Samples were stored at −80 °C for further analysis.
2.4.2. RNA Extraction, Library Preparation, and Sequencing
Total RNA was extracted following the protocol of RNAprep Pure Kit DP432 (TIANGEN Biotech, Beijing, China) and the integrity of the extracted RNA was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). All RNA samples were evaluated using Qsep 1 instrument for their stability and 3.5 μg of total RNA was used for the construction of RNA libraries by MGIEasy mRNA Library Prep kit After the construction of the library, the library was initially quantified by Qubit2.0 Fluorometer, then diluted to 1.5 ng/μL, and the insert size of the library is detected by Agilent 2100 bioanalyzer. After the insert size meets the expectation, qRT-PCR is used to accurately quantify the effective concentration of the library (the effective concentration of the library is higher than that of 2 nM) to ensure the quality of the library. After the library is qualified, the different libraries are pooled according to the effective concentration and the target amount of data off the machine, then sequenced by the Illumina NovaSeq 6000 [19].
2.4.3. Transcriptome Assembly
The image data measured by the high-throughput sequencer are converted into sequence data (reads) by CASAVA base recognition. Raw data (raw reads) of fastq format were first processed through in-house Perl scripts. At the same time, Q20, Q30, and GC content of the clean data were calculated. After the clean reads were obtained, the Trinity software (v2.6.6) was used to assemble the clean reads into the reference transcriptome [20,21].
2.4.4. Differential Expression Analysis
Differential expression analysis of two groups was performed using the DESeq2 R package (1.20.0) with a filter threshold of adjusted q-value < 0.05 and |log2FoldChange| > 2 [22]. The resulting p-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. FPKM values between the biological replications of resistant and susceptible groups were analyzed for each gene. [23].
2.4.5. GO and KEGG Enrichment Analysis of Differentially Expressed Genes (DEGs)
GOseq (1.10.0) and KOBAS (v2.0.12) software were used for GO function enrichment analysis and KEGG pathway enrichment analysis of differential gene sets. Differential gene sets are the gene set obtained by significant difference analysis and annotated to the GO or KEGG database [24,25].
2.5. Validation of DEG’s by Real-Time Quantitative PCR (RT-qPCR) Analysis
Extraction of total RNA was performed by using a modified CTAB method (Tiangen Biotech, Dalian, China; code: FP204). The first strand of the reverse transcribed cDNA was synthesized using the specifications of the Monad first-strand cDNA Synthesis Kit. The primers were designed using NCBI’s primer designing tool (Primer-BLAST). The ABI7500 quantitative PCR instrument was adopted to perform real-time fluorescence quantitative PCR. The Cha-Actin (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7830083/ accessed on 12 July 2022) gene was selected as a reference gene in the study. All data were obtained from three biological repetitions and experiments were repeated three times. The RT-qPCR analysis was performed by Norminkoda Biotechnology Co., Ltd. (Wuhan, China) (https://doi.org/10.3390/horticulturae8070594, accessed on 12 July 2022). The primers used are listed in Supplementary Table S1.
3. Results
3.1. Quantification of Disease Resistance
To determine the disease resistance level among two Ping’ou hybrid hazelnut varieties Dawei (A) and Qiuxiang (B), a disease index was calculated. At 3 DPI, the plastic bags were removed and clear visual symptoms of disease in hazelnut buds were observed (Figure 1A–D). The degree of infection in Qiuxiang (B) fruit bud was severe as compared to Dawei (A). The hazelnut variety Dawei showed comparatively fewer disease symptoms and only 28% disease index whereas 85% disease index was recorded in Qiuxiang (B) variety (Figure 1E). Disease index observation clearly indicates that Dawei has more ability to resist the disease as compared to Qiuxiang. Therefore, we further explored the resistance mechanism at transcriptome level in the two varieties at 3DPI in response to B. cinerea.
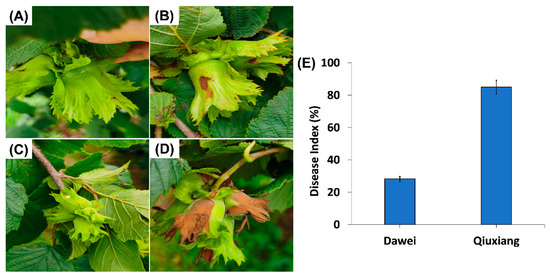
Figure 1.
Disease occurrence in two hazelnut varieties infected by B. cinerea Z9. (A,C) are non-inoculated controls for Dawei and Qiuxiang, (B) B. cinerea Z9 infected Dawei shows fewer symptoms, (D) B. cinerea Z9 infected Qiuxiang showing severe disease symptoms at 3 DPI and (E) Disease index (%) in Dawei and Qiuxiang.
3.2. Overview of Transcriptome Analysis and Differentially Expressed Genes
The bud samples from four treatments including A-CK (Dawei—control), A (Dawei—B. cinerea infected), B-CK (Qiuxiang—control) and B (Qiuxiang—B. cinerea infected) were subjected to high throughput Illumina sequencing to obtain an overview of immunity-related gene expression profiles between resistant and susceptible hazelnut varieties. The transcriptome analysis of A-CK, A, B-CK, and B in three replicates generated a total raw data ranging from 20362836 to 24855221 reads from which 20125481 to 24699286 of clean reads were assembled with the GC content ranging from 47.26% to 47.87% (Supplementary Table S2). Each sample possessed up to 97.76 and 93.77 base score in Q20 and Q30, respectively.
The differential gene analysis was performed through pair-wise comparison in four comparison groups (A vs. A-CK, B vs. B-CK, A-CK vs. B-CK, and A vs. B) that generated a total of 11,945 (3120, 2205, 4385 and 2235, respectively) differentially expressed genes among which 7671 (1873, 1571, 2629 and 1598, respectively) were up regulated and 4274 (1247, 634, 1756 and 637, respectively) were down regulated (Table 1). The pattern of up and down regulated genes in different comparison groups was also visualized through volcano plots (>2-fold change, False discovery rate < 0.05). (Supplementary Figure S2). The number of up regulated differentially expressed genes (DEGs) were higher in each comparison group as compared to down regulated DEGs.

Table 1.
Differentially expressed genes identified in four comparison groups of inoculated and un-inoculated disease resistant Dawei (A) and susceptible Qiuxiang (B) hazelnut varieties. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea).
The expression levels of all DEGs were presented in a heat map. The color differences indicate higher expression (red color) and low expression (green color). To visualize the common and unique DEGs in each comparison groups, a Venn diagram was generated which showed that only 38 and 2 up and down regulated common DEGs exist, respectively. Whereas 788, 654, 1491, 526, up- and 832, 173, 1252, and 201 down regulated DEGs were uniquely expressed in A vs. A-CK, B vs. B-CK, A-CK vs. B-CK and A vs. B respectively (Figure 2A–C).
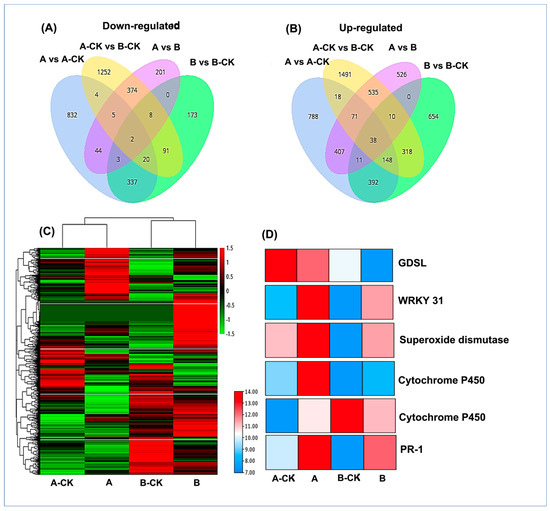
Figure 2.
(A,B) show the distribution of up and down regulating differentially expressed genes (DEGs) in different comparison groups of resistant Dawei (A) and susceptible Qiuxiang (B) of hazelnuts after B. cinerea Z9 infection. (C) Heat map showing the expression level of DEGs in all groups. Red color indicates higher expression and green color indicates lower expression. (D) Commonly expressed potential key genes involve in resistance to B. cinerea. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea).
3.3. Potential Key Genes Involved in Detection, Progression, and Resistance to B. cinerea
In all pair-wise comparisons, a total of 40 DEGs were commonly identified (Figure 2A,B). Among these genes, 5 genes involved in immunity related pathways were identified with differential expressing in resistant (Dawei) and susceptible (Qiuxiang) samples. One GDSL esterase (Cluster-22704.0), which plays a role in early detection of pathogen invasion, was highly expressed in resistant Dawei before and after B. cinerea infection as compared to susceptible Qiuxiang variety. A Pathogenesis related protein 1 (Cluster-35363.7914) was identified with an increase in expression after B. cinerea infection in both varieties. Superoxide dismutase gene (Cluster-1547.0) an important scavenger of oxidative stress also expressed in both varieties, but it showed a higher expression in Dawei after B. cinerea infection. Two putative cytochrome-P450 genes (Cluster-35363.28232 and Cluster-35363.28235) also showed higher expression in the resistant variety Dawei which was further increased after B. cinerea infection, whereas in the susceptible Qiuxiang the expressions of these genes were down regulated after B. cinerea infection. Along with these genes, a transcription factor, WRKY31 (Cluster-35363.3092), involved in various biosynthetic pathways was identified with increasing expression in both varieties but the highest expression was seen in Dawei post B. cinerea infection. The over-expression of these identified key genes clearly signifies their role in inducing resistance against B. cinerea infection in Dawei variety (Figure 2D).
3.4. GO and KEGG Pathway Enrichment of DEGs
All DEGs were classified into different sub-groups belonging to three main groups i.e., biological processes (BP), cellular components (CC), and Molecular functions (MF) through Gene ontology (GO) annotation. In A vs. A-CK, A vs. A-CK, B vs. B-CK, A-CK vs. B-CK, and A vs. B, DEGs were classified into 4, 3, 1, and 6 subgroups of BP, 2, 1, 1, 5 subgroups of CC and 7, 5, 3, 4 subgroups of MF (Figure 3). In the A vs. A-CK BP, cytoplasm and oxidoreductase activity were identified as highly enriched pathways (Figure 3A), whereas, in B vs. B-CK multiple processes were highly enriched including ribosome biogenesis, translation activity, ribosomes, and their structural constituents and structural molecular activity but the greatest number of genes expressed in this group belongs to oxidoreductase activity (Figure 3B). A-CK vs. B-CK was revealed as the least enriched group in terms of GO annotation as all genes fall in a total of 5 but important pathways including photosynthesis, thylakoid, kinase activity oxidoreductase activity, and transferase activity (Figure 3C). Furthermore, the A vs. B group was found to be the most enriched group in terms of pathways in which highly expressing pathways were biosynthesis process, cellular nitrogen compound metabolism, protein-containing complex, organelle, and intracellular processes (Figure 3D). Surprisingly the oxidoreductase activity pathway which is the most enriched pathway in B vs. B-CK and A-CK vs. B-CK comparison groups was not expressed in A vs. B. It indicates that the immune response was more active in B (susceptible-Qiuxiang variety).
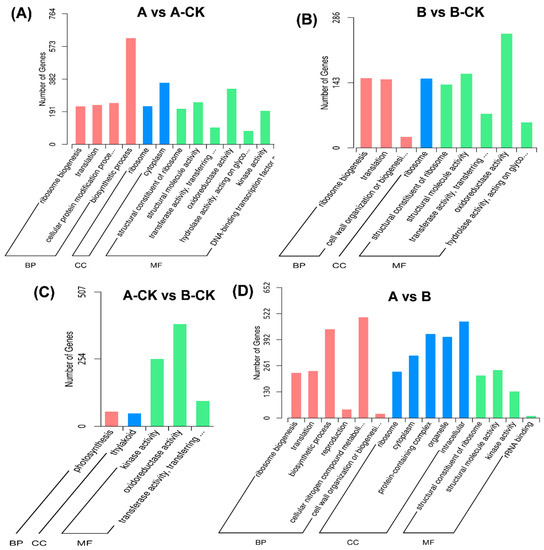
Figure 3.
Gene Ontology (GO) classifications of differentially expressed genes. The differentially expressed genes were grouped into three hierarchically stretched GO terms, biological process (BP), cellular component (CC), and molecular functions (MF). (A) Comparison between A and A-CK, (B) Comparison between B and B-CK, (C) Comparison between A-CK and B-CK, (D) Comparison between A and B. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs provides information on pathways and functions of genes associated with resistant (A) and susceptible (B) hazelnut molecular mechanisms after Botrytis cinerea Z9 infection. The KEGG enrichment classified the DEGs into 20 highly enriched pathways (p-value < 0.05) including, phenylpropanoid biosynthesis, phenylalanine metabolism, TOLL-like receptors signaling pathway, flavonoid biosynthesis, drug metabolism-cytochrome P450 and plant hormone and signal transduction in all comparison groups (Figure 4).
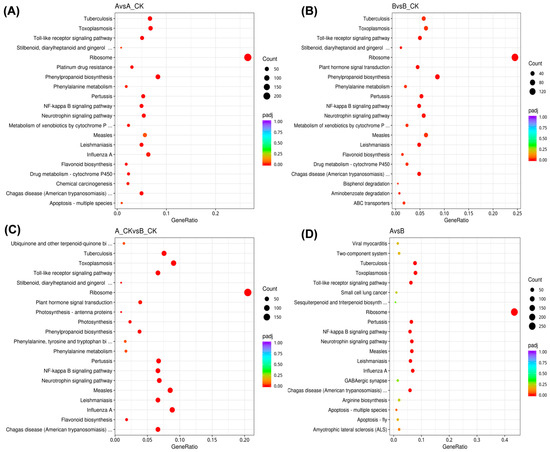
Figure 4.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment scatter plot. (A) Comparison between A and A-CK, (B) Comparison between B and B-CK, (C) Comparison between A-CK and B-CK, (D) Comparison between A and B. The vertical axis represents the path name, and the horizontal axis represents the path factor corresponding to the Rich factor. The size of the q-value is represented by the color of the point. The smaller the q-value, the closer the color is to the red color. The number of differential genes included in each pathway are expressed by the size of the point. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea).
3.5. Candidate Genes Involved in Oxidoreductase Activity
By exploring the DEGs involved in oxidoreductase pathway in the resistant (Dawei) and susceptible (Qiuxiang) hazelnut varieties, a total of 1116 genes in all pairwise comparisons were identified. Among them, two protein kinase genes acting as a receptor to initiate signaling pathway were highly expressed. Serine/threonine-protein kinase, SERK-2 (Cluster-35363.22630) and CERK-1 (Cluster-35363.27904) were found to be highly expressed in Dawei specially after B. cinerea infection, whereas their expression in Qiuxiang was relatively low (Figure 5A).
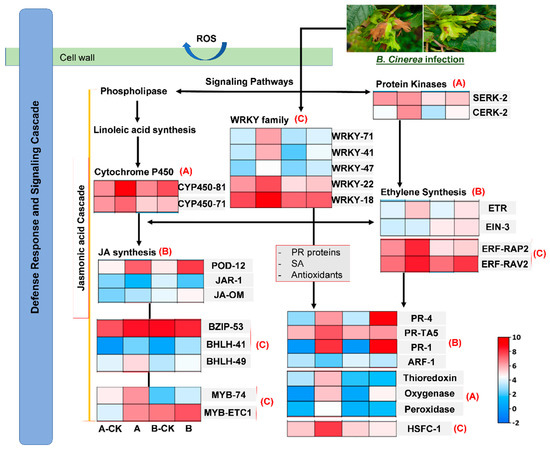
Figure 5.
Expression profile of potential key genes partaking in resistance of hazelnut plant against B. cinerea infection. Heatmaps were generated with LOG2 transformation of FPKM values for each treatment through TB-tool software. (A) expression changes of genes involve in oxidoreductase pathway, (B) expression changes of genes involve in Phytohormone and signaling pathway and (C) expression changes in key TFs. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea).
Two cytochrome P450 genes (Cluster-35363.16825 and Cluster-35363.24208) expressed higher in both varieties but the expression was highest in resistant variety after B. cinerea infection (Figure 5A). Three antioxidant enzyme associated proteins, Peroxidase (Cluster-36501.0), Thioredoxin (Cluster-35363.1884) and oxygenase (Cluster-35363.26972) were down regulated in CK plants (Treatment A-CK and B-CK) and only a slight increase in expression was observed in Qiuxiang after B. cinerea infection. Whereas these genes were highly expressed in Dawei after B. cinerea infection (Figure 5A).
3.6. Key Genes Partaking in Phytohormone Biosynthesis
Plants are sessile organisms which rely on different compounds and metabolites to defend themselves from multiple pest and pathogens. One such group of compounds that regulate signaling networks are phytohormones including, Auxins, JA, SA, ET. Through pairwise analysis of DEGs, total 101 DEGs involved in phytohormone biosynthesis were revealed in resistant (Dawei) and susceptible (Qiuxiang) hazelnut varieties. We detected one auxin-responsive protein ARF-1 (Cluster-35363.12293) highly expressed in order of CK-B > B > CK-A > A (Figure 5B).
Similarly, one ethylene receptor gene (Cluster-35363.24745) was identified with higher expression in both varieties after B. cinerea infection. Interestingly, we detected one Ethylene insensitive-3 protein EIN-3 (Cluster-35363.16105) higher-expressing in susceptible (Qiuxiang) variety as compared to Dawei. This gives the base evidence of Qiuxiang being susceptible as the high-accumulation of EIN-3 is known to exhibit enhanced disease susceptibility by compromising in pathogen associated molecular pattern (PAMP) [26] (Figure 5B).
Then, we found three pathogenesis related proteins (PR), PR-1 (Cluster-35363.1936), PR-4 (Cluster-35363.2053) and PR-TA5 (Cluster-35363.10932) involved in SA signaling pathway. PR-1 and PR-2 were found to be higher expressed after B. cinerea infection in both varieties of hazelnut. Whereas PR-TA5 was seen to be expressed in all treatments but the highest expression was recorded in resistant Dawei after B. cinerea infection (Figure 5B).
JA is considered the most important phytohormone in defense mechanism and is responsible for induction of antioxidant enzyme, POD [26]. One POD-12 gene (Cluster-35363.20218) was identified involved in JA synthesis cascade which showed higher expression after B. cinerea infection in both varieties. Similarly, one JA amino synthetase gene JAR-1 (Cluster-35363.13467) showed increased expression after B. cinerea infection, but the highest expression was again recorded in resistant Dawei. In addition, another JA-related gene, Jasmonate O-methyltransferase J-Om (Cluster-35363.4911) was found with an increased expression after infection in both varieties (Figure 5B).
3.7. Botrytis Cinerea Induced Expression Changes in Key Transcription Factors
We screened our 13 transcription factors (TFs) with differential expression profiles belonging to important families involved in multiple physiological and defense regulating pathways. Five WRKY genes (WRKY-71 Cluster-35363.15256, WRKY-41 Cluster-35363.26912, WRKY-47 Cluster-35363.13044, WRKY-22 Cluster-35363.12679 and WRKY-18 Cluster-35363.14548) were identified with increasing trend of expression after B. cinerea infection and the highest expression was recorded in Dawei. Susceptible Qiuxiang variety also showed a slight increase in expression as compared to control, but the expression was considerably low when compared with resistant Dawei (Figure 5C).
Two Ethylene responsive TFs, ERFs (Cluster-35363.16260 and Cluster-35363.15767) were found with contrasting trend of expression. Cluster-35363.16260 (ERF-RAP2) showed a slight increase in Qiuxiang whereas it was highly expressed in Dawei after B. cinerea infection. Similarly, Cluster-35363.15767 (ERF-RAV2) also showed increased expression after B. cinerea infection with no significant difference among susceptible and resistant varieties. This indicates that ethylene can be a potential regulator of defense response in hazelnut plant against B. cinerea (Figure 5C).
Two BHLH TFs were identified with considerable expression changes, of which BHLH-49 (Cluster-35363.9932) largely expressed upon B. cinerea infection in Dawei resistant plants. While BHLH-41 (Cluster-5598.1) was up regulated in both varieties upon B. cinerea infection. On the other hand, one BZIP-58 gene (Cluster-35363.7106) had highest expression in all treatments with no considerable transcriptional changes between CK and B. cinerea infected plants of both varieties (Figure 5C).
Two MYB genes showed increasing expression, of which MYB-74 (Cluster-35363.10719) showed highly increased expression in resistant Dawei plants upon B. cinerea infection. While MYB-ETC1 (Cluster-35363.22755) showed an increasing expression pattern in Dawei upon B. cinerea infection as compared to CK, the expression was seen to be high in control treatment of susceptible Qiuxiang variety and no remarkable change in expression was recorded upon B. cinerea infection (Figure 5C).
Moreover, we found one TF related to heat shock protein, HSFC-1 (Cluster-35363.20093) with remarkably increased expression in resistant Dawei variety after B. cinerea infection, whereas comparatively lower expression was recorded in susceptible Qiuxiang variety. This indicates its positive role is inducing resistance against fungal pathogens (Figure 5C).
Taken together the above results, suggest that B. cinerea infection induce multiple transcriptional changes for early detection and initiation of signaling pathways by contrasting expressional participation of genes and TFs, which at the end produce phytohormones and several enzymes to minimize the spread of infection and develop resistance against B. cinerea.
3.8. Expression Pattern and Validation of Immunity Related DEGs by Real-Time Quantitative PCR (RT-qPCR)
The real-time fluorescence quantitative PCR analysis was performed to validate the expression of 14 randomly selected genes in resistant Dawei (A) and susceptible Qiuxiang (B) varieties of hazelnut after B. cinerea Z9 infection (A and B) and in their relative controls (A-CK and B-CK). The q-RT-PCR results were found to be consistent with the RNA-seq data (Figure 6).
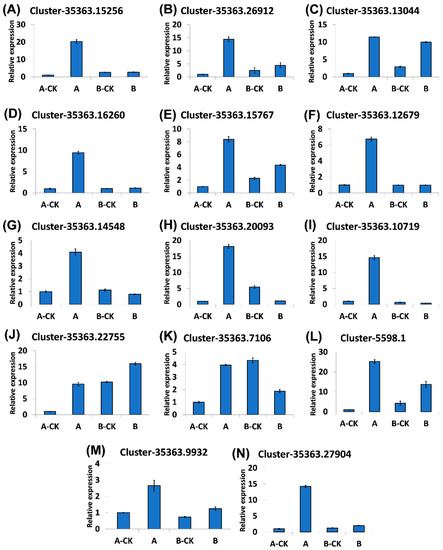
Figure 6.
Validation of immunity related genes by Real-Time Quantitative PCR in both resistant and susceptible varieties. (A) WRKY-71, (B) WRKY-41, (C) WRKY-47, (D) ERF-1, (E) ERF-RAV2, (F) WRKY-22, (G) WRKY-18, (H) HSTF-1, (I) MYB-74, (J) MYP-ETC1, (K) bZIP-53, (L) bHLH-41, (M) bHLH-49 (N) CERK-1. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea). Error bars indicates standard deviation.
4. Discussion
Modern omics technologies have facilitated the development of an efficient platform to understand the molecular and functional mechanisms in plants regarding their response and resistance against biotic and abiotic stresses [13]. Botrytis cinerea is one of the destructive pathogens of hazelnut causing yield and quality loss. In this study, high throughput sequencing helped us to directly detect and identify genetic and defense mechanisms regulating plant responses to curb the spread of B. cinerea in symptomatic and asymptomatic plants [3]. Besides, the omics studies, especially transcriptomic studies can provide a list of candidate genes partaking an important role in plant resistance, which can develop efficient strategies to generate crop resistance in an accurate, inexpensive, and rapid way [27,28]. Higher expression of these genes can activate the signal transduction pathways which in turn initiate the defense pathways through oxidant and anti-oxidant biochemical enzyme production, defensive metabolite, and phytohormone signaling [28]. A gene-to-gene relationship between host and pathogen occurs in which the pathogen’s a-virulence genes are recognized by host plants’ resistance genes to initiate a hypersensitive response at the infection site and cause programmed cell death to stop further spread of infection to other cells and limit the pathogen growth [29]. However, this approach is least effective in the case of necrotrophic pathogens [30,31]. So, identifying the most promising genes through transcriptomic study and utilizing it to develop transgenic plants is a better way to manage B. cinerea infection in hazelnut [3]. Transgenic plants can produce several enzymes, toxic proteins and proteinase inhibitors that can hydrolyze key fungal compounds to retard their growth in a plant [32]. But some plant species have developed innate resistance ability through a higher expression of immunity-associated genes, Hazelnut variety Dawei is one of them. So here we did an extensive study to determine the genetic responses in Dawei which made it resistant to B. cinerea and compared it with the susceptible hazelnut variety Qiuxiang. Through our study, we found out major expression differences in some transcripts between resistant and susceptible hazelnut varieties which can be the reason of causing immunity against B. cinerea.
Through transcriptomic analysis of resistant Dawei and susceptible Qiuxiang hazelnut varieties three days post B. cinerea infection, we obtained 11,945 DEGs by performing pairwise comparison of Dawei (A) and Qiuxiang (B) in four groups (A vs. A-CK, A vs. A-CK, B vs. B-CK, A-CK vs. B-CK and A vs. B). More up regulated DEGs as compared to down regulated DEGs was obtained. Some previous studies also reported more up regulated genes against plant disease and pest attack [33,34,35]. The overall gene ontology (GO) analysis revealed higher proportion of DEGs in functions related to biological, cellular, and molecular processes and most of them were up regulated.
In our study, we noticed that the comparison group A-CK vs. B-CK possessed a higher number of DEGs, but they were categorized into limited but most important pathways including, photosynthesis, thylakoid, kinase activity, oxidoreductase activity, and transferase activity. The activation of these molecular processes indicates that the transcription difference in resistant Dawei and susceptible Qiuxiang occur even without fungal infection which is future enhanced and improved after fungal infection in A vs. B as this combination revealed the most diverse pathway enrichment among all groups. Resistant Dawei when compared to its relative control (A-CK) also showed diverse classification of DEGs in 13 groups belonging to cellular, biological, and molecular processes whereas the susceptible variety Qiuxiang when compared to its relative control (B-CK) characterized DEGs into only 9 subgroups related to biological, cellular, and molecular functions. Biosynthetic processes, oxidoreductase activity, and kinase activity are the most important processes detected, and as these GO terms can be linked to detoxification processes in plants, so, the up regulation of these DEGs in B. cinerea infected plants especially in resistant Dawei can be responsible for disease resistance. Oxidoreductase in a large group of enzymes that are involved in REDOX reaction in plants. They are an important component of inducing resistance in plants [36,37]. Some previous studies reported the oxidoreductase enzyme activity in inducing plant resistance in different plant species [36,38].
The KEGG analysis also revealed that the higher enrichment and up regulation of DEGs occur in metabolic and detoxification pathways in all pairwise comparisons which include TOLL-like receptor signaling pathway, drug metabolisms, phytohormone signaling, phenylpropanoid biosynthesis. There was not much difference in KEGG enrichment among all comparison groups except in resistant Dawei vs. susceptible Qiuxiang in which the gene ratio was relatively low and absence of genes belonging to major signaling pathways like phytohormone and phenylpropanoid signaling. These results are consistent with some previous studies by [39,40,41]. Drug resistance–Cytochrome P450 promotes plant growth and protects it from abiotic and biotic infections through activation of detoxification and biosynthetic pathways [33]. Cytochrome P450 was reported to be involved in JA synthesis, phytoalexin biosynthesis and hormone metabolism [42].
Key Genes Mutually Participating in Resistance Pathways
Plant pathogens boost the expression of phytohormone and pathogen-resistant related genes [43]. Plant recognizes the initiation of fungal attack and counters it by producing reactive oxygen species and effector proteins which aid in the activation of the signaling pathway in systemic resistance and initiates the expression of defensive genes [44]. In this study we identified two such signaling genes, SERK-2 (Cluster-35363.22630) and CERK-1 (Cluster-35363.27904) showing a higher response after B. cinerea infection specially in resistant variety (Dawei). It was reported previously that some kinases and lipases are dependent on ET pathway and responsible for protecting plant from pest and pathogens [33]. Their accumulation also resulted in retarded growth of Pseudomonas syringae and are responsible for linoleic acid metabolism for the synthesis of JA [45].
In our study we discovered that different pathways activate and interacts with each other through involvement of multiple genes together at the same time. We found higher expression of some genes involved in oxidoreductase pathway which also have function in biosynthesis of phytohormone related compounds, like cytochrome P450 genes (Cluster-35363.16825 and Cluster-35363.24208), that are involved in conversion of linoleic acid to JA. CYP450 partakes in protection of plant from several biotic and abiotic stresses through detoxification and biosynthetic pathways. So far, the involvement of CYP450 in metabolic pathways including allene oxide synthesis in JA pathway is validated [33,46].
JA and ET biosynthesis aid in the production of multiple defensive proteins and enzymes like PR proteins and antioxidant enzyme encoded genes [47]. We identified three highly expressing PR genes (PR-1 (Cluster-35363.1936), PR-4 (Cluster-35363.2053) and PR-TA5 (Cluster-35363.10932) and three antioxidant related genes (Peroxidase (Cluster-36501.0), Thioredoxin (Cluster-35363.1884) and oxygenase (Cluster-35363.26972)). Peroxidases are important enzymes involve in different physiological and metabolic activities apart from ROS scavenging, like, lignin and suberin production to increase the barrier structure and stop invasion of pathogens [47]. It was reported that different stress conditions increases the ROS level in plant which results in an enhanced expression of thioredoxin and oxygenase genes to participate in oxidoreductase pathway [37,48]. Apart from that, our results also imply that other phytohormones including, auxin, SA and ET were involved in plant response to B. cinerea infection. The expression of ET receptors, EIN-3, PR- proteins and auxin related genes are the evidence of the hormonal cascade in defense. PR transcript accumulation due to ET and SA signaling have also been reported previously in Arabidopsis [49].
Expression of various TF families indicated that TFs plays an essential role in resistance response of hazelnut against B. cinerea (Figure 5C). Expression accumulation of large number of WRKY transcripts reflect their important role in resistant related pathways. The accumulation of ERF TFs signifies the importance of ET signaling in response to B. cinerea infection. Nakayasu, et al. [50] reported increased plant defenses in tomato upon overexpression of ERF TF JRE4. Apart from that, the expression changes in BHLH, BZIP, MYB and HSTF TFs with increased expression in resistant variety Dawei presents them as immunity related TFs. Our finding of these TFs was in consistent with previous findings by [47], Li, et al. [51,52].
The results of the comparative analysis between two hazelnut varieties, one susceptible to B. cinerea (Qiuxiang) and the other resistant (Dawei), showed that Dawei had a higher number and expression of immunity-related genes. This suggests that these genes are instrumental in providing innate resistance against B. cinerea infection in Dawei hazelnuts.
5. Conclusions
The results of the study comparing the resistance of two hazelnut varieties, Dawei and Qiuxiang, against B. cinerea infection is a significant contribution to the field of plant immunity. With the help of high-throughput technology and a focus on the molecular and genetic aspects of immunity, the research community has made great strides in understanding the mechanisms of plant resistance and susceptibility. This study specifically highlights the fact that resistance to B. cinerea is not controlled by a single gene, but rather, involves the coordinated expression of multiple genes. The identification of these genes in this study opens new avenues for future research and has implications for the development of more resistant cultivars through transgenic technology. In conclusion, this study emphasizes the importance of continued research in this field to gain a better understanding of the complex processes involved in plant immunity, and how these processes can be harnessed to protect crops against devastating pathogens like B. cinerea.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f14030493/s1, Supplementary Figure S1. Bagging of the wounded flower buds after being inoculated with B. cinerea Z9. (A) disease-resistant variety Dawei (A). (B) Disease susceptible variety Qiuxiang (B). Supplementary Figure S2. Volcano plot showing DEGs (>2-fold change, FDR< 0.05) between four comparison groups of resistant Dawei (A) and susceptible Qiuxiang (B) of hazelnuts after Botrytis cinerea Z9 infection. The scatter in the figure represents each gene. Red indicates upregulated genes, green indicates downregulated genes, and blue are common genes. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea). Supplementary Table S1. Primers used for RT-qPCR analysis. Supplementary Table S2. Summary of reads in Dawei (A) and Qiuxiang (B) variety of hazelnut transcriptome. Treatments: A-CK (Dawei-un-inoculated control), A (Dawei- inoculated with B. cinerea), B-CK (Qiuxiang-un-inoculated control), B (Qiuxiang- inoculated with B. cinerea).
Author Contributions
Conceptualization, J.S., Y.Z., Z.Z., J.H. and L.C.; Formal analysis, Y.Z., Z.Z., J.H. and L.C.; Funding acquisition, J.S.; Investigation, Y.Z. and L.C.; Methodology, Y.Z. and Z.Z.; Resources, L.C.; Software, Y.Z.; Supervision, Z.Z.; Validation, Z.Z. and L.C.; Visualization, Z.Z.; Writing—original draft, J.S. and J.H.; Writing—review & editing, J.S. and J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset supporting the conclusions of this article is available in the NCBI SRA repository under the project number PRJNA798644 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA798644 (accessed on 5 August 2022)).
Conflicts of Interest
The authors declare that they have no conflict of interest for the publication of the manuscript.
References
- ABAWI, G.; Moktan, K.; Stewart, C.; Hadad, R.; Jones, L. 2010 Northeastern Division Meeting Abstracts. Phytopathology 2012, 101, S257. [Google Scholar]
- Choquer, M.; Fournier, E.; Kunz, C.; Levis, C.; Pradier, J.-M.; Simon, A.; Viaud, M. Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 2007, 277, 1–10. [Google Scholar] [CrossRef]
- AbuQamar, S.; Moustafa, K.; Tran, L.S. Mechanisms and strategies of plant defense against Botrytis cinerea. Crit. Rev. Biotechnol. 2017, 37, 262–274. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Legard, D.; Xiao, C.; Mertely, J.; Chandler, C. Effects of plant spacing and cultivar on incidence of Botrytis fruit rot in annual strawberry. Plant Dis. 2000, 84, 531–538. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Antico, C.J.; Colon, C.; Banks, T.; Ramonell, K.M. Insights into the role of jasmonic acid-mediated defenses against necrotrophic and biotrophic fungal pathogens. Front. Biol. 2012, 7, 48–56. [Google Scholar] [CrossRef]
- Audenaert, K.; De Meyer, G.B.; Höfte, M.M. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002, 128, 491–501. [Google Scholar] [CrossRef]
- Newman, M.-A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 2013, 4, 139. [Google Scholar] [CrossRef]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef]
- Eckardt, N.A. BIK1 Function in Plant Growth and Defense Signaling; American Society of Plant Biologists: Rockville, MD, USA, 2011. [Google Scholar]
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E. Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 2012, 24, 3530–3557. [Google Scholar] [CrossRef]
- Smith, J.E.; Mengesha, B.; Tang, H.; Mengiste, T.; Bluhm, B.H. Resistance to Botrytis cinerea in Solanum lycopersicoides involves widespread transcriptional reprogramming. BMC Genom. 2014, 15, 334. [Google Scholar] [CrossRef]
- Son, G.H.; Wan, J.; Kim, H.J.; Nguyen, X.C.; Chung, W.S.; Hong, J.C.; Stacey, G. Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant-Microbe Interact. 2012, 25, 48–60. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef]
- Abuqamar, S.; Ajeb, S.; Sham, A.; Enan, M.R.; Iratni, R. A mutation in the expansin-like A 2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in A rabidopsis thaliana. Mol. Plant Pathol. 2013, 14, 813–827. [Google Scholar] [CrossRef]
- Walz, A.; Zingen-Sell, I.; Loeffler, M.; Sauer, M. Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum. Plant Pathol. 2008, 57, 453–458. [Google Scholar] [CrossRef]
- Li, Z.K.; Chen, B.; Li, X.X.; Wang, J.P.; Zhang, Y.; Wang, X.F.; Yan, Y.Y.; Ke, H.F.; Yang, J.; Wu, J.H. A newly identified cluster of glutathione S-transferase genes provides Verticillium wilt resistance in cotton. Plant J. 2019, 98, 213–227. [Google Scholar] [CrossRef]
- Hansen, K.D.; Brenner, S.E.; Dudoit, S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010, 38, e131. [Google Scholar] [CrossRef]
- Chin, C.-S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef]
- Jiang, Z.; Dong, X.; Zhang, Z. Network-based comparative analysis of Arabidopsis immune responses to Golovinomyces orontii and Botrytis cinerea infections. Sci. Rep. 2016, 6, 19149. [Google Scholar] [CrossRef]
- Dean, R.A.; Lichens-Park, A.; Kole, C. Genomics of Plant-Associated Fungi and Oomycetes: Dicot Pathogens; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Greenberg, J.T. Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Biol. 1997, 48, 525–545. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Le Hénanff, G.; Profizi, C.; Courteaux, B.; Rabenoelina, F.; Gérard, C.; Clément, C.; Baillieul, F.; Cordelier, S.; Dhondt-Cordelier, S. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 2013, 64, 4877–4893. [Google Scholar] [CrossRef]
- Armijo, G.; Schlechter, R.; Agurto, M.; Muñoz, D.; Nuñez, C.; Arce-Johnson, P. Grapevine pathogenic microorganisms: Understanding infection strategies and host response scenarios. Front. Plant Sci. 2016, 7, 382. [Google Scholar] [CrossRef]
- Batool, R.; Umer, M.J.; Wang, Y.; He, K.; Shabbir, M.Z.; Zhang, T.; Bai, S.; Chen, J.; Wang, Z. Myco-Synergism Boosts Herbivory-Induced Maize Defense by Triggering Antioxidants and Phytohormone Signaling. Front. Plant Sci. 2022, 13, 790504. [Google Scholar] [CrossRef]
- Sasaki, K.; Yuichi, O.; Hiraga, S.; Gotoh, Y.; Seo, S.; Mitsuhara, I.; Ito, H.; Matsui, H.; Ohashi, Y. Characterization of two rice peroxidase promoters that respond to blast fungus-infection. Mol. Genet. Genom. 2007, 278, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-T.; Gou, L.-J.; Zeng, H.; Zhou, G.; Dong, W.-R.; Cui, Y.; Cai, Q.; Chen, Y.-X. Inhibitory Effect and Mechanism of Dill Seed Essential Oil on Neofusicoccum parvum in Chinese Chestnut. Separations 2022, 9, 296. [Google Scholar] [CrossRef]
- Meng, F.; Lv, R.; Cheng, M.; Mo, F.; Zhang, N.; Qi, H.; Liu, J.; Chen, X.; Liu, Y.; Ghanizadeh, H. Insights into the molecular basis of biocontrol of Botrytis cinerea by Clonostachys rosea in tomato. Sci. Hortic. 2022, 291, 110547. [Google Scholar] [CrossRef]
- Raufa, B.; Mazhar, R.; Javed, A.; Tehmeena, M.; Shehzad, M.; Tariq, S.; Munis, F.; Chaudhary, H. Biocontrol potential of Bacillus gibsonii and Brevibacterium frigoritolerans in suppression of Fusarium stalk rot of maize: A sustainable approach. Asian J. Agric. Biol. 2019, 7, 320–333. [Google Scholar]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- Ge, Y.; Chen, Y.; Li, C.; Zhao, J.; Wei, M.; Li, X.; Yang, S.; Mi, Y. Effect of sodium nitroprusside treatment on shikimate and phenylpropanoid pathways of apple fruit. Food Chem. 2019, 290, 263–269. [Google Scholar] [CrossRef]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef]
- Ramos Aguila, L.C.; Ashraf, H.J.; Sánchez Moreano, J.P.; Akutse, K.S.; Bamisile, B.S.; Lu, L.; Li, X.; Lin, J.; Wu, Q.; Wang, L. Genome-Wide Identification and Characterization of Toll-like Receptors (TLRs) in Diaphorina citri and Their Expression Patterns Induced by the Endophyte Beauveria bassiana. J. Fungi 2022, 8, 888. [Google Scholar] [CrossRef]
- Jun, X.; Wang, X.-y.; Guo, W.-z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar]
- AbuQamar, S.F.; Moustafa, K.; Tran, L.-S.P. ‘Omics’ and plant responses to Botrytis cinerea. Front. Plant Sci. 2016, 7, 1658. [Google Scholar] [CrossRef]
- Noctor, G.; Reichheld, J.-P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.B. Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 2004, 9, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Batool, R.; Umer, M.J.; Wang, Y.; He, K.; Zhang, T.; Bai, S.; Zhi, Y.; Chen, J.; Wang, Z. Synergistic effect of Beauveria bassiana and Trichoderma asperellum to induce maize (Zea mays L.) defense against the Asian corn borer, Ostrinia furnacalis (Lepidoptera, Crambidae) and larval immune response. Int. J. Mol. Sci. 2020, 21, 8215. [Google Scholar] [CrossRef] [PubMed]
- Batool, R.; Umer, M.J.; Shabbir, M.Z.; Wang, Y.; Ahmed, M.A.; Guo, J.; He, K.; Zhang, T.; Bai, S.; Chen, J. Seed Myco-priming improves crop yield and herbivory induced defenses in maize by coordinating antioxidants and Jasmonic acid pathway. BMC Plant Biol. 2022, 22, 554. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Zarate, S.I.; Kempema, L.A.; Walling, L.L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007, 143, 866–875. [Google Scholar] [CrossRef]
- Nakayasu, M.; Shioya, N.; Shikata, M.; Thagun, C.; Abdelkareem, A.; Okabe, Y.; Ariizumi, T.; Arimura, G.I.; Mizutani, M.; Ezura, H. JRE 4 is a master transcriptional regulator of defense-related steroidal glycoalkaloids in tomato. Plant J. 2018, 94, 975–990. [Google Scholar] [CrossRef]
- Li, R.; Zhang, J.; Li, J.; Zhou, G.; Wang, Q.; Bian, W.; Erb, M.; Lou, Y. Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. Elife 2015, 4, e04805. [Google Scholar] [CrossRef]
- Shen, X.-J.; Wang, Y.-Y.; Zhang, Y.-X.; Guo, W.; Jiao, Y.-Q.; Zhou, X.-A. Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3958. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).