Linking Microbial Decomposition to Dissolved Organic Matter Composition in the Revegetation of the Red Soil Erosion Area
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Analysis of Soil Chemical Properties
2.3. Molecular Analysis of Soil DOM
2.4. Analysis of Soil Microbial Community
2.5. Data Analysis
3. Results
3.1. Characterization of DOM Chemodiversity
3.2. Characterization of the Microbial Community
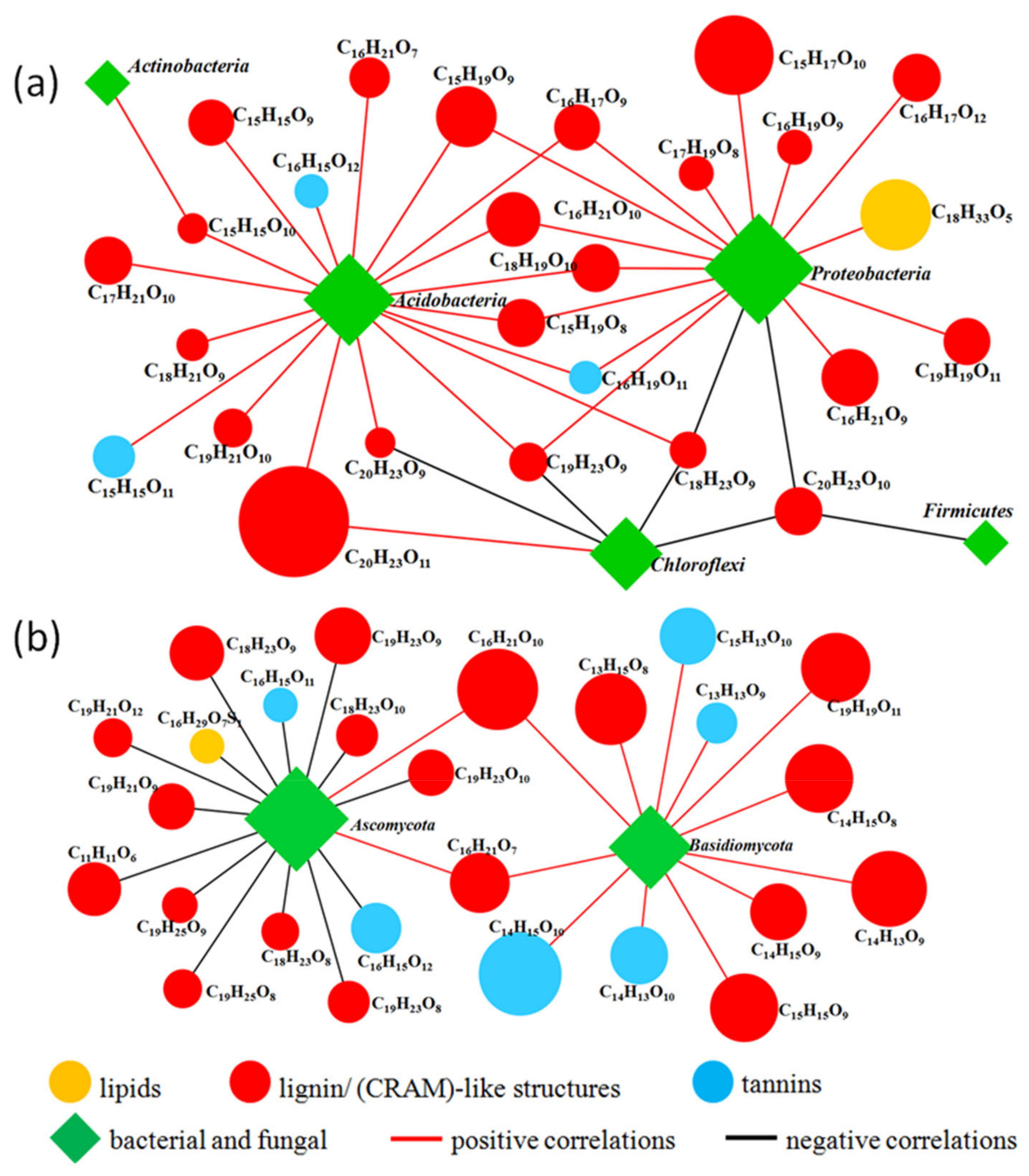
3.3. Characterizing the Relationship between Soil Microbial Communities, DOM Molecular Composition, and Soil Properties
4. Discussion
4.1. DOM Molecules Become More Complex and Difficult to Degrade with Restoration Age
4.2. Different Changes of Soil Bacteria and Fungi during Vegetation Restoration
4.3. Linkages between Soil Microbial Communities, DOM Molecular Compositions, and Soil Properties
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, P.; Raza, S.; Zamanian, K.; Ullah, S.; Kuzyakov, Y.; Virto, I.; Zhou, J. Inorganic carbon losses by soil acidification jeopardize global efforts on carbon sequestration and climate change mitigation. J. Clean. Prod. 2021, 315, 128036. [Google Scholar]
- Deng, L.; Liu, G.B.; Shangguan, Z.P. Land-use conversion and changing soil carbon stocks in China’s ‘Grain-for-Green’Program: A synthesis. Glob. Chang. Biol. 2014, 20, 3544–3556. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, H.; Wu, Y.; Ai, Z.; Zhang, J.; Liu, G.; Xue, S. Effects of natural vegetation restoration on dissolved organic matter (DOM) biodegradability and its temperature sensitivity. Water Res. 2021, 191, 116792. [Google Scholar] [CrossRef] [PubMed]
- Battin, T.J.; Luyssaert, S.; Kaplan, L.A.; Aufdenkampe, A.K.; Richter, A.; Tranvik, L.J. The boundless carbon cycle. Nat. Geosci. 2009, 2, 598–600. [Google Scholar] [CrossRef]
- Roulet, N.; Moore, T.R. Environmental chemistry: Browning the waters. Nature 2006, 444, 283–284. [Google Scholar] [CrossRef]
- Wang, J.J.; Liu, Y.; Bowden, R.D.; Lajtha, K.; Simpson, A.J.; Huang, W.L.; Simpson, M.J. Long-term nitrogen addition alters the composition of soil-derived dissolved organic matter. ACS Earth Space Chem. 2019, 4, 189–201. [Google Scholar] [CrossRef]
- Kalbitz, K.; Kaiser, K. Ecological aspects of dissolved organic matter in soils. Geoderma 2003, 113, 177–178. [Google Scholar] [CrossRef]
- Zhang, K.; Cheng, X.; Shu, X.; Liu, Y.; Zhang, Q. Linking soil bacterial and fungal communities to vegetation succession following agricultural abandonment. Plant Soil 2018, 431, 19–36. [Google Scholar] [CrossRef]
- Lu, J.; Li, S.; Wu, X.; Liang, G.; Gao, C.; Li, J.; Jin, D.; Wang, B.; Zhang, M.; Zheng, F.; et al. The dominant microorganisms vary with aggregates sizes in promoting soil carbon accumulation under straw application. Arch. Agron. Soil Sci. 2021, 69, 1–17. [Google Scholar] [CrossRef]
- Tian, Q.; Taniguchi, T.; Shi, W.Y.; Li, G.; Yamanaka, N.; Du, S. Land-use types and soil chemical properties influence soil microbial communities in the semiarid Loess Plateau region in China. Sci. Rep. 2017, 7, 45289. [Google Scholar] [CrossRef]
- Bourbonniere, R.A.; Creed, I.F. Biodegradability of dissolved organic matter extracted from a chronosequence of forest-floor materials. J. Plant Nutr. Soil Sci. 2006, 169, 101–107. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, Y.H.; Zhang, Z.T.; Huang, W.L.; Li, L.P.; Li, J.; Liu, J.; Zheng, Y.; Mo, J.M.; Zhang, W.; et al. Dissolved organic matter characteristics in soils of tropical legume and non-legume tree plantations. Soil Biol. Biochem. 2020, 148, 107880. [Google Scholar] [CrossRef]
- Guo, X.P.; Chen, H.Y.H.; Meng, M.J.; Biswas, S.R.; Ye, L.X.; Zhang, J.C. Effects of land use change on the composition of soil microbial communities in a managed subtropical forest. For. Ecol. Manag. 2016, 373, 93–99. [Google Scholar] [CrossRef]
- Deng, Q.; Cheng, X.L.; Hui, D.F.; Zhang, Q.; Li, M.; Zhang, Q.F. Soil microbial community and its interaction with soil carbon and nitrogen dynamics following afforestation in central China. Sci. Total Environ. 2016, 541, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Guo, J.; Yang, Z.; Huang, Z.; Chen, G.; Yang, Y. Rapid accumulation of carbon on severely eroded red soils through afforestation in subtropical China. For. Ecol. Manag. 2013, 300, 53–59. [Google Scholar] [CrossRef]
- Wang, X.; Li, S.; Huang, S.; Cui, Y.; Fu, H.; Li, T.; Zhao, W.; Yang, X. Pinus massoniana population dynamics: Driving species diversity during the pioneer stage of ecological restoration. Glob. Ecol. Conserv. 2021, 27, e01593. [Google Scholar] [CrossRef]
- Gao, S.; DeLuca, T.H. Wood biochar impacts soil phosphorus dynamics and microbial communities in organically-managed croplands. Soil Biol. Biochem. 2018, 126, 144–150. [Google Scholar] [CrossRef]
- Jones, D.L.; Willett, V.B. Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil. Soil Biol. Biochem. 2006, 38, 991–999. [Google Scholar] [CrossRef]
- Huang, H.; Jia, Y.; Sun, G.X.; Zhu, Y.G. Arsenic speciation and volatilization from flooded paddy soils amended with different organic matters. Environ. Sci. Technol. 2012, 46, 2163–2168. [Google Scholar] [CrossRef]
- Li, X.M.; Sun, G.X.; Chen, S.; Fang, Z.; Yuan, H.Y.; Shi, Q.; Zhu, Y.G. Molecular chemodiversity of dissolved organic matter in paddy soils. Environ. Sci. Technol. 2018, 52, 963–971. [Google Scholar] [CrossRef]
- Li, X.M.; Chen, Q.; He, C.; Shi, Q.; Chen, S.C.; Reid, B.J.; Zhu, Y.G.; Sun, G.X. Organic carbon amendments affect the chemodiversity of soil dissolved organic matter and its associations with soil microbial communities. Environ. Sci. Technol. 2018, 53, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Zhu, X.; Shen, M.; Luo, J.; Zhou, S.; Li, L.; Shi, Q.; Zhou, D.; Zhang, S.; Chen, J.; et al. Decarbonylation reaction of saturated and oxidized tar from pyrolysis of low aromaticity biomass boost reduction of hexavalent chromium. Chem. Eng. J. 2019, 360, 1042–1050. [Google Scholar] [CrossRef]
- Chen, Q.L.; An, X.L.; Li, H.; Su, J.Q.; Ma, Y.B.; Zhu, Y.G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef]
- Hu, H.Y.; Zhang, Y.L.; Zhu, Z.P.; Huang, C.F.; Wang, S.Z.; Zhou, C.F. Changes in soil enzyme activity and microbial diversity at different vegetation restoration stages in eroded red soil. Chin. J. Appl. Environ. Biol. 2021, 27, 734–741. (In Chinese) [Google Scholar] [CrossRef]
- Smith, C.R.; Sleighter, R.L.; Hatcher, P.G.; Lee, J.W. Molecular characterization of inhibiting biochar water-extractable substances using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Environ. Sci. Technol. 2013, 47, 13294–13302. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Chen, W.; Wang, J.; Du, N.; Li, Q.; Wei, G. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 2018, 6, 146. [Google Scholar] [CrossRef]
- Hansell, D.A. Recalcitrant dissolved organic carbon fractions. Annu. Rev. Mar. Sci. 2013, 5, 421–445. [Google Scholar] [CrossRef]
- Vorobev, A.; Sharma, S.; Yu, M.; Lee, J.; Washington, B.J.; Whitman, W.B.; Ballantyne, F.; Medeiros, P.M.; Moran, M.A. Identifying labile DOM components in a coastal ocean through depleted bacterial transcripts and chemical signals. Environ. Microbiol. 2018, 20, 3012–3030. [Google Scholar] [CrossRef]
- Osterholz, H.; Singer, G.; Wemheuer, B.; Daniel, R.; Simon, M.; Niggemann, J.; Dittmar, T. Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 2016, 10, 1717–1730. [Google Scholar] [CrossRef]
- Riedel, T.; Biester, H.; Dittmar, T. Molecular fractionation of dissolved organic matter with metal salts. Environ. Sci. Technol. 2012, 46, 4419–4426. [Google Scholar] [CrossRef]
- Šantl-Temkiv, T.; Finster, K.; Dittmar, T.; Hansen, B.M.; Thyrhaug, R.; Nielsen, N.W.; Karlson, U.G. Hailstones: A window into the microbial and chemical inventory of a storm cloud. PLoS ONE 2013, 8, e53550. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wullschleger, S.D.; Liang, L.; Graham, D.E.; Gu, B. Effects of warming on the degradation and production of low molecular-weight labile organic carbon in an Arctic tundra soil. Soil Biol. Biochem. 2016, 95, 202–211. [Google Scholar] [CrossRef]
- Arrieta, J.M.; Mayol, E.; Hansman, R.L.; Herndl, G.J.; Dittmar, T.; Duarte, C.M. Dilution limits dissolved organic carbon utilization in the deep ocean. Science 2015, 348, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, L.; Zhou, Y.; Wei, Q.; Chung, K.H.; Zhao, S.; Xu, C.; Shi, Q. Transformation of nitrogen compounds in deasphalted oil hydrotreating: Characterized by electrospray ionization Fourier transform-ion cyclotron resonance mass spectrometry. Energy Fuels 2013, 27, 2952–2959. [Google Scholar] [CrossRef]
- Dubenok, N.N.; Lebedev, A.V.; Gemonov, A.V. Hydrological Role of Forest Stands in a Small Catchment Basin. Russ. Agric. Sci. 2021, 47, 323–327. [Google Scholar] [CrossRef]
- Thieme, L.; Graeber, D.; Hofmann, D.; Bischoff, S.; Schwarz, M.T.; Steffen, B.; Meyer, U.N.; Kaupenjohann, M.; Wilcke, W.; Michalzik, B.; et al. Dissolved organic matter characteristics of deciduous and coniferous forests with variable management: Different at the source, aligned in the soil. Biogeosciences 2019, 16, 1411–1432. [Google Scholar] [CrossRef]
- Bai, L.L.; Cao, C.C.; Wang, C.H.; Xu, H.C.; Zhang, H.; Slaveykova, V.I.; Jiang, H.L. Toward Quantitative Understanding of the Bioavailability of Dissolved Organic Matter in Freshwater Lake during Cyanobacteria Blooming. Environ. Sci. Technol. 2017, 51, 6018–6026. [Google Scholar] [CrossRef]
- Pharand, B.; Carisse, O.; Benhamou, N. Cytological aspects of compost-mediated induced resistance against Fusarium crown and root rot in tomato. Phytopathology 2002, 92, 424–438. [Google Scholar] [CrossRef]
- Rodríguez, J.; Elissetche, J.P.; Valenzuela, S. Tree endophytes and wood biodegradation. In Endophytes of Forest Trees; Springer: Dordrecht, The Netherlands, 2011; pp. 81–93. [Google Scholar]
- Osono, T. Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can. J. Microbiol. 2006, 52, 701–716. [Google Scholar] [CrossRef]
- Schmidt, S.K.; Nemergut, D.R.; Darcy, J.L.; Lynch, R. Do bacterial and fungal communities assemble differently during primary succession? Mol. Ecol. 2014, 23, 254–258. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.B.; Xue, S.; Wang, G.L. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 2016, 97, 40–49. [Google Scholar] [CrossRef]
- Liu, G.; Chen, L.; Shi, X.; Yuan, L.Y.; Yuan Lock, T.R.; Kallenbach, R.L.; Yuan, Z. Changes in rhizosphere bacterial and fungal community composition with vegetation restoration in planted forests. Land Degrad. Dev. 2019, 30, 1147–1157. [Google Scholar] [CrossRef]
- Li, W.Q.; Huang, Y.X.; Chen, F.S.; Liu, Y.Q.; Lin, X.F.; Zong, Y.Y.; Wu, G.Y.; Yu, Z.R.; Fang, X.M. Mixing with broad-leaved trees shapes the rhizosphere soil fungal communities of coniferous tree species in subtropical forests. For. Ecol. Manag. 2021, 480, 118664. [Google Scholar] [CrossRef]
- Guo, Y.; Hou, L.; Zhang, Z.; Zhang, J.; Cheng, J.; Wei, G.; Lin, Y. Soil microbial diversity during 30 years of grassland restoration on the Loess Plateau, China: Tight linkages with plant diversity. Land Degrad. Dev. 2019, 30, 1172–1182. [Google Scholar] [CrossRef]
- Bautista-Cruz, A.; del Castillo, R.F. Soil changes during secondary succession in a tropical montane cloud forest area. Soil Sci. Soc. Am. J. 2005, 69, 906–914. [Google Scholar] [CrossRef]
- Zhu, C.; Tian, G.L.; Luo, G.W.; Kong, Y.L.; Guo, J.J.; Wang, M.; Guo, S.W.; Ling, N.; Shen, Q.R. N-fertilizer-driven association between the arbuscular mycorrhizal fungal community and diazotrophic community impacts wheat yield. Agric. Ecosyst. Environ. 2018, 254, 191–201. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Venturini, A.M.; Meyer, K.M.; Klein, A.M.; Tiedje, J.M.; Bohannan, B.J.; Nüsslein, K.; Tsai, S.M.; Rodrigues, J.L. Differential response of Acidobacteria subgroups to forest-to-pasture conversion and their biogeographic patterns in the western Brazilian Amazon. Front. Microbiol. 2015, 6, 1443. [Google Scholar] [CrossRef]
- Chen, Z.; Maltz, M.R.; Zhang, Y.; O’Brien, B.J.; Neff, M.; Wang, Y.; Cao, J. Plantations of cinnamomum camphora (Linn) presl with distinct soil bacterial communities mitigate soil acidity within polluted locations in southwest China. Forests 2021, 12, 657. [Google Scholar] [CrossRef]
- Liu, T.; Wu, X.; Li, H.; Ning, C.; Li, Y.; Zhang, X.; He, J.; Filimonenko, E.; Chen, S.; Chen, X.; et al. Soil quality and r–K fungal communities in plantations after conversion from subtropical forest. Catena 2022, 219, 106584. [Google Scholar] [CrossRef]
- Zumsteg, A.; Luster, J.; Göransson, H.; Smittenberg, R.H.; Brunner, I.; Bernasconi, S.M.; Zeyer, J.; Frey, B. Bacterial, archaeal and fungal succession in the forefield of a receding glacier. Microb. Ecol. 2012, 63, 552–564. [Google Scholar] [CrossRef]
- Boye, K.; Noël, V.; Tfaily, M.M.; Bone, S.E.; Williams, K.H.; Bargar, J.R.; Fendorf, S. Thermodynamically controlled preservation of organic carbon in floodplains. Nat. Geosci. 2017, 10, 415–421. [Google Scholar] [CrossRef]
- Follett, C.L.; Repeta, D.J.; Rothman, D.H.; Xu, L.; Santinelli, C. Hidden cycle of dissolved organic carbon in the deep ocean. Proc. Natl. Acad. Sci. USA 2014, 111, 16706–16711. [Google Scholar] [CrossRef] [PubMed]
- Roth, V.-N.; Dittmar, T.; Gaupp, R.; Gleixner, G. The molecular composition of dissolved organic matter in forest soils as a function of pH and temperature. PLoS ONE 2015, 10, e0119188. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liu, Y.; Liu, C.; Liu, Y.; Malak, M.T. Compositional changes of dissolved organic carbon during its dynamic desorption from hyporheic zone sediments. Sci. Total Environ. 2019, 658, 16–23. [Google Scholar] [CrossRef]
- Zhou, L.; Zhou, Y.Q.; Hu, Y.; Cai, J.; Liu, X.; Bai, C.G.; Tang, X.M.; Zhang, Y.L.; Jang, K.S.; Spencer, R.G.M.; et al. Microbial production and consumption of dissolved organic matter in glacial ecosystems on the Tibetan Plateau. Water Res. 2019, 160, 18–28. [Google Scholar] [CrossRef]
- Bi, B.; Yuan, Y.; Zhang, H.; Wu, Z.; Wang, Y.; Han, F. Rhizosphere soil metabolites mediated microbial community changes of Pinus sylvestris var. mongolica across stand ages in the Mu Us Desert. Appl. Soil Ecol. 2022, 169, 104222. [Google Scholar] [CrossRef]
- Zhou, M.; Hu, H.; Wang, J.; Zhu, Z.; Feng, Y. Nitric Acid Rain Increased Bacterial Community Diversity in North Subtropical Forest Soil. Forests 2022, 13, 1349. [Google Scholar] [CrossRef]
- Mao, H.; Wang, K.; Wang, Z.; Peng, J.; Ren, N. Metabolic function, trophic mode, organics degradation ability and influence factor of bacterial and fungal communities in chicken manure composting. Bioresour. Technol. 2020, 302, 122883. [Google Scholar] [CrossRef]
- Blanchet, M.; Pringault, O.; Panagiotopoulos, C.; Lefèvre, D.; Charriere, B.; Ghiglione, J.F.; Fernandez, C.; Aparicio, F.K.; Marassé, C.; Catala, P.; et al. When riverine dissolved organic matter (DOM) meets labile DOM in coastal waters: Changes in bacterial community activity and composition. Aquat. Sci. 2017, 79, 27–43. [Google Scholar] [CrossRef]
- Melillo, J.M.; Aber, J.D.; Linkins, A.E.; Ricca, A.; Fry, B.; Nadelhoffer, K.J. Carbon and nitrogen dynamics along the decay continuum: Plant litter to soil organic matter. Plant Soil 1989, 115, 189–198. [Google Scholar] [CrossRef]
- Yang, Q.; Li, R.; Zhang, W.; Zheng, W.; Wang, Q.; Chen, L.; Guan, X.; Xu, M.; Wang, S. Decomposition of harvest residue needles of different needle ages in a Chinese fir (Cunninghamia lanceolata) plantation. Plant Soil 2018, 423, 273–284. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, S.; Wang, S.; Luo, L.; Cao, D.; Christie, P. Molecular-scale investigation with ESI-FT-ICR-MS on fractionation of dissolved organic matter induced by adsorption on iron oxyhydroxides. Environ. Sci. Technol. 2016, 50, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Mesfioui, R.; Love, N.G.; Bronk, D.A.; Mulholland, M.R.; Hatcher, P.G. Reactivity and chemical characterization of effluent organic nitrogen from wastewater treatment plants determined by Fourier transform ion cyclotron resonance mass spectrometry. Water Res. 2012, 46, 622–634. [Google Scholar] [CrossRef]
- Xu, C.; Chen, H.; Sugiyama, Y.; Zhang, S.; Li, H.P.; Ho, Y.F.; Chuang, C.Y.; Schwehr, K.A.; Kaplan, D.I.; Yeager, C.; et al. Novel molecular-level evidence of iodine binding to natural organic matter from Fourier transform ion cyclotron resonance mass spectrometry. Sci. Total Environ. 2013, 449, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, M.P.; Das, S.B.; Longnecker, K.; Charette, M.A.; Kujawinski, E.B. Molecular characterization of dissolved organic matter associated with the Greenland ice sheet. Geochim. Cosmochim. Acta 2010, 74, 3768–3784. [Google Scholar] [CrossRef]
- Chen, H.; Stubbins, A.; Perdue, E.M.; Green, N.W.; Helms, J.R.; Mopper, K.; Hatcher, P.G. Ultrahigh resolution mass spectrometric differentiation of dissolved organic matter isolated by coupled reverse osmosis-electrodialysis from various major oceanic water masses. Mar. Chem. 2014, 164, 48–59. [Google Scholar] [CrossRef]
- Ohno, T.; He, Z.; Sleighter, R.L.; Honeycutt, C.W.; Hatcher, P.G. Ultrahigh resolution mass spectrometry and indicator species analysis to identify marker components of soil-and plant biomass-derived organic matter fractions. Environ. Sci. Technol. 2010, 44, 8594–8600. [Google Scholar] [CrossRef] [PubMed]
- LaRowe, D.E.; Van Cappellen, P. Degradation of natural organic matter: A thermodynamic analysis. Geochim. Cosmochim. Acta 2010, 75, 2030–2042. [Google Scholar] [CrossRef]
- Noriega-Ortega, B.E.; Wienhausen, G.; Mentges, A.; Dittmar, T.; Simon, M.; Niggemann, J. Does the chemodiversity of bacterial exometabolomes sustain the chemodiversity of marine dissolved organic matter? Front. Microbiol. 2019, 10, 215. [Google Scholar] [CrossRef] [PubMed]
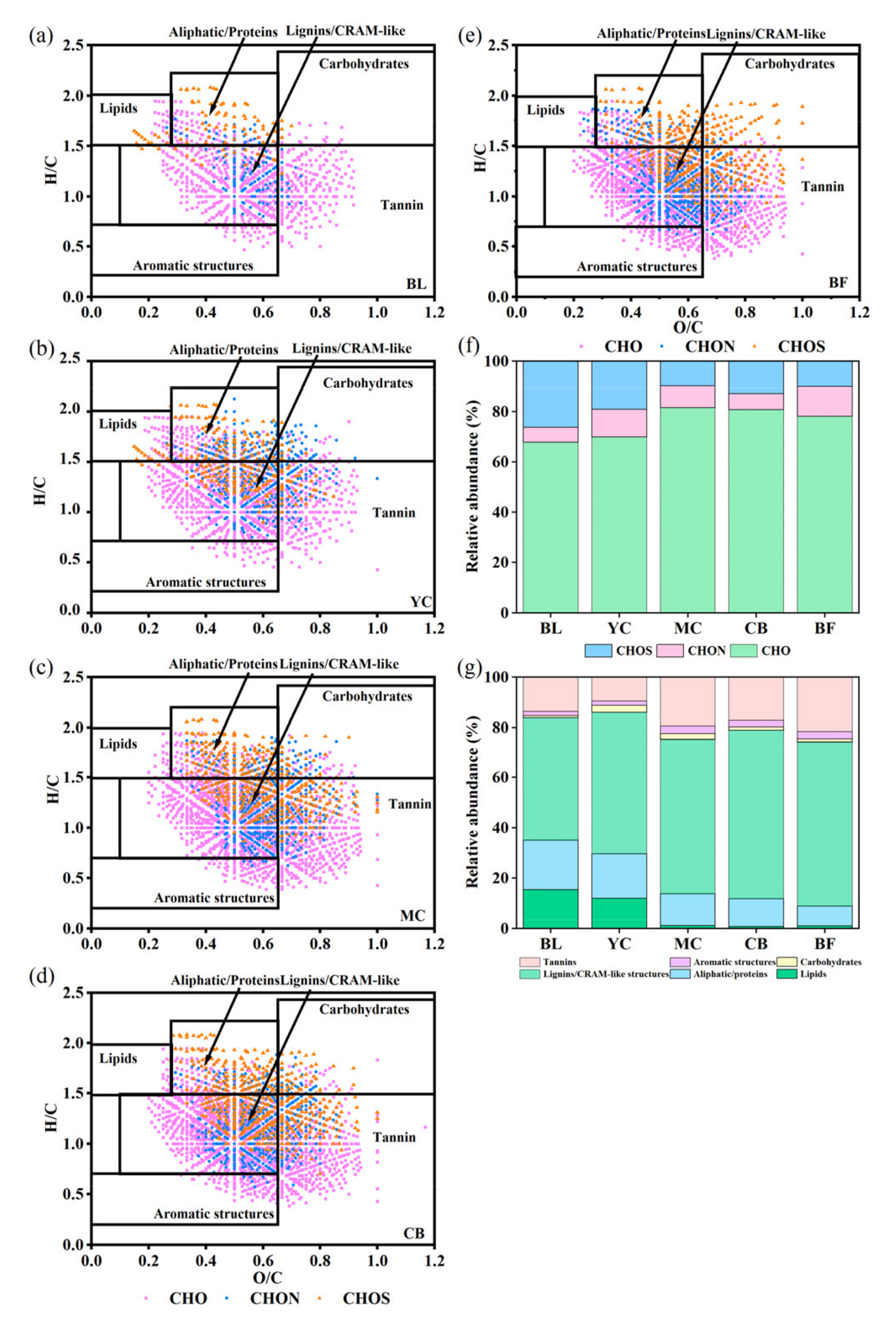
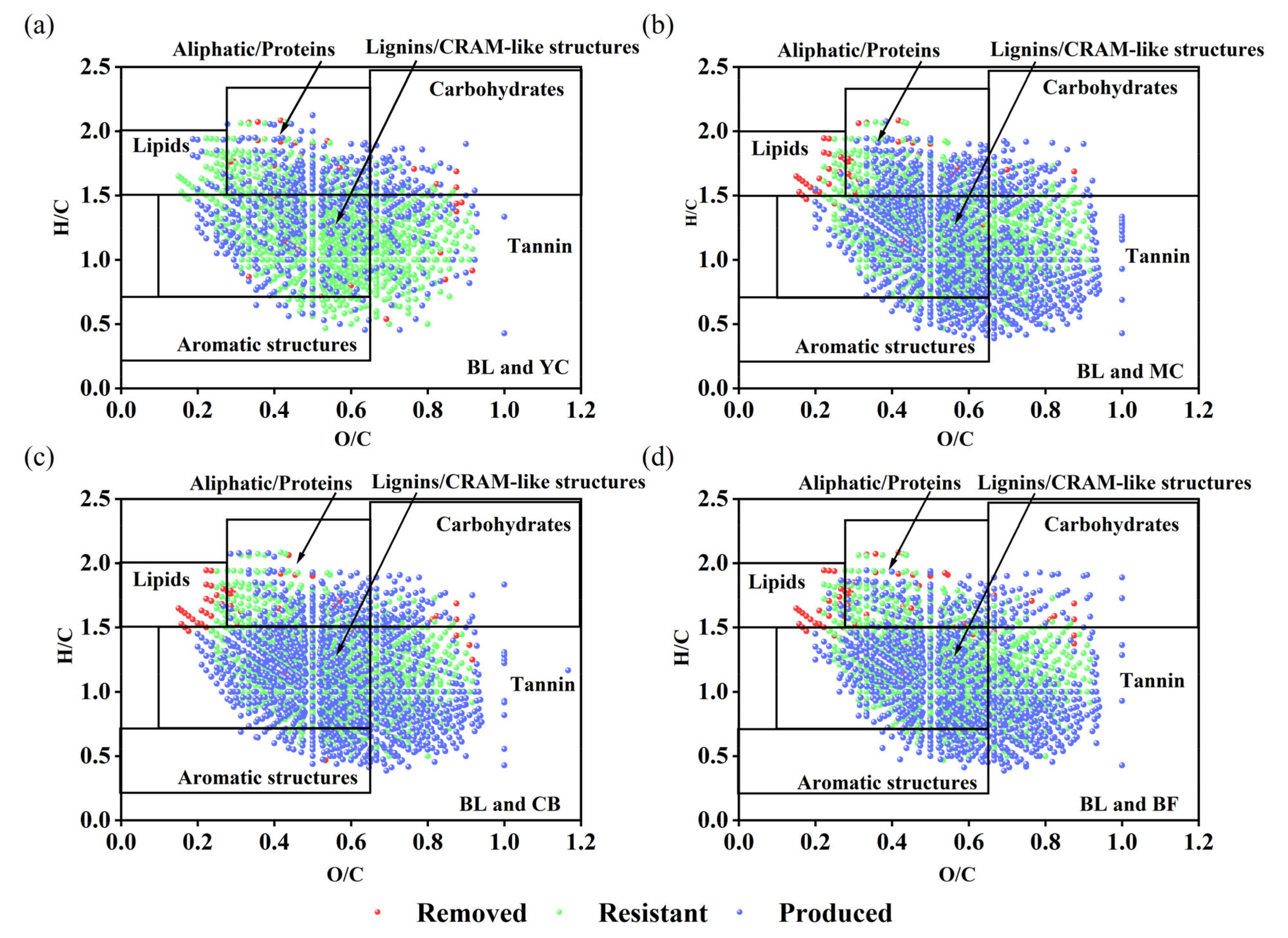
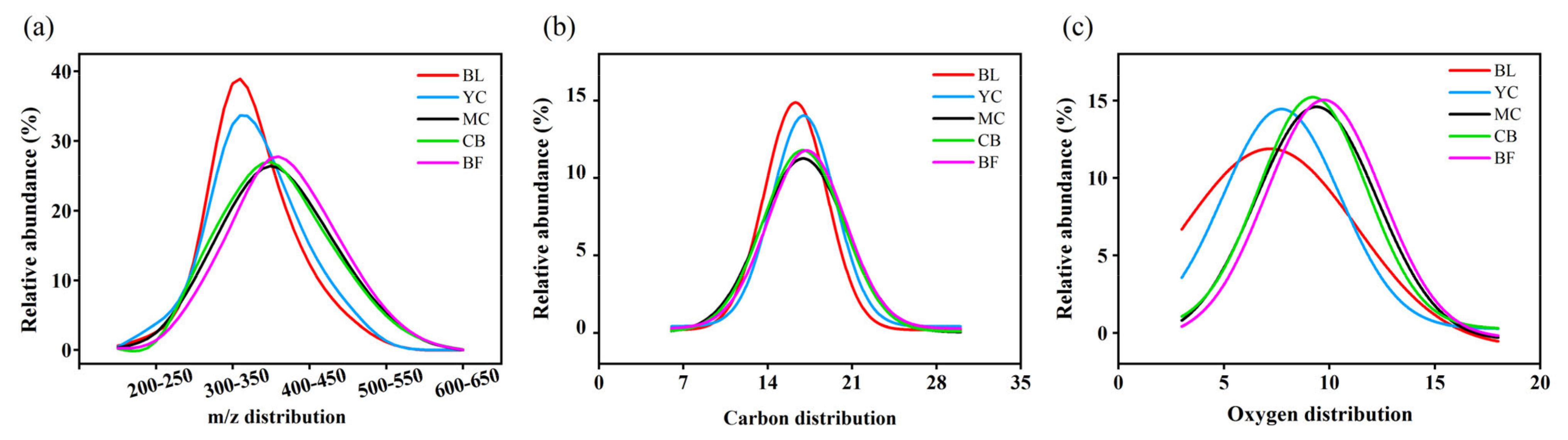
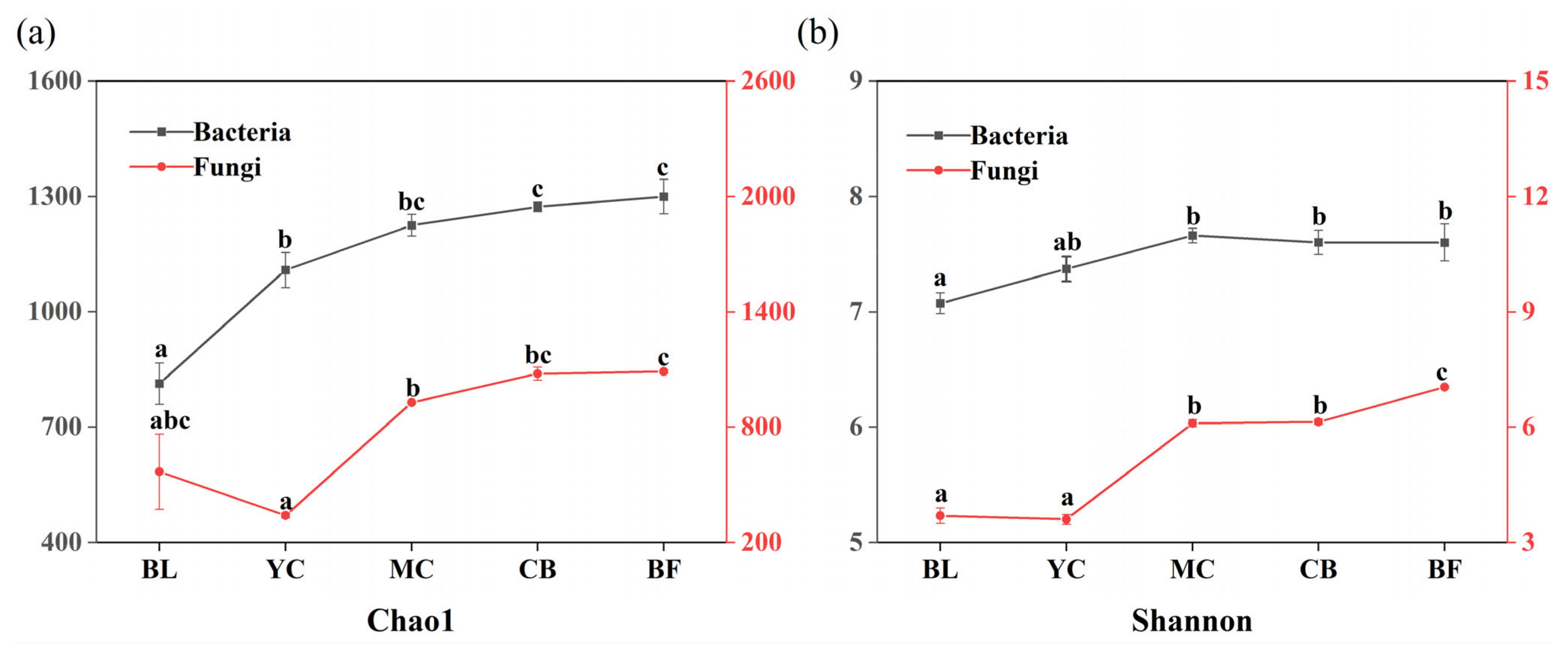



| Site | DBE | H/C | O/C | m/z | Shannon Index | Pielou Index | NOSC | H/C > 1.5 | H/C < 1.5 |
|---|---|---|---|---|---|---|---|---|---|
| BL | 6.82 | 1.23 | 0.54 | 356.90 | 6.18 | 0.88 | −0.11 | 35.81% | 64.19% |
| YC | 6.56 | 1.29 | 0.53 | 369.20 | 6.85 | 0.91 | −0.16 | 32.43% | 67.57% |
| MC | 7.70 | 1.20 | 0.57 | 395.20 | 7.31 | 0.93 | 0.02 | 16.17% | 83.83% |
| CB | 8.00 | 1.18 | 0.56 | 400.38 | 7.21 | 0.91 | 0.01 | 13.26% | 86.74% |
| BF | 8.19 | 1.16 | 0.57 | 399.34 | 7.28 | 0.93 | 0.06 | 10.20% | 89.80% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Hu, H.; Heal, K.; Sohi, S.; Tigabu, M.; Qiu, W.; Zhou, C. Linking Microbial Decomposition to Dissolved Organic Matter Composition in the Revegetation of the Red Soil Erosion Area. Forests 2023, 14, 270. https://doi.org/10.3390/f14020270
Chen W, Hu H, Heal K, Sohi S, Tigabu M, Qiu W, Zhou C. Linking Microbial Decomposition to Dissolved Organic Matter Composition in the Revegetation of the Red Soil Erosion Area. Forests. 2023; 14(2):270. https://doi.org/10.3390/f14020270
Chicago/Turabian StyleChen, Wenxin, Huaying Hu, Kate Heal, Saran Sohi, Mulualem Tigabu, Weijuan Qiu, and Chuifan Zhou. 2023. "Linking Microbial Decomposition to Dissolved Organic Matter Composition in the Revegetation of the Red Soil Erosion Area" Forests 14, no. 2: 270. https://doi.org/10.3390/f14020270
APA StyleChen, W., Hu, H., Heal, K., Sohi, S., Tigabu, M., Qiu, W., & Zhou, C. (2023). Linking Microbial Decomposition to Dissolved Organic Matter Composition in the Revegetation of the Red Soil Erosion Area. Forests, 14(2), 270. https://doi.org/10.3390/f14020270








