Identification of Key Genes for Oleoresin Biosynthesis in High and Low Oleoresin-Yielding Slash Pine Based on Transcriptome Analysis
Abstract
1. Introduction
2. Materials and Methods
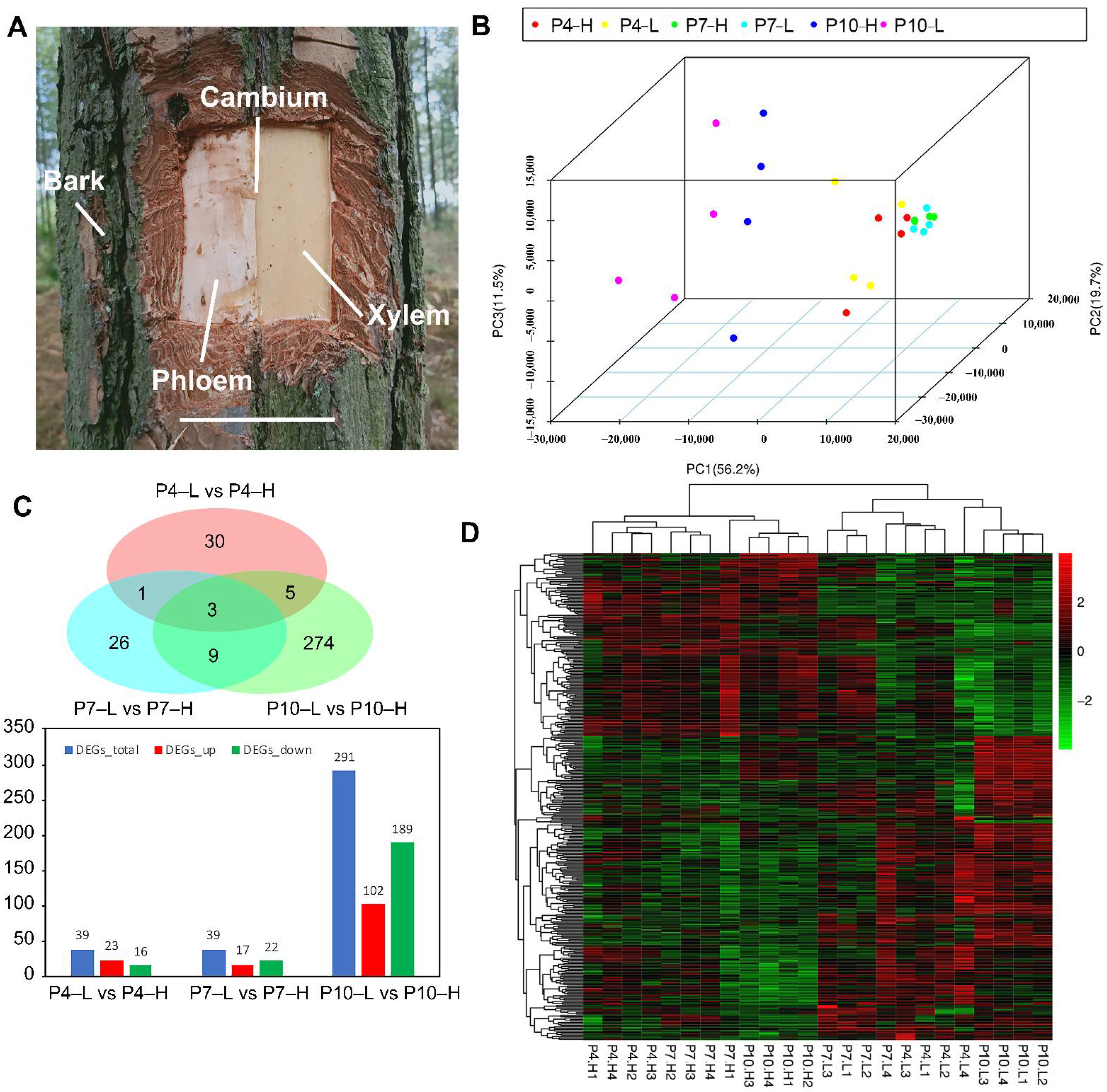
2.1. Plant Materials and Tissue Sampling for RNA-Seq
2.2. GC-MS Analysis of Oleoresin Compositions
2.3. cDNA Library Construction
2.4. Clustering and Sequencing
2.5. Sequence Data Analysis and Annotation
2.6. Differential Expression Analysis
2.7. GO and KEGG Enrichment ANALYSIS
2.8. Plant RNA Extraction and Real-Time RT-PCR
2.9. WGCNA Analysis
2.10. Yeast One-Hybrid Assay
3. Results
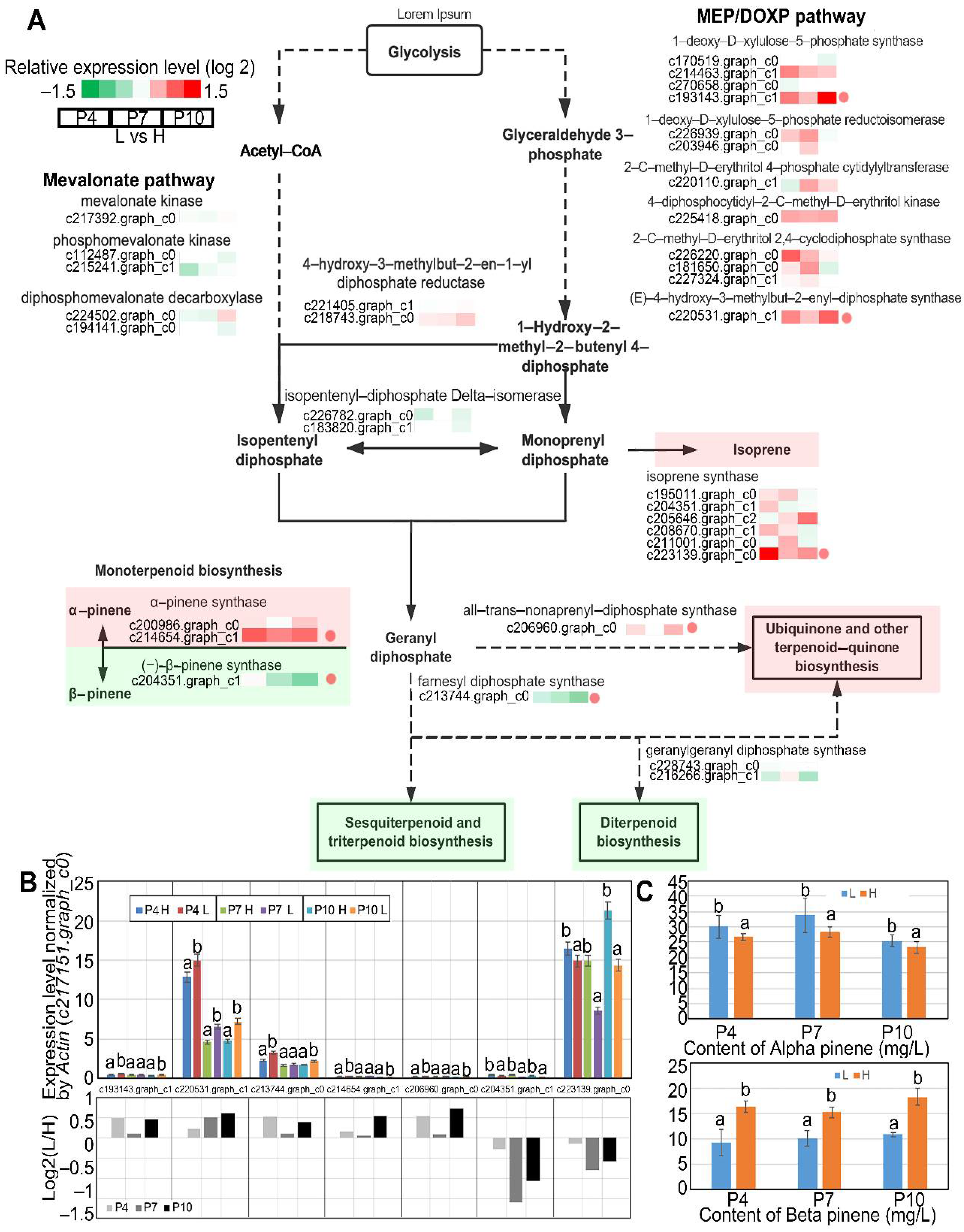
3.1. Compositions of Oleoresin Compounds in High- and Low-Yielding Slash Pine
3.2. RNA-Seq Analysis
3.3. Functional Annotation
3.4. DEGs Related to Terpenoids Biosynthesis
3.5. Co-Expressed Modules Related to the Total Oleoresin Biosynthesis
3.6. Screening of Genes Related to Monoterpene Synthesis in High and Low Oleoresin-Yielding Slash Pine
3.7. Transcription Factors Involved in the Regulation of Pine Oleoresin Biosynthesis
3.8. Identification of the Candidate Genes Targeted by c215396.Graph_c0
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, M.A.; Croteau, R.B. Resin-based defenses in conifers. Trends Plant Sci. 1999, 4, 184–190. [Google Scholar] [CrossRef]
- Pichersky, E.; Gershenzon, J. The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 2002, 5, 237–243. [Google Scholar] [CrossRef]
- Neis, F.A.; de Costa, F.; de Almeida, M.R.; Colling, L.C.; Junkes, C.F.D.O.; Fett, J.P.; Fett-Neto, A.G. Resin exudation profile, chemical composition, and secretory canal characterization in contrasting yield phenotypes of Pinus elliottii Engelm. Ind. Crop. Prod. 2019, 132, 76–83. [Google Scholar] [CrossRef]
- Loreto, F.; Pinelli, P.; Manes, F.; Kollist, H. Impact of ozone on monoterpene emissions and evidence for an isoprene-like antioxidant action of monoterpenes emitted by Quercus ilex leaves. Tree Physiol. 2004, 24, 361–367. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Yeh, S. ISOPRENE EMISSION FROM PLANTS. Annu. Rev. Plant Biol. 2001, 52, 407–436. [Google Scholar] [CrossRef]
- Ramawat, K.G.; Mérillon, J.M. Natural products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar] [CrossRef]
- Sharon, H. Book Reviews. Act. Learn. High. Educ. 2001, 2, 81–82. [Google Scholar] [CrossRef]
- Demyttenaere, J.; De Kimpe, N. Biotransformation of terpenes by fungi: Study of the pathways involved. J. Mol. Catal. B Enzym. 2001, 11, 265–270. [Google Scholar] [CrossRef]
- Chatzivasileiou, A.O.; Ward, V.; Edgar, S.M.; Stephanopoulos, G. Two-step pathway for isoprenoid synthesis. Proc. Natl. Acad. Sci. USA 2018, 116, 506–511. [Google Scholar] [CrossRef]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef]
- Liang, P.-H. Reaction kinetics, catalytic mechanisms, conformational changes, and inhibitor design for prenyltransferases. Biochemistry 2009, 48, 6562–6570. [Google Scholar] [CrossRef]
- Liang, P.-H.; Ko, T.-P.; Wang, A.H.-J. Structure, mechanism and function of prenyltransferases. JBIC J. Biol. Inorg. Chem. 2002, 269, 3339–3354. [Google Scholar] [CrossRef]
- Gershenzon, J.; Kreis, W. Biochemistry of Terpenoids: Monoterpenes, Sesquiterpenes, Diterpenes, Sterols, Cardiac Glycosides and Steroid Saponins. Biochem. Plant Second. Metab. 2018, 2, 218–294. [Google Scholar] [CrossRef]
- Schmidt, A.; WaݶChtler, B.; Temp, U.; Krekling, T.; SeݩGuin, A.; Gershenzon, J. A Bifunctional Geranyl and Geranylgeranyl Diphosphate Synthase Is Involved in Terpene Oleoresin Formation in Picea abies. Plant Physiol. 2009, 152, 639–655. [Google Scholar] [CrossRef]
- Trapp, S.; Croteau, R. DEFENSIVE RESIN BIOSYNTHESIS IN CONIFERS. Annu. Rev. Plant Biol. 2001, 52, 689–724. [Google Scholar] [CrossRef]
- Trapp, S.C.; Croteau, R.B. Genomic Organization of Plant Terpene Synthases and Molecular Evolutionary Implications. Genetics 2001, 158, 811–832. [Google Scholar] [CrossRef]
- Yi, W.; Utilization, Y.A.O.F.K.L.O.F.P.C.A.; Xiaolong, Y.; Mei, H.; Jiang, L.; Juan, W. Transcriptome and gene expression analysis revealed mechanisms for producing high oleoresin yields from Simao pine (Pinus kesiya var. langbianensis). Plant Omics 2018, 11, 42–49. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, Z.; Wei, Y.; Shen, D.; Feng, Z.; Hong, S. Genome-Wide Identification of Differentially Expressed Genes Associated with the High Yielding of Oleoresin in Secondary Xylem of Masson Pine (Pinus massoniana Lamb) by Transcriptomic Analysis. PLoS ONE 2015, 10, e0132624. [Google Scholar] [CrossRef]
- Mei, L.; Li, Z.; Yan, Y.; Wen, Z.; Wen, X.; Yang, Z.; Feng, Y. Identification and functional study of oleoresin terpenoid biosynthesis-related genes in masson pine (Pinus massoniana L.) based on transcriptome analysis. Tree Genet. Genomes 2020, 16, 53. [Google Scholar] [CrossRef]
- Rodrigues, K.; Azevedo, P.; Sobreiro, L.; Pelissari, P.; Fett-Neto, A. Oleoresin yield of Pinus elliottii plantations in a subtropical climate: Effect of tree diameter, wound shape and concentration of active adjuvants in resin stimulating paste. Ind. Crop. Prod. 2008, 27, 322–327. [Google Scholar] [CrossRef]
- Susaeta, A.; Peter, G.F.; Hodges, A.W.; Carter, D.R. Oleoresin tapping of planted slash pine (Pinus elliottii Engelm. var. elliottii) adds value and management flexibility to landowners in the southern United States. Biomass-Bioenergy 2014, 68, 55–61. [Google Scholar] [CrossRef]
- Falkenhagen, E.R. Relationships between some genetic parameters and test environments in open-pollinated families of Pinus elliottii in South Africa. Theor. Appl. Genet. 1989, 77, 857–866. [Google Scholar] [CrossRef]
- Barnett, J.P.; Sheffield, R.M. Slash Pine: Characteristics, History, Status and Trends. Slash Pine: Still Growing and Growing! In Proceedings of the Slash Pine Symposium, Asheville, NC, USA, April 23–25 2002. [Google Scholar]
- Lange, B.M. The Evolution of Plant Secretory Structures and Emergence of Terpenoid Chemical Diversity. Annu. Rev. Plant Biol. 2015, 66, 139–159. [Google Scholar] [CrossRef]
- Yi, M.; Jia, T.; Dong, L.; Zhang, L.; Leng, C.; Liu, S.; Lai, M. Resin yield in Pinus elliottii Engelm is related to the resin flow rate, resin components and resin duct characteristics at three locations in southern China. Ind. Crop. Prod. 2020, 160, 113141. [Google Scholar] [CrossRef]
- Cunningham, A. Pine resin tapping techniques used around the world. Pine Resin Biol. Chem. Appl. 2012, 661, 1–8. [Google Scholar]
- Hodges, A.W.; Johnson, J.D. Borehole Oleoresin Production from Slash Pine. South. J. Appl. For. 1997, 21, 108–115. [Google Scholar] [CrossRef][Green Version]
- Junkes, C.F.D.O.; Duz, J.V.V.; Kerber, M.R.; Wieczorek, J.; Galvan, J.L.; Fett, J.P.; Fett-Neto, A.G. Resinosis of young slash pine (Pinus elliottii Engelm.) as a tool for resin stimulant paste development and high yield individual selection. Ind. Crop. Prod. 2019, 135, 179–187. [Google Scholar] [CrossRef]
- Junkes, C.F.D.O.; Júnior, A.T.D.A.; de Lima, J.C.; de Costa, F.; Füller, T.; de Almeida, M.R.; Neis, F.A.; Rodrigues-Corrêa, K.C.D.S.; Fett, J.P.; Fett-Neto, A. Resin tapping transcriptome in adult slash pine (Pinus elliottii var. elliottii). Ind. Crop. Prod. 2019, 139, 111545. [Google Scholar] [CrossRef]
- Lai, M.; Zhang, L.; Lei, L.; Liu, S.; Jia, T.; Yi, M. Inheritance of resin yield and main resin components in Pinus elliottii Engelm. at three locations in southern China. Ind. Crop. Prod. 2019, 144, 112065. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Liu, C.; Wang, B.; Li, Z.; Peng, Z.; Zhang, J. TsNAC1 Is a Key Transcription Factor in Abiotic Stress Resistance and Growth. Plant Physiol. 2017, 176, 742–756. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, J.; Luan, Q. Genetic and correlation analysis of oleoresin chemical components in slash pine. Genet. Mol. Res. 2016, 15, 1–12. [Google Scholar] [CrossRef]
- Roberds, J.H.; Strom, B.L.; Hain, F.P.; Gwaze, D.P.; McKeand, S.; Lott, L.H. Estimates of genetic parameters for oleoresin and growth traits in juvenile loblolly pine. Can. J. For. Res. 2003, 33, 2469–2476. [Google Scholar] [CrossRef]
- Karanikas, C.; Walker, V.; Scaltsoyiannes, A.; Comte, G.; Bertrand, C. High vs. low yielding oleoresin Pinus halepensis Mill. trees GC terpenoids profiling as diagnostic tool. Ann. For. Sci. 2010, 67, 412. [Google Scholar] [CrossRef]
- Bai, Q.; Zhang, Q.; Cai, Y.; Lian, H.; Liu, W.; Luo, M.; Zeng, L.; He, B. Genome-wide association study of terpenoids in resin reveals candidate genes for resin yield in Pinus massoniana. Dendrobiology 2020, 84, 109–121. [Google Scholar] [CrossRef]
- Estévez, J.M.; Cantero, A.; Reindl, A.; Reichler, S.; León, P. 1-Deoxy-d-xylulose-5-phosphate Synthase, a Limiting Enzyme for Plastidic Isoprenoid Biosynthesis in Plants. J. Biol. Chem. 2001, 276, 22901–22909. [Google Scholar] [CrossRef]
- Hsieh, M.-H.; Goodman, H.M. Functional evidence for the involvement of Arabidopsis IspF homolog in the nonmevalonate pathway of plastid isoprenoid biosynthesis. Planta 2005, 223, 779–784. [Google Scholar] [CrossRef]
- Hsieh, M.-H.; Chang, C.-Y.; Hsu, S.-J.; Chen, J.-J. Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis. Plant Mol. Biol. 2008, 66, 663–673. [Google Scholar] [CrossRef]
- Querol, J.; Campos, N.; Imperial, S.; Boronat, A.; Rodríguez-Concepción, M. Functional analysis of the Arabidopsis thaliana GCPE protein involved in plastid isoprenoid biosynthesis. FEBS Lett. 2002, 514, 343–346. [Google Scholar] [CrossRef]
- Simkin, A.J.; Guirimand, G.; Papon, N.; Courdavault, V.; Thabet, I.; Ginis, O.; Bouzid, S.; Giglioli-Guivarc’H, N.; Clastre, M. Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 2011, 234, 903–914. [Google Scholar] [CrossRef]
- Cordier, H.; Karst, F.; Bergès, T. Heterologous expression in Saccharomyces cerevisiae of an Arabidopsis thaliana cDNA encoding mevalonate diphosphate decarboxylase. Plant Mol. Biol. 1999, 39, 953–967. [Google Scholar] [CrossRef]
- Henry, L.K.; Gutensohn, M.; Thomas, S.T.; Noel, J.P.; Dudareva, N. Orthologs of the archaeal isopentenyl phosphate kinase regulate terpenoid production in plants. Proc. Natl. Acad. Sci. USA 2015, 112, 10050–10055. [Google Scholar] [CrossRef]
- Sasaki, K.; Ohara, K.; Yazaki, K. Gene expression and characterization of isoprene synthase from Populus alba. FEBS Lett. 2005, 579, 2514–2518. [Google Scholar] [CrossRef]
- Phillips, M.A.; Wildung, M.R.; Williams, D.C.; Hyatt, D.C.; Croteau, R. cDNA isolation, functional expression, and characterization of (+)-α-pinene synthase and (−)-α-pinene synthase from loblolly pine (Pinus taeda): Stereocontrol in pinene biosynthesis. Arch. Biochem. Biophys. 2003, 411, 267–276. [Google Scholar] [CrossRef]
- Lu, S.; Xu, R.; Jia, J.-W.; Pang, J.; Matsuda, S.P.; Chen, X.-Y. Cloning and Functional Characterization of a β-Pinene Synthase from Artemisia annua That Shows a Circadian Pattern of Expression. Plant Physiol. 2002, 130, 477–486. [Google Scholar] [CrossRef]
- Bohlmann, J.; Steele, C.L.; Croteau, R. Monoterpene Synthases from Grand Fir (Abies grandis). J. Biol. Chem. 1997, 272, 21784–21792. [Google Scholar] [CrossRef]
- Block, A.; Fristedt, R.; Rogers, S.; Kumar, J.; Barnes, B.; Barnes, J.; Elowsky, C.G.; Wamboldt, Y.; Mackenzie, S.A.; Redding, K.; et al. Functional Modeling Identifies Paralogous Solanesyl-diphosphate Synthases That Assemble the Side Chain of Plastoquinone-9 in Plastids. J. Biol. Chem. 2013, 288, 27594–27606. [Google Scholar] [CrossRef]
- Ksas, B.; Becuwe, N.; Chevalier, A.; Havaux, M. Plant tolerance to excess light energy and photooxidative damage relies on plastoquinone biosynthesis. Sci. Rep. 2015, 5, 10919. [Google Scholar] [CrossRef]
- Sallaud, C.; Rontein, D.; Onillon, S.; Jabès, F.; Duffé, P.; Giacalone, C.; Thoraval, S.; Escoffier, C.; Herbette, G.; Leonhardt, N.; et al. A Novel Pathway for Sesquiterpene Biosynthesis from Z,Z-Farnesyl Pyrophosphate in the Wild Tomato Solanum habrochaites. Plant Cell 2009, 21, 301–317. [Google Scholar] [CrossRef]
- Akhtar, T.A.; Matsuba, Y.; Schauvinhold, I.; Yu, G.; Lees, H.A.; Klein, S.E.; Pichersky, E. The tomato cis-prenyltransferase gene family. Plant J. 2012, 73, 640–652. [Google Scholar] [CrossRef]
- Brock, N.L.; Tudzynski, B.; Dickschat, J.S. Biosynthesis of Sesqui- and Diterpenes by the Gibberellin Producer Fusarium fujikuroi. ChemBioChem 2011, 12, 2667–2676. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.; Coman, D.; Beck, G.; Barja, M.V.; Colinas, M.; Graf, A.; Welsch, R.; Rütimann, P.; Bühlmann, P.; Bigler, L.; et al. Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 2015, 209, 252–264. [Google Scholar] [CrossRef]
- Yijiang, H.; Bangping, C.; Yichi, L. A Study on the Relationships between Chlorophyll Content and Resin Productivity of Masson Pine. J. Fujian Coll. For. 1998, 18, 58. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Farrell, R.E. Transcription and the Organization of Eukaryotic Genes. In RNA Methodologies; Elsevier Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- De Boer, K.; Tilleman, S.; Pauwels, L.; Bossche, R.V.; De Sutter, V.; Vanderhaeghen, R.; Hilson, P.; Hamill, J.D.; Goossens, A. apetala2/ethylene response factor and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 2011, 66, 1053–1065. [Google Scholar] [CrossRef]
- Montiel, G.; Zarei, A.; Körbes, A.P.; Memelink, J. The Jasmonate-Responsive Element from the ORCA3 Promoter from Catharanthus roseus is Active in Arabidopsis and is Controlled by the Transcription Factor AtMYC2. Plant Cell Physiol. 2011, 52, 578–587. [Google Scholar] [CrossRef]
- Wang, C.-T.; Liu, H.; Gao, X.-S.; Zhang, H.-X. Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Rep. 2010, 29, 887–894. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, M.; Zhang, L.; Cheng, Z.; Hu, R.; Gao, Y.; Jin, C.; Yuan, S.; Sun, S.; Lai, M. Identification of Key Genes for Oleoresin Biosynthesis in High and Low Oleoresin-Yielding Slash Pine Based on Transcriptome Analysis. Forests 2022, 13, 1337. https://doi.org/10.3390/f13081337
Yi M, Zhang L, Cheng Z, Hu R, Gao Y, Jin C, Yuan S, Sun S, Lai M. Identification of Key Genes for Oleoresin Biosynthesis in High and Low Oleoresin-Yielding Slash Pine Based on Transcriptome Analysis. Forests. 2022; 13(8):1337. https://doi.org/10.3390/f13081337
Chicago/Turabian StyleYi, Min, Lu Zhang, Zishan Cheng, Rong Hu, Yuan Gao, Cangfu Jin, Shenggui Yuan, Shiwu Sun, and Meng Lai. 2022. "Identification of Key Genes for Oleoresin Biosynthesis in High and Low Oleoresin-Yielding Slash Pine Based on Transcriptome Analysis" Forests 13, no. 8: 1337. https://doi.org/10.3390/f13081337
APA StyleYi, M., Zhang, L., Cheng, Z., Hu, R., Gao, Y., Jin, C., Yuan, S., Sun, S., & Lai, M. (2022). Identification of Key Genes for Oleoresin Biosynthesis in High and Low Oleoresin-Yielding Slash Pine Based on Transcriptome Analysis. Forests, 13(8), 1337. https://doi.org/10.3390/f13081337





