Abstract
The mechanism of Picea wilsonii adapting to abiotic stress remains largely unknown. NAC (NAM, ATAF and CUC) transcription factors play significant roles in plant response to adverse environments. In this study, based on our previous RNA-seq, we analyzed the expression patterns of PwNAC38, and revealed its functions in the process of PwNAC38-mediated stress responses. An open-reading frame, encoding PwNAC38 protein with 330 amino acids, was isolated from the cDNA library, a process which can be induced by drought, salt and ABA treatment. Subcellular localization and yeast experiments showed that PwNAC38 was a nuclear-localized transcription factor, and could form homodimers. The full length of PwNAC38 showed transcriptional activity, while the truncated segments, C-PwNAC38 (156–330 aa) and N-PwNAC38 (1–156 aa), did not. The constitutive expression of PwNAC38 (OE lines) in Arabidopsis did not exert influence on the growth of transgenic plants under normal conditions, whereas transgenic seedlings showed higher survival rates, and the seeds had stronger vigor and a higher germination rate under drought and salt stress. The seed germination and root growth of PwNAC38 OE lines were significantly inhibited in the presence of ABA, suggesting the hypersensitivity of PwNAC38 to ABA treatment. Physiological assays showed that the activity of antioxidant enzymes, such as SOD and POD, increased, and that the accumulation of superoxide anion decreased, in OE lines under stress conditions. Moreover, overexpression of PwNAC38 significantly improved drought and salt tolerance in apple calli. A qRT-PCR assay showed that overexpression of PwNAC38 in Arabidopsis promoted the expression of drought or ABA-responsive genes ATHB-7, ANAC019, ERD1, DREB2A, RD29A, ABI5 and NCED3. Taken together, our results revealed that PwNAC38 is positively involved in plants’ response to drought and salt stress by enhancing ROS scavenging efficiency, and is partially dependent on the ABA signaling pathway.
1. Introduction
Adverse environmental conditions, such as drought and salt stress, are the main limiting factors that restrict plant growth and distribution [1]. Plants have evolved a series of strategies to respond to abiotic stress at the molecular and cellular levels, thus improving the expression of a variety of stress-induced genes [2,3]. These target genes can be classified into two groups—functional genes and structural genes [4]—in which transcription factors (TFs) cooperatively or separately regulate the transcription of key genes, to constitute gene regulation networks [5].
Transcription factors are a class of essential regulators with transcriptional activation or inhibition functions that consist of specifically binding to the cis-acting elements in the promoter of target genes [6]. NAC (NAM, ATAF1/2 and CUC2) proteins constitute a major plant-specific transcription factor family, which is characterized by a structural variable C-terminal transcriptional activation domain and a relatively conserved N-terminal DNA-binding domain [7]. In the past decade, the functional roles of NAC TFs in various processes have been extensively studied, and many have been reported to be involved in the process of plant growth and development [2], senescence [8], formation of secondary walls [9], biotic stress response [10], and the transcription factors that participate in regulating plant responses to abiotic stress—which has become a research hotspot in recent years.
In addition, NAC TFs have been shown to be involved in abiotic stress responses in a variety of plants [6]. For example, overexpression of CarNAC4 in Arabidopsis significantly improves survival rates in transgenic plants under drought and salt conditions, which indicates that CarNAC4 is a positive regulator of drought and salt tolerance in chickpea [11]. Under cold stress, a CaNAC064 loss-of-function mutant in pepper showed higher MDA (malondialdehyde, a product of lipid peroxidation) content and electrolyte leakage rate than the control groups, which indicates that RNAi plants confer more severe cell damage. Furthermore, CaNAC064 modulates plant cold-tolerance by interacting with low-temperature-induced haplo-proteinase proteins [12].
OsNTL3 is constitutively expressed in rice, and shows higher transcript abundance under heat stress. Hypersensitivity to heat stress is conferred by osntl3 knockout mutants, and OsNTL3 can directly bind to the promoter of OsZIP74 to regulate its expression under heat stress [13]. In Thellungiella halophila, TsNAC1 has been proved to serve as a collaborator of a homeodomain transcription factor, TsHD1; co-expression of two TFs accelerated the translation of heat shock proteins and the transcription level of drought responsive genes, thus promoting plant tolerance to drought and high-temperature stress [14]. MusaSNAC1, a banana NAC TF, can induce stomatal closure under drought condition by regulating H2O2 content in guard cells, thus inhibiting water loss and reducing transpiration efficiency, to enable plants to adapt to adverse environmental conditions [15].
Picea wilsonii (Picea wilsonii Mast.) belongs to temperate coniferous forests and is the endemic species in northern China [16,17]. With the frequent occurrence of natural disasters, the distribution of Picea wilsonii is becoming more limited, and the mechanism of Picea wilsonii response to abiotic stress is still largely unknown. Our previous RNA-seq analysis showed that a number of genes were induced by drought and salt stress in Picea wilsonii seedlings, among which the abiotic-stress-responsive TFs and other structure genes constituted the main components [18]. For example, PwNAC2 was induced significantly by drought and salt treatments in the PwNAC2pro:GUS transgenic Arabidopsis plant, and overexpressing PwNAC2 transgenic Arabidopsis showed increased drought and salt stress tolerance without adverse effect on plant development [19], while the other NAC TF PwNAC30 was considered as a negative regulator in response to abiotic stress [20]. Our previous study established that PwNAC38 belongs to the NAC transcription factor family. In this study, we further investigated its role in plant response to abiotic stress, by transferring into Arabidopsis and apple callus. The research will provide a theoretical basis for revealing the mechanism of stress resistance in Picea wilsonii.
2. Materials and Methods
2.1. RNA Extraction and Gene Expression Analysis
14-day-old seedlings of Arabidopsis were collected for gene expression analysis. After immersing in 10% PEG solution for 0, 3, 6 and 12 h total, RNA was extracted using an RNA extraction kit (Kangwei Century Biotechnology Co., Ltd., Beijing, China). First strand cDNA was generated by reverse transcription kit (ABM, Beijing, China). Using SYBR Green Master Mix enzymes (ABI, Vernon, CA, USA), qRT-PCR assays were then conducted. Relative expression levels were calculated by the 2−∆∆CT method, with three biological replicates. ACTIN in Arabidopsis was chosen as the reference gene; the specific reaction program was set according to the reference instruction manual.
2.2. In Silico Analysis
The cDNA sequence of PwNAC38 was obtained from a previous RNA-seq database; it was then imported into DNAMAN software to gain the corresponding CDS. The NCBI database (https://www.ncbi.nlm.nih.gov/ (accessed on 1 November 2018)) was used for homologous sequences BLAST analysis, which were subsequently analyzed by ClustalX to conduct comparative analysis. The neighbor-joining method on MEGA5 was used to construct the phylogenetic tree.
2.3. Vector Construction and Acquisition of Overexpression and Mutant Lines
The One Step Cloning Kit C112 (Vazyme Biotech Co., Ltd., Nanjing, China) was used for vector construction. The total 5 µL reaction system was listed as follows: 5 × CE II Buffer 1 µL, Exnase®II 0.5 µL, linearized vectors 1 µL, DNA products 1.5 µL and deionized water 1 µL. After incubating at 37 °C for 30 min, the recombinant vectors were transformed into Trans T1 chemically competent cells (TransGen Biotech, Beijing, China), followed by Colony PCR.
A pCAMBIA1205 vector, driven by a 35S promotor, was used to overexpress PwNAC38 in Arabidopsis. We transformed the agrobacterium strains, GV3101, harboring PwNAC38-pCAMBIA1205, into plants using floral dipping and sowed seeds in an MS selective medium with hygromycin resistance. T3 seedlings without trait segregation were regarded as homozygous lines.
At1g69490 was chosen as a homologous protein of PwNAC38 in Arabidopsis by sequence alignment. The SALK number of At1g69490 with 005010 on the T-DNA Express website (http://signal.salk.edu/cgi-bin/tdnaexpress, (accessed on 1 November 2018)) was used for genetic analysis.
2.4. Transcriptional Activation and Y2H Assays
We constructed full-length (pGBKT7-PwNAC38) and N-terminal (pGBKT7-PwNAC38-1-156) or C-terminal (pGBKT7-PwNAC38-156-330) truncated sequences of PwNAC38 into pGBKT7 vectors. Recombinant plasmids were transformed into yeast AH109-competent cells, and grown in an SD/-Trp medium. The grown yeast strains were subsequently transferred into an SD/-Trp-His-Ade+X-α-gal selective medium, to detect the expression of reporter genes.
For the Y2H assays, pGADT7+pGBKT7, pGADT7-RecT+pGBKT7-53, pGADT7-PwNAC38+pGBKT7, pGBKT7-PwNAC38-1-156+pGADT7 and pGADT7-PwNAC38+pGBKT7-PwNAC38-1-156 plasmids were transformed into AH109-competent cells. The yeast cells were grown in an SD/-Leu-Trp medium, and then transferred into SD/-Trp/Ade/Leu/Ura with X-α-gal.
2.5. Subcellular Localization
The subcellular localization assays were performed as previously described [21]. In short, a pCAMBIA1205-GFP-PwNAC38 vector was used to conduct subcellular localization. Recombinant plasmid was transformed into GV3101 cells, and then infiltrated into Nicotiana benthamiana leaves. A TCS SP8 (Leica, Wetzlar, Germany) fluorescence microscope was used to detect GFP signals.
2.6. Salt and Drought Treatments
For the seed germination experiment, seeds of WT, OE and at1g69490 lines were sown in an MS medium with different concentrations of NaCl (100/150/200 mM) and mannitol (100/200/300 mM); vernalization was processed in a refrigerator at 4 °C for 2 days, and then transferred into an incubator (21 °C, 16 h light/8 h dark photoperiod, with 70% relative humidity). The germination percentage referred to the final germination condition, and the germination rate referred to the germination condition within a set time. Germinated seedlings were counted every day for 7 days. Seeds were considered to be germinated when radicles were at least 2 mm long.
For root growth experiment, seeds of WT, OE and at1g69490 lines were sown in an MS medium for 4 days; the germinated seeds were subsequently transferred to an MS medium, with different concentrations of NaCl (100/150/200 mM) and mannitol (100/200/300 mM), for 14 days, and were vertically cultivated, to observe the growth of the taproots.
For survival assays, seeds of each line, after germination in an MS medium, were transformed into soil. The 2-week-old seedlings were used to conduct dehydration treatment for 14–20 days, followed by rewatering for 3 days. Survival rates and flowering time were recorded during this process. The plants with severe curling, wilting, browning and, finally, stunted growth were identified as dead plants. Plants after premature bolting and flower bud differentiation/inflorescence began to scatter, which was identified as the signal of flowering. For salt treatment, 2-week-old seedlings of each line grown in the ground were watered once every 3 days, with 200 mM NaCl solutions. Survival rates and phenotypic differences were recorded during this process. The plants with severe wilting and yellowing rosette leaves were identified as dead plants. All experiments had three duplications.
2.7. Biochemical Analysis
Nitro Blue Tetrazolium (NBT) and Diaminobenzidine (DAB) staining assays were conducted as previously described [22]. Briefly, 95% ethanol was used as a decoloring agent to elute chlorophyll. The antioxidant enzymes and proline content were measured by SOD, CAT and Pro detection kits (Jiancheng Bioengineering Institute, Nanjing, China).
2.8. ABA Treatment
The procedure of germination and root elongation experiments was mentioned in Section 2.6 above. Seeds of each line were sown on MS and MS + 0.5 or 1.0 µM ABA, to observe growth state. Observation of stomatal aperture was conducted as previously described [21]. Briefly, the leaves from 4-week-old seedlings of each line were chosen and immersed in MES-KCl buffer, then placed under normal light conditions for 2.5 h, to open up all pores. The leaves were subsequently transferred into an MES-KCl buffer with 0 or 10 μM ABA for another 2.5 h, and observed for stomatal aperture under a biomicroscope (DM2500).
2.9. Measurement of Fresh Weight for Transgenic Apple Calli
Calli of the apple (Royal Gala) was used for the phenotyping experiment. PwNAC38 was cloned into pSuper1300 to generate a 35S:PwNAC38-GFP construct. We transformed the agrobacterium EHA105 harboring 35S:PwNAC38-GFP into WT plants, as previously described [23]. The positive transgenic calli and WT materials were subcultured in an MS medium with appropriate concentration of 2,4-D (2,4-dichlorophenoxyacetic acid) and 6-BA, until they developed into tender and vigorous calli, then the similar-sized calli were placed in an MS medium with NaCl or PEG6000. The growth state of each group was observed, and the fresh weight was measured after 14 days.
2.10. Statistical Analysis
SPSS software version 20.0 (SPSS Corp, Chicago, IL, USA) was used for variance analysis. Duncan’s multiple range test was performed for ANOVA analysis, with p < 0.05 designated as a significant difference. Sigma Plot 10.0 was used for drawing.
3. Results
3.1. Analysis of PwNAC38 Gene
Our previous RNA-seq analysis indicated that PwNAC38 was significantly induced by PEG, salt and ABA treatment. The open-reading frame of PwNAC38 encoded 330 amino acids. Based on the results of BLAST, 13 NAC proteins in other plant species were chosen to conduct multiple sequence alignment. The results suggested that the N-terminal part of these NAC TFs was highly conserved, while the C-terminal part was relatively variable (Figure 1A). Phylogenetic analysis indicated that there was extensive homology of PwNAC38 and CiNAC in Cunninghamia lanceolate (Figure 1B).
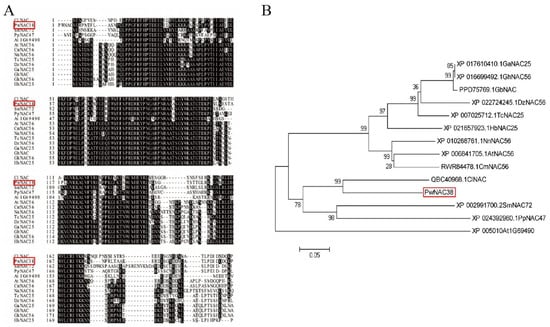
Figure 1.
Multiple sequence alignment (A) and phylogenetic analysis (B) of PwNAC38.
For subcellular localization of PwNAC38, the CDS of PwNAC38 was fused with GFP in the pCAMBIA1205 vector, then the pCAMBIA1205 empty vector and pCAMBIA1205-PwNAC38-GFP were transformed into leaves of tobacco. As was shown in Figure 2A, GFP signals were non-specifically detected in the whole cell for the empty vector, while PwNAC38-GFP signals were only detected in the nucleus, suggesting that PwNAC38 may function in the nucleus.
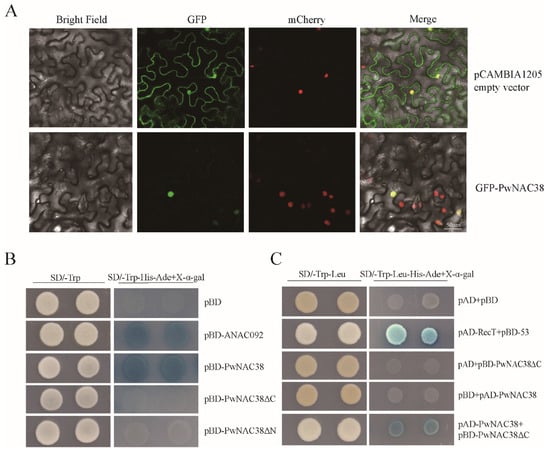
Figure 2.
Functional verification of PwNAC38. (A) Subcellular localization assay. Transgenic tobacco with nucleus-located mCherry expression was used as the positive control. The bar in the image represents 50 μm. (B) Yeast trans-acting activity assay. pBD-ANAC092 was used as the positive control, and an empty pBD vector was used as the negative control. (C) Yeast two-hybrid assay for PwNAC38 forming homodimers. pAD-RectT+pBD-53 were used as the positive controls.
To test the transcriptional activity of PwNAC38, yeast assays were used. Transformed yeast cells expressing different recombinant vectors grew well in the SD/-Trp selective medium. However, the growth of yeast cells containing pGBKT7-PwNAC38-156-330 (pBD-PwNAC38ΔN), pGBKT7-PwNAC38-1-156 (pBD-PwNAC38ΔC) and pGBKT7 (pBD) was inhibited to a large extent, while pGBKT7-PwNAC38 and the positive control were less affected in the SD/-Trp-His-Ade medium, indicating that only the full length of PwNAC38 had transcriptional activity (Figure 2B). We also conducted the Y2H assay, to verify whether PwNAC38 could form homodimers. As shown in Figure 2C, all yeast cells grew well in the SD/-Trp-Leu medium, but only the pGADT7-PwNAC38 (pAD-PwNAC38) + pBD-PwNAC38ΔC co-transformed yeast cells could grow in the SD/-Trp/Ade/Leu/Ura selective medium and activate the expression of β-galactosidase (Figure 2C). These observations confirmed that PwNAC38 can form a homodimer.
3.2. Transgenic Arabidopsis and Drought Phenotype Experiments
To determine the function of PwNAC38 in plant response to drought and salt stress, we heterologously overexpressed PwNAC38 in Arabidopsis. Three independent T3 homozygous lines (OE1-3) were selected by qRT-PCR assays (Figure S1) and used for further experiments. We first observed the phenotype of each group under normal conditions. No significant differences were observed in root length, germination and survival rates between WT, at1g69490 and OE lines. In addition, flowering time, plant height, pod length and rosette leaf morphology also displayed a nearly uniform tendency for the WT, at1g69490 and OE groups (Figure S2), suggesting that overexpression of PwNAC38 caused no impact on plant growth and development process.
Next, seeds of each line were sown in an MS medium containing a specific concentration of mannitol, which was used to simulate the drought stress. The germination rates and percentages of the OE lines were significantly higher than the WT and at1g69490 lines when the medium was supplied with mannitol (Figure 3A,B). In particular, in the medium with 200 mM mannitol, the germination rate of the OE lines reached nearly 50% after sowing for 3 days, in contrast to less than 10% of the WT lines (Figure S3C). Similar results were obtained in the root length experiment: transgenic seedlings showed higher root growth rate in 100, 200 and 300 mM mannitol media (Figure 3C), which were an average 6–8 mm longer than the WT and at1g69490 lines (Figure 3D).
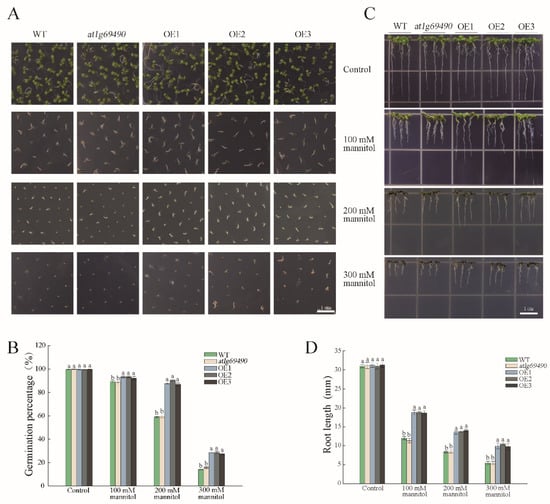
Figure 3.
Overexpression of PwNAC38 significantly improved drought tolerance in seed germination and root elongation in Arabidopsis. Observation (A) and statistics (B) of germination percentage under mannitol treatment. Observation (C) and statistics (D) of root length under mannitol treatment. The significantly different mean values at p ≤ 0.05 were labeled by different letters.
To investigate the effect of PwNAC38 on drought stress response during the adult stage, three-week-old seedlings grown in soil were used to process the drought experiment. The growth state of each line showed no significant difference before treatment. After 18-day dehydration treatment, the WT and at1g69490 seedlings were severely damaged, most of which displayed curled rosette, and even died. In contrast, the transgenic lines retained normal growth state, and showed slightly wilted phenotype (Figure 4A). After dehydration, all samples were immediately rewatered for 3 days, and nearly 90% of the OE seedlings recovered from the adverse environment, while only 10% of the WT or at1g69490 seedlings survived under drought stress (Figure 4B). Moreover, the transgenic plants maintained 50% relative water content (RWC) after 2 h dehydration, in contrast to 23% for the control groups (Figure 4C). Taken together, we conclude that overexpression of PwNAC38 could significantly improve the drought tolerance of plants.
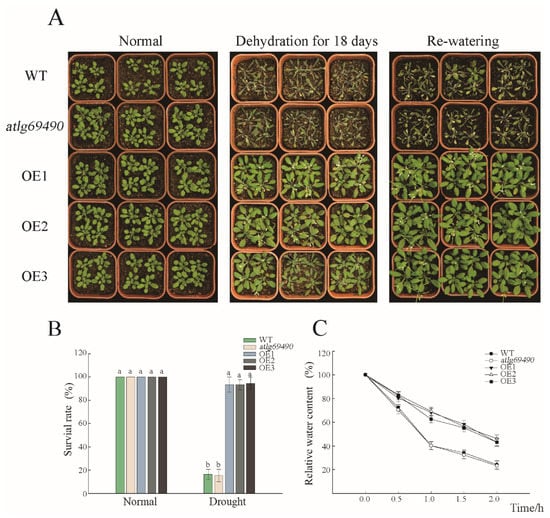
Figure 4.
Overexpression of PwNAC38 significantly improved the drought tolerance of Arabidopsis. (A) Phenotypes of different PwNAC38 transgenic lines under dehydration treatment. (B) Statistics of survival rates after drought treatment. (C) Relative water content (RWC) of isolated leaves under dehydration treatment. WT: Wild type; OE line 1, 2, 3: Overexpressing plants; at1g69490: Mutant plants. The significantly different mean values at p ≤ 0.05 were labeled by different letters.
3.3. PwNAC38 Positively Regulated Plant Salt Tolerance
We subsequently investigated the performance of overexpressing PwNAC38 transgenic Arabidopsis under salt stress. The germination rates and percentages of the OE lines were significantly higher than the WT and at1g69490 lines in the medium containing a different concentration of NaCl (Figure 5A,B), among which the final germination percentage of transgenic plants exceeded 60%, in contrast to less than 40% of the control groups under the 200 mM NaCl treatment (Figure S4D). Similarly, the root growth of the OE lines was less affected under salt treatment. For example, in the MS medium with 100 mM NaCl, the root length of the OE lines was calculated by almost 20 mm, but the control groups were nearly 12 mm (Figure 5C,D).
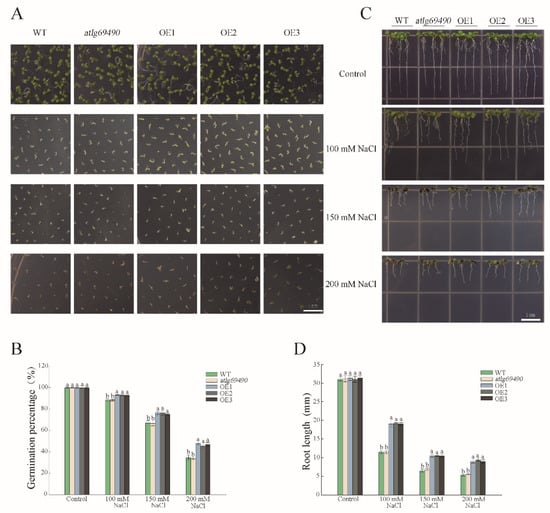
Figure 5.
Overexpression of PwNAC38 significantly improved salt tolerance in seed germination and root elongation. (A,B) Statistics of germination percentage after salt treatment. (C,D) Statistics of seedling root length after salt treatment. The significantly different mean values at p ≤ 0.05 were labeled by different letters.
We performed the phenotype assays of transgenic Arabidposis grown on saline soil, to determine whether PwNAC38 conferred plant salt tolerance. The growth situation of each line displayed significant differences after 7 days of salt treatment: in the WT and at1g69490 lines, most of the rosette leaves were severely wilting or even dead, but the transgenic lines were mildly curled (Figure 6A). The survival rates of the control groups fell to 80% after 20 days, while the OE lines maintained nearly 90% (Figure 6B). Based on the results above, the PwNAC38 OE lines exhibited enhanced survival rates under salt stress. Moreover, no obvious difference was detected between the WT and at1g69490 lines under both drought and salt conditions, implying that at1g69490 could function redundantly with homologous genes in Arabidopsis. Therefore, at1g69490 single mutant lines were not used for the following experiment.

Figure 6.
Overexpression of PwNAC38 significantly improved salt tolerance in the adult phase. (A) Phenotype experiments of different lines after NaCl treatment. (B) Statistics of survival rates under salt stress. The significantly different mean values at p ≤ 0.05 were labeled by different letters.
3.4. Overexpression of PwNAC38 Caused Susceptibility to ABA
As ABA is frequently involved in plant resistance to abiotic stress, we wondered whether the expression of PwNAC38 was mediated by the ABA signal. As shown in Figure 7A, germination of PwNAC38-OE seeds was prominently inhibited in the presence of exogenous ABA; the final germination rates of the OE lines were 10% lower than WT. Moreover, application of increasing concentrations of ABA strongly suppressed the root elongation of the OE lines, especially under 0.5 μM ABA treatment groups, in which the average root length of the transgenic plants was 15 mm as compared to 27 mm of the WT lines (Figure 7C,D). To further evaluate the effect of ABA on the plant physiological process, we observed the stomatal aperture after exogenous application of ABA. There was no significant difference of stomatal aperture under normal illumination; after imposing 10 μM ABA, however, the stomatal aperture of the OE lines was reduced to 1 μM, but was nearly 3 μM in the WT (Figure 7E,F), indicating that PwNAC38-OE plants were more responsive to the ABA signal.
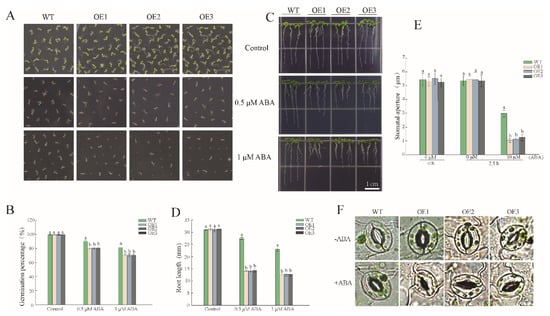
Figure 7.
Overexpression of PwNAC38 improved the sensitivity of transgenic Arabidopsis to ABA. (A,B) Statistics of germination rate under ABA treatment. (C,D) Statistics of seedling root length under ABA treatment. (E,F) Representative epidermal stoma taken by biomicroscope (DM2500, Leica) treated with or without ABA. The significantly different mean values at p ≤ 0.05 were labeled by different letters.
3.5. ROS Scavenging Efficiency Was Enhanced in Transgenic Arabidopsis
DAB and NBT staining were used to detect the accumulation of ROS under drought and salt conditions. There was no significant difference of O2 and H2O2 content observed between the WT and OE lines under normal conditions, while they massively accumulated in the WT under drought and salt stress compared to the OE lines (Figure 8A,B). To further prove these results, we also measured the activity of antioxidant enzymes under stress. The content of SOD and CAT in transgenic lines were respectively 1.32 and 1.36 times higher than control groups exposed to drought stress, and 1.52 and 2.2 times higher under salt stress (Figure 8C,D). Similarly, Proline (Pro) was also enriched in the OE lines after drought and salt treatments, the content of which was about 50 μg/g higher than the WT (Figure 8F). In addition, the content of MDA was nearly 75 nmol/mg in the OE lines, in contrast to 120 nmol/mg in the WT lines under stress conditions (Figure 8E), indicating a lighter degree of membrane lipid oxidation in PwNAC38-OE. Collectively, these results proved that overexpression of PwNAC38 conspicuously improved the ROS scavenging capacity of plants, by improving the activity of antioxidant enzymes and decreasing the accumulation of MDA.
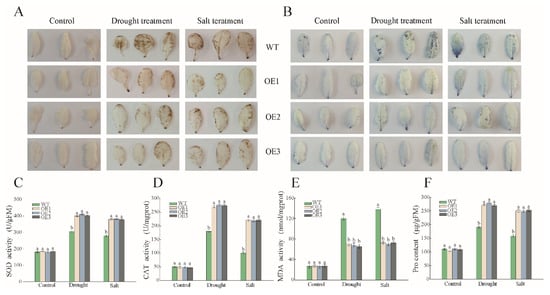
Figure 8.
Drought and salt-induced changes in ROS elimination in WT and OE plants. (A) In vivo DAB staining to detect H2O2 content. (B) In vivo NBT staining to detect the content of O2 (C) SOD activity. (D) CAT activity. (E) MDA activity. (F) Proline content. The significantly different mean values at p ≤ 0.05 were labeled by different letters.
3.6. Overexpression of PwNAC38 Improved Drought and Salt Tolerance of Transgenic Apple Calli
To investigate whether PwNAC38 can function in the regulation of plant resistance to abiotic stress in woody plants, we heterogeneously expressed PwNAC38 in the apple calli of ‘Wanglin’, and detected its expression level by using qRT-PCR assays (Figure 9).
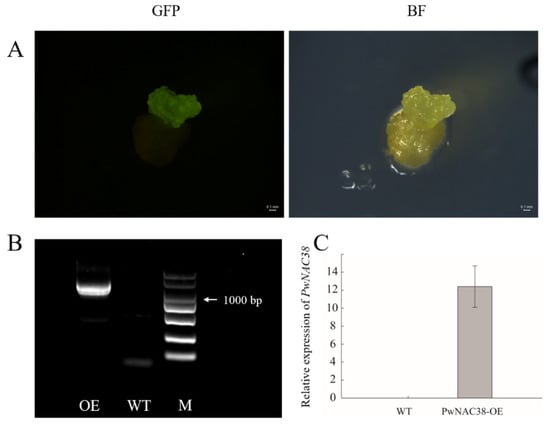
Figure 9.
Identification of overexpressing PwNAC38 in apple calli. (A) Detection of PwNAC38-GFP fusion protein under GFP channel and Bright field. (B) Identification of PwNAC38 in transgenic apple calli. (C) Relative expression of PwNAC38 in transgenic apple calli.
The transgenic apple calli overexpressing PwNAC38 (PwNAC38-OE) and WT plants were placed in the MS medium with different concentrations of PEG6000 and NaCl, to imitate adversity stress. After two weeks’ treatment, the fresh weight of the OE lines was significantly higher than that of the control groups, under both drought and salt conditions, with slighter etiolation phenotype, which indicated the improved growth rate and abiotic stress tolerance of the transgenics (Figure 10). Collectively, our findings imply that PwNAC38 is an essential regulator in plants responding to drought and salt stress.
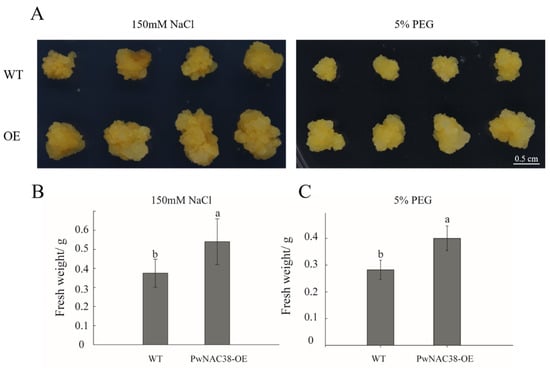
Figure 10.
Overexpression of PwNAC38 significantly improved drought and salt tolerance in apple calli. (A) Phenotype experiments of WT and OE lines after PEG6000 and NaCl treatments. (B) Fresh weight of WT and OE lines after NaCl treatment. (C) Fresh weight of WT and OE lines after PEG6000 treatment.
3.7. The Expression Level of Stress-Responsive Genes in PwNAC38 OE Lines under PEG Treatment
Due to the hypersensitivity of PwNAC38 to dehydration signals, and the potential involvement of the ABA pathway, we used a qRT-PCR assay to compare the expression levels of some drought-responsive and ABA-mediated genes, including ATHB-7, ANAC019, ERD1, DREB2A, RD29A, ABI5, ABF3 and NCED3. As shown in Figure 11, no significant differences were detected under control conditions, but these stress-responsive genes showed higher mRNA levels in OE lines after PEG6000 treatment, among which the relative expression of DREB2A reached a peak after 12-h treatment, and ERD1 was detected to be five times higher than in the WT groups (Figure 11D). Moreover, the key ABA-synthesis NCED3 and ABA-responsive ABF3 were also transcriptionally abundant in the presence of PEG6000 (Figure 11F,H). These results indicate that PwNAC38 increases drought tolerance in plants, via activation of the expression of stress-related and ABA dependent genes.
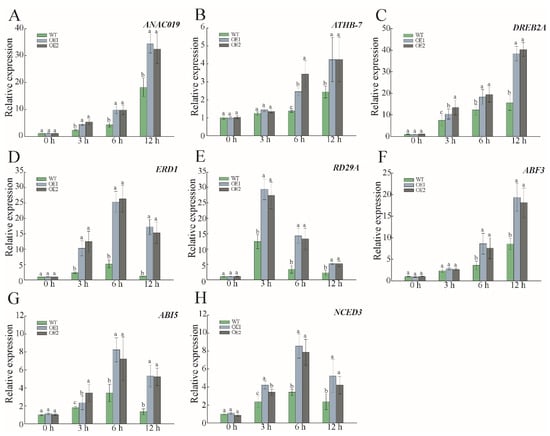
Figure 11.
Relative expression of downstream genes in WT and PwNAC38 transgenic lines under simulated drought stress by qRT-PCR. (A–E) Expression patterns of some drought responsive genes. (F–H) Expression patterns of some ABA responsive genes. The significantly different mean values at p ≤ 0.05 were labeled by different letters.
4. Discussion
4.1. PwNAC38 Is a Nucleus-Located Transcription Factor That Can Form Homodimers
NAC is a key component of plant transcription factor families. In the present study, the comparison of ORFs revealed that the NAM binding domain had little diversity between PwNAC38 and its homologs, while the C-terminal of PwNAC38 was relatively variable, which was in accordance with the typical characteristics of NAC TFs [24]. Many NAC TFs have been reported to have transcription activity. For example, TaNAC30 in wheat [25], ONAC122 and ONAC131 in rice [26], MaNAC1 and MaNAC6 in banana [27], MdNAC1 in apple [28] and CpNAC1 in pawpaw [29] have all been proved to function as transcriptional activators to regulate the processes of plant development and metabolism. However, in a recent study, some TFs were reported have no transcription activation activity, which may have been attributable to the interference of the NAC Repression Domain. In soybean, for example, the N-terminal of GmNAC20 harbored a 35-amino-acid active repression domain named NARD, and the deletion of NARD recovered transcription activation activity [6]. Intriguingly, in our assays, the full length of PwNAC38 had transcriptional activity but the truncated segments could not activate the expression of reporter genes. One possible explanation was that the functional completeness of PwNAC38 protein was destroyed due to segmentation. Further deletion experiments are required, to find out the core sequences that determined the transcription activation activity of PwNAC38.
Previous studies have indicated that NAC TFs generally work as nucleus-located proteins. For example, OsNAC6 in rice [3], MaNAC6 in banana [27] and DkNAC7 in persimmon [30] have all been confirmed to be located in the nucleus. Here, we found that PwNAC38 was also located in the nucleus, to participate in abiotic stress responses. But some NAC TFs can shuttle between cytoplasm and nucleus to play different roles under certain environmental conditions, thus contributing to the expression of spatio–temporal specificity [31]. In rice, OsNTL3 was detected in the cytomembrane under normal conditions, but the OsNTL3-GFP signals were largely observed in the nucleus after heat treatment [32]. Moreover, the tolerance of the plant to heat stress was solely dependent on the abundance of OsNTL3 in the nucleus, which implied that the location of the TF was of great significance to its functional performance. Accordingly, it will be of interest to investigate whether the location of PwNAC38 changes under abiotic stress treatments.
NAC TFs can also form homodimers or heterodimers to cause different degrees of impact on downstream genes [33]. A previous study showed that BnaNAC55 could interact with itself in vivo to regulate the expression of ROS- and defense-related genes [34]. OsNACs may function by forming homodimers and/or heterodimers through yeast two-hybrid screening of the rice cDNA library [5]. In the present study, we found that PwNAC38 forms homodimers by Y2H assays, but whether or not the physical interactions have an impact on binding affinity and the transcription activity of PwNAC38, to target genes, requires further verification.
4.2. PwNAC38 Functions as a Positive Regulator to Drought and Salt Stress via ROS and ABA Pathways
NAC TFs involved in abiotic stress responses have been reported to conduct different roles. For example, overexpression of TaNAC2 in Arabidopsis significantly improves drought stress tolerance [35]. Transgenic soybean overexpressing GmNAC20 displays enhanced salt and cold-stress-resistance, while GmNAC11 is only highly responsive to salt signals [36]. Our previous studies suggested that PwNAC11, an abiotic stress-responsive TF in Picea wilsonii, improves drought tolerance in transgenic Arabidopsis by interacting with DREB2A and ABF3, and cooperatively activating the expression of ERD1 [21]. Overexpression of PwNAC30 remarkably inhibited the growth and viability of transgenic Arabidopsis under drought and salt conditions [20]. These studies indicated that different member of NAC families in Picea wilsonii performed their specific functions in the presence of abiotic stress. In this study, PwNAC38 OE lines displayed higher germination and root elongation rates during seeding stages under both salt and mannitol treatments. Meanwhile, the adult plants showed greater drought and salt tolerance than WT groups, especially in drought phenotype experiments; the survival rates of the OE lines were 70% higher than the control groups. qRT-PCR showed that some drought-responsive genes, such as ATHB-7, ANAC019, ERD1, DREB2A and RD29A, were significantly induced in PwNAC38 OE lines. These results demonstrated that PwNAC38 is a positive regulator in response to drought and salt stress in plants, probably by regulating the downstream drought-responsive and ABA-regulated genes.
ROS homeostasis is a vital prerequisite for enabling plants’ survival under abiotic stress, as excessive accumulation of ROS may destroy the cell membrane and protein structures, and even cause DNA damage and cell death [37]. Accordingly, ROS content is an ideal parameter to reflect plant tolerance of drought and salt stress. Further testing of antioxidant enzymes activities demonstrated that PwNAC38 transgenic Arabidopsis had a higher ROS scavenging efficiency, with stronger activity of SOD and POD, enriched with Pro and less accumulation of MDA, suggesting a slighter degree of membrane lipid peroxidation present in PwNAC38 OE lines compared with WT, and that PwNAC38 had the ability to accelerate elimination of ROS under stress treatment.
It has been reported that the stress response of NAC TFs is precisely mediated by phytohormones signaling. Expression of PwNAC38 in Picea wilsonii is strongly induced by ABA treatments [38]. In this paper, we further confirmed that PwNAC38 participates in the ABA pathway. After exogenous application of ABA, germination rates and root elongation of OE lines were notably inhibited; meanwhile, the degree of stomatal closure was more obvious in transgenic plants. ABA signals were transmitted from the roots to the aerial parts of plants under stress conditions, followed by a decrease of the opening degree of stomatal aperture to minimize water loss from evaporation; synthesis of osmoprotectant and antioxidant enzymes was also promoted, to maintain cell turgor at the same time. These processes were usually regulated by TFs; overexpression of Aeluropus lagopoides TF AlNAC1 significantly increased endogenous ABA content in transgenic Arabidopsis, thus reducing stomatal aperture and eventually reducing water loss [22]. Our studies indicated that PwNAC38 was involved in ABA-induced stomatal closure, and further qRT-PCR assays implied that two pivotal genes in the ABA pathway, named ABI5 and NCED3, were highly expressed under PEG treatments, among which the NCED3 played a vital role for ABA biosynthesis in drought stress. The key ABA-synthesis NCED3 was substantially induced in vasculature; loss-of-function of nced3 mutant displayed a decreased ABA accumulation level and drought stress tolerance [39]. We thus speculated that PwNAC38 exerted its function as a regulator at least partially through the ABA pathway: on the one hand, it increased plants’ sensitivities to ABA signals, and initiated a series of physiological processes; on the other hand, overexpression of PwNAC38 promoted the transcription of ABA-synthesis genes to increase the content of endogenous ABA, thus improving the abiotic stress tolerance of plants.
4.3. The Effects of PwNAC38 on Plant Growth and Functional Similarity among Plant Species
It should be noted that some genetic evidence was uncovered in our study to confirm the complex functional similarity of PwNAC38 among species. Firstly, based on the consistent results of phenotypic experiments, both in Arabidopsis and apple calli, we confirmed that the positive regulation of PwNAC38 under environmental stress was very similar in both herbaceous and woody plants, which also provides a theoretical basis for practical application. Secondly, we investigated the functional similarity between PwNAC38 and its homologous proteins in Arabidopsis and screened homozygous at1g69490 seeds, and the loss-of-function mutants of PwNAC38 homologue in Arabidopsis, and restored it with PwNAC38. Unfortunately, the at1g69490 mutant showed no phenotypic difference under both salt and drought treatments compared to WT, not excluding the functional redundancy of At1G69490 in vivo. In fact, this is not a rare phenomenon in the functional redundancy of genes, especially for the TF superfamily. Regulation of TFs in plants is elaborately controlled by internal and external signals to maintain the homeostasis. Once the expression of one kind of TF is inhibited or overproduced, the adverse effect could be weakened by the compensatory regulation of homologous genes [40]. For instance, none of the single mutations of anac019, anac055, anac072 caused visible phenotype difference in Arabidopsis, while the anac019 anac055 anac072 triple mutant apparently accelerated leaf senescence [41].
PwNAC38 also had a 57% homology with AtNAP in Arabidopsis. AtNAP was reported to be substantially up-regulated in senescent leaves; overexpression of AtNAP also accelerated leaf aging by specifically binding to the CACGTAAGT motifs on the SAG13 promoter [42], which is an indictor gene of the aging process. Moreover, in the face of abiotic stress, many plants have evolved a unique strategy of stress-escape response, to enable them to have complete life cycles through the adaptation of flowering time. The transgenic Arabidopsis, with overexpressing WRKY71, displayed higher survival rates towards salt treatments, while knockdown of wrky71 postponed florescence at the same time [43]. In rice, overexpression of OsWOX13 enhanced drought resistance of transgenic plants, but the early flowering phenotype of the OE lines was also detected under dehydration treatments [44]. In our study, constitutive expression of PwNAC38 caused inconspicuous florescence changes under normal conditions, while it promoted floral initiation under both drought and salt conditions. However, it remains unknown whether the stress-escape response mediated by PwNAC38 is influenced by other external factors, and whether similar senescence regulation exists in Picea wilsonii.
5. Conclusions
The PwNAC38 gene encodes 336 amino acids with 1011 bp ORF. It functions as a nucleus-located TF with transactivating activity and can form homodimer by itself. Overexpression of PwNAC38 significantly improved tolerance and adaption to drought and salt stress both in Arabidopsis and apple calli, including the promotion of seed germination, evident root elongation and higher survival rates under adverse environmental conditions. The drought-responsive genes ATHB-7, ANAC019, ERD1, DREB2A and RD29A, and ABA-responsive genes such as ABI5 and NCED3, were highly expressed in PwNAC38 OE lines. Moreover, PwNAC38 transgenic Arabidopsis showed enhanced ROS scavenging efficiency and susceptibility to ABA. Collectively, PwNAC38 positively participates in the processes of plant response to drought and salt stress, partially depending on the ABA signaling pathway.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13081304/s1. Figure S1: Identification of transgenic PwNAC38 plants of Arabidopsis thaliana. Figure S2: Effects of overexpression PwNAC38 on growth and development of Arabidopsis. Figure S3: Statistics of germination rates within 7 days after different concentrations of mannitol treatment. Figure S4: Statistics of germination rates within 7 days after different concentrations of NaCl treatment. Table S1: Primers used in this experiment.
Author Contributions
Conceptualization, L.Z., Y.C. and M.Y.; methodology, M.Y. and J.L.; software, M.Y. and Y.C.; validation, J.H. and J.Z.; investigation, M.Y., M.Z., J.L., J.Z. and J.H.; resources, L.Z. and Y.C.; data analysis, M.Y. and J.L.; writing—original draft preparation, M.Y. and J.L.; writing—review and editing, L.Z. and Y.C.; supervision, L.Z.; project administration, L.Z.; funding acquisition, Y.C. and L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Funds for the Central Universities (2021ZY01), the China Postdoctoral Science Foundation (2020M680403) and a grant from the Agricultural Ministry of China (2016ZX08009-003).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Kazuko, Y.S.; Kazuo, S. Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005, 169, 785–797. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Shinozaki, K.Y. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Niwa, M.; Kawai, Y.; Nakamura, N.; Futaki, S. The Structure of the Promoter Region for Rat Inducible Nitric Oxide Synthase Gene. Life Sci. 1997, 61, 45–49. [Google Scholar] [CrossRef]
- Jeong, J.S.; Park, Y.T.; Jung, H.; Park, S.; Kim, J. Rice NAC proteins act as homodimers and heterodimers. Plant Biotechnol. Rep. 2009, 3, 127–134. [Google Scholar] [CrossRef]
- Hao, Y.; Song, Q.; Chen, H.; Zou, H.; Wei, W.; Kang, X.; Ma, B.; Zhang, W.; Zhang, J.; Chen, S. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 2010, 232, 1033–1043. [Google Scholar] [CrossRef]
- Phan, T.L.; Rie, N.; Kazuko, Y.; Kazuo, S. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 2010, 1, 32–39. [Google Scholar] [CrossRef]
- Yang, S.; Seo, P.J.; Yoon, H.; Park, C. The Arabidopsis NAC Transcription Factor VNI2 Integrates Abscisic Acid Signals into Leaf Senescence via the COR/RD Genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Lee, C.H.; Ye, Z. Global Analysis of Direct Targets of Secondary Wall NAC Master Switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Christianson, J.A.; Dennis, E.S.; Llewellyn, D.J.; Wilson, I.W. ATAF NAC transcription factors: Regulators of plant stress signaling. Plant Signal. Behav. 2010, 5, 428–432. [Google Scholar] [CrossRef]
- Yu, X.W.; Liu, Y.M.; Wang, S.; Yuan, T.; Zhankui, W.; Yingjie, S.; Hui, P.; Abudoukeyumu, M.; Ze, W.; Hua, Z.; et al. CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 2016, 35, 613–627. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, H.; Liu, S.; Wang, X.; Zhang, Y.; Meng, Y.; Luo, D.; Chen, R. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.M.; Mi-Jeong, P.; Joon, S.P.; Jin-Su, S.; Hie-Joon, K.; Chung-Mo, P. Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 2012, 448, 353–363. [Google Scholar] [CrossRef]
- Liu, C.; Ma, H.Z.; Zhou, J.; Li, Z.X.; Peng, Z.H.; Guo, F.; Zhang, J.R. TsHD1 and TsNAC1 cooperatively play roles in plant growth and abiotic stress resistance of Thellungiella halophile. Plant J. 2019, 99, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.; Himanshu, T.; Ganapathi, T.R. A banana NAC transcription factor (MusaSNAC1) impart drought tolerance by modulating stomatal closure and H2O2 content. Plant Mol. Biol. 2018, 96, 457–471. [Google Scholar] [CrossRef]
- Lu, M. Monitoring Radial Growth of Three Conifer Species in the Eastern Qilian Mountains. Master’s Thesis, Lanzhou University, Lanzhou, China, 2016. [Google Scholar]
- Ma, H. Functional Traits of Picea wilsonii Based on Tree Age and Altitude. Master’s Thesis, Shanxi Agricultural University, Jinzhong, China, 2019. [Google Scholar]
- Guo, Y.X.; Zhang, H.H.; Yuan, Y.H.; Cui, X.Y.; Zhang, L.Y. Identification and characterization of NAC genes in response to abiotic stress conditions in Picea wilsonii using transcriptome sequencing. Biotechnol. Biotec. Eq. 2020, 34, 93–103. [Google Scholar] [CrossRef]
- Zhang, H.H.; Cui, X.Y.; Guo, Y.X.; Luo, C.B.; Zhang, L.Y. Picea wilsonii transcription factor NAC2 enhanced plant tolerance to abiotic stress and participated in RFCP1-regulated flowering time. Plant Mol. Biol. 2018, 98, 471–493. [Google Scholar] [CrossRef]
- Liang, K.H.; Wang, A.B.; Yuan, Y.H.; Miao, Y.H.; Zhang, L.Y. Picea wilsonii NAC Transcription Factor PwNAC30 Negatively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Mol. Biol. Rep. 2020, 38, 554–571. [Google Scholar] [CrossRef]
- Yu, M.X.; Liu, J.L.; Du, B.S.; Zhang, M.J.; Wang, A.B.; Zhang, L.Y. NAC Transcription Factor PwNAC11 Activates ERD1 by Interaction with ABF3 and DREB2A to Enhance Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6952. [Google Scholar] [CrossRef]
- Joshi, P.S.; Parinita, A.; Agarwal, P.K. Overexpression of AlNAC1 from recretohalophyte Aeluropus lagopoides alleviates drought stress in transgenic tobacco. Environ. Exp. Bot. 2021, 181, 104277. [Google Scholar] [CrossRef]
- Jiang, H.; Zhou, L.; Gao, H.; Wang, X.; Li, Z.; Li, Y. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol. 2022, 189, 2044–2060. [Google Scholar] [CrossRef] [PubMed]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The No Apical Meristem Gene of Petunia Is Required for Pattern Formation in Embryos and Flowers and Is Expressed at Meristem and Primordia Boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Wang, B.; Wei, J.; Song, N.; Wang, N.; Zhao, J.; Kang, Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J. Integr. Plant Biol. 2018, 60, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, H.; Li, D.; Huang, L.; Hong, Y.; Ding, X.S.; Nelson, R.S.; Zhou, X.; Song, F. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Chen, L.; Xie, H. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef]
- Jia, D.F.; Gong, X.Q.; Li, M.J.; Li, C.; Sun, T.T.; Ma, F.W. Overexpression of a Novel Apple NAC Transcription Factor Gene, MdNAC1, Confers the Dwarf Phenotype in Transgenic Apple (Malus domestica). Genes 2018, 9, 229. [Google Scholar] [CrossRef]
- Fu, C.C.; Han, Y.C.; Fan, Z.Q.; Jian-Ye, C.; Wei-Xin, C.; Wang-Jin, L.; Jian-Fei, K. The Papaya Transcription Factor CpNAC1 Modulates Carotenoid Biosynthesis through Activating Phytoene Desaturase Genes CpPDS2/4 during Fruit Ripening. J. Agric. Food Chem. 2016, 64, 5454–5463. [Google Scholar] [CrossRef]
- Jin, R.; Zhu, Q.G.; Shen, X.Y.; Wang, M.M.; Jamil, W.; Grierson, D.; Yin, X.R.; Chen, K.S. DkNAC7, a novel high-CO2/hypoxia-induced NAC transcription factor, regulates persimmon fruit de-astringency. PLoS ONE 2018, 13, e0194326. [Google Scholar] [CrossRef]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; Van Aken, O.; De Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A Membrane-Bound NAC Transcription Factor, ANAC017, Mediates Mitochondrial Retrograde Signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2020, 18, 1317–1329. [Google Scholar] [CrossRef]
- Sun, L.; Yang, Z.T.; Song, Z.T.; Wang, M.J.; Sun, L.; Lu, S.J.; Liu, J.X. The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J. 2013, 76, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.F.; Wang, C.; Yan, J.L.; Guo, X.H.; Wu, F.F.; Yang, B.; Deyholos, M.K.; Jiang, Y. Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol. Biol. 2016, 92, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.G.; Zhang, H.Y.; Qian, X.Y.; Li, A.; Zhao, G.Y.; Jing, R.L. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.S.; Yuan, Y.; Wu, Q.; Wang, J.; Li, J.G.; Zhao, M.L. LcEIL2/3 are involved in fruitlet abscission via activating genes related to ethylene biosynthesis and cell wall remodeling in litchi. Plant J. 2020, 103, 1338–1350. [Google Scholar] [CrossRef]
- An, J.; Zhang, X.; Liu, Y.; Wang, X.; You, C.; Hao, Y. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 2021, 72, 1460–1472. [Google Scholar] [CrossRef]
- Truong, H.A.; Lee, S.; Trịnh, C.S.; Lee, W.J.; Chung, E.; Hong, S.; Lee, H. Overexpression of the HDA15 Gene Confers Resistance to Salt Stress by the Induction of NCED3, an ABA Biosynthesis Enzyme. Front. Plant Sci. 2021, 12, 640443. [Google Scholar] [CrossRef]
- Han, D.G.; Du, M.; Zhou, Z.Y.; Wang, S.; Li, T.M.; Han, J.X.; Xu, T.L.; Yang, G.H. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef]
- Zhang, K.W.; Gan, S.S. An Abscisic Acid-AtNAP Transcription Factor-SAG113 Protein Phosphatase 2C Regulatory Chain for Controlling Dehydration in Senescing Arabidopsis Leaves. Plant Physiol. 2012, 158, 961–969. [Google Scholar] [CrossRef]
- Yu, Y.C.; Wang, L.; Chen, J.C.; Liu, Z.; Chung-Mo, P.; Xiang, F.N. WRKY71 Acts Antagonistically Against Salt-Delayed Flowering in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Pham-Thi, M.; Sug, K.J.; Songhwa, C.; Mi, J.K.; Gang-Seob, L.; Dong-Eun, K.; Jong-Joo, C.; Ik, S.S.; Hie, N.B.; Yeon-Ki, K. A WUSCHEL Homeobox Transcription Factor, OsWOX13, Enhances Drought Tolerance and Triggers Early Flowering in Rice. Mol. Cells 2018, 41, 781–798. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).