Brown Bear Food-Probability Models in West-European Russia: On the Way to the Real Resource Selection Function
Abstract
:1. Introduction
2. Materials and Methods
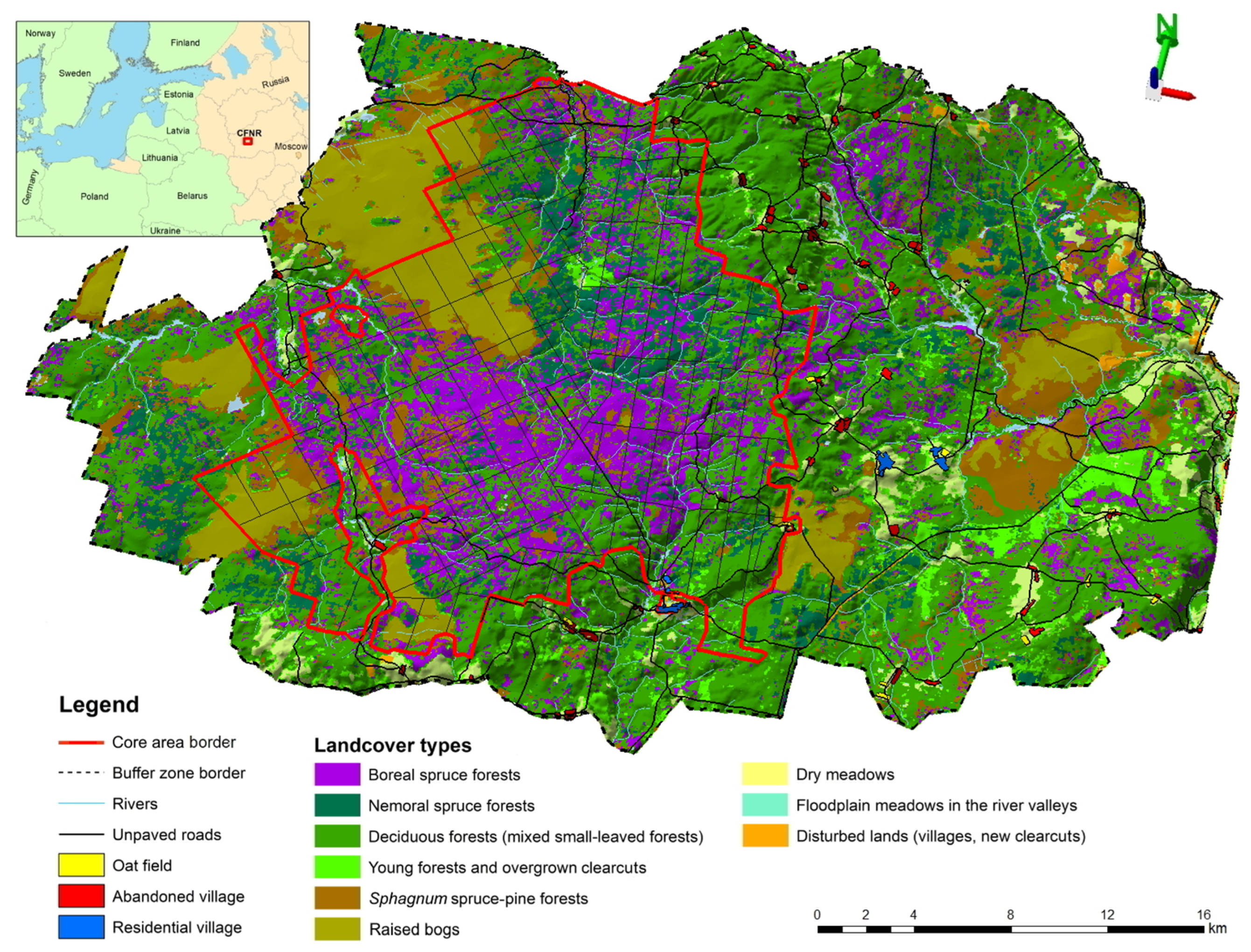
2.1. Study Area
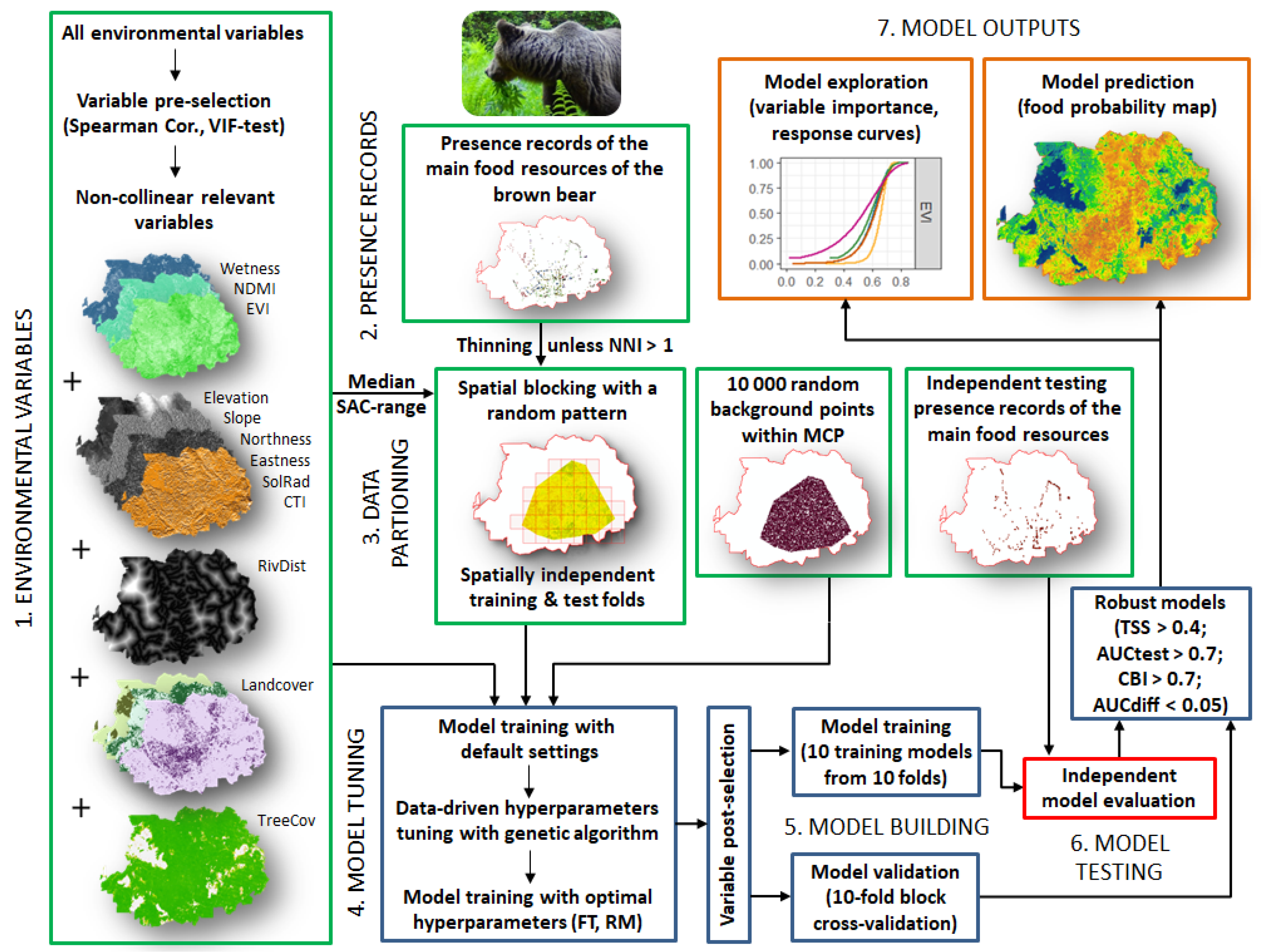
2.2. Food-Probability Models
2.3. Presence Records of Brown Bear Food Resources
2.4. Environmental Variables
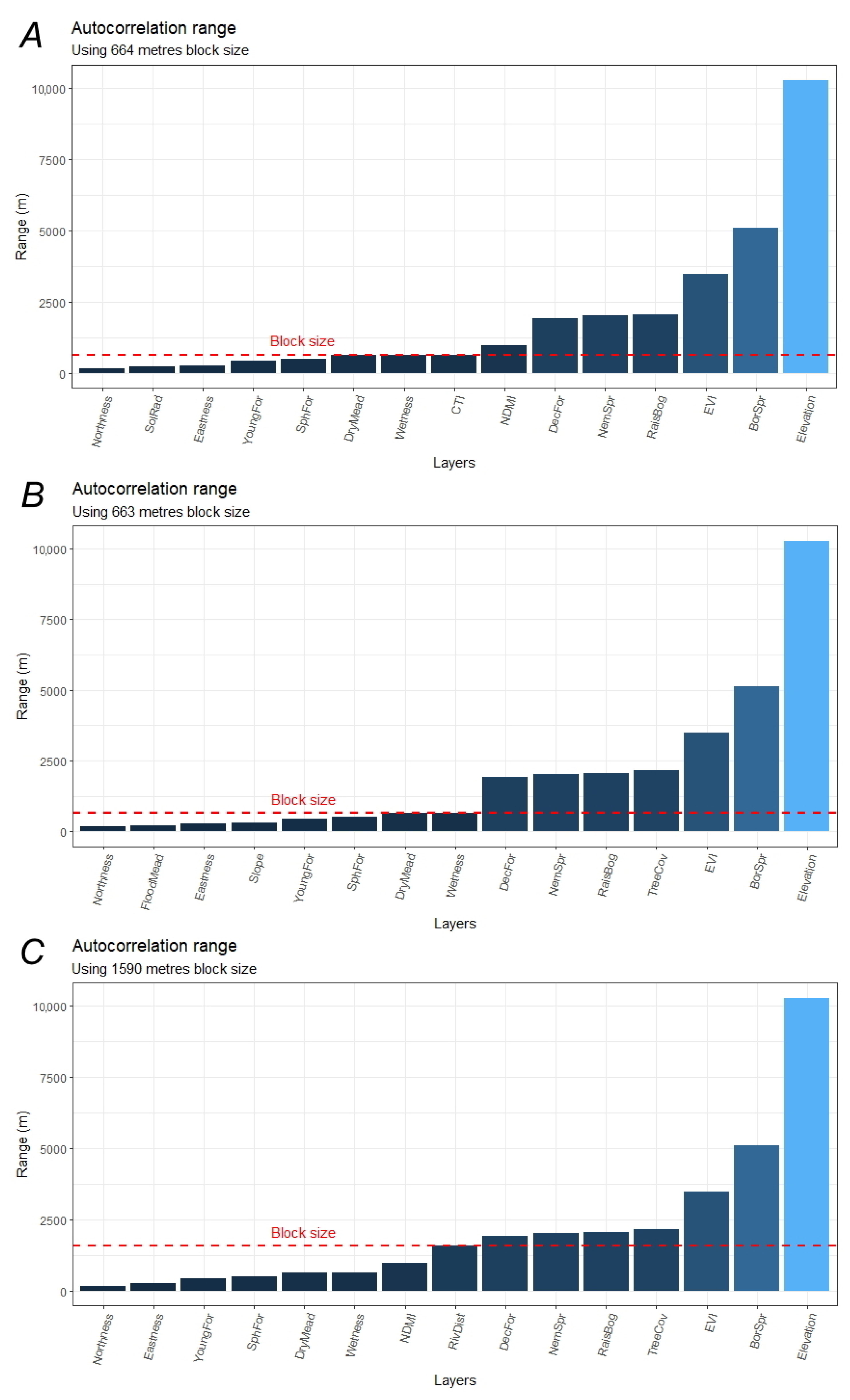
2.5. Model Tuning and Calibration (Training)
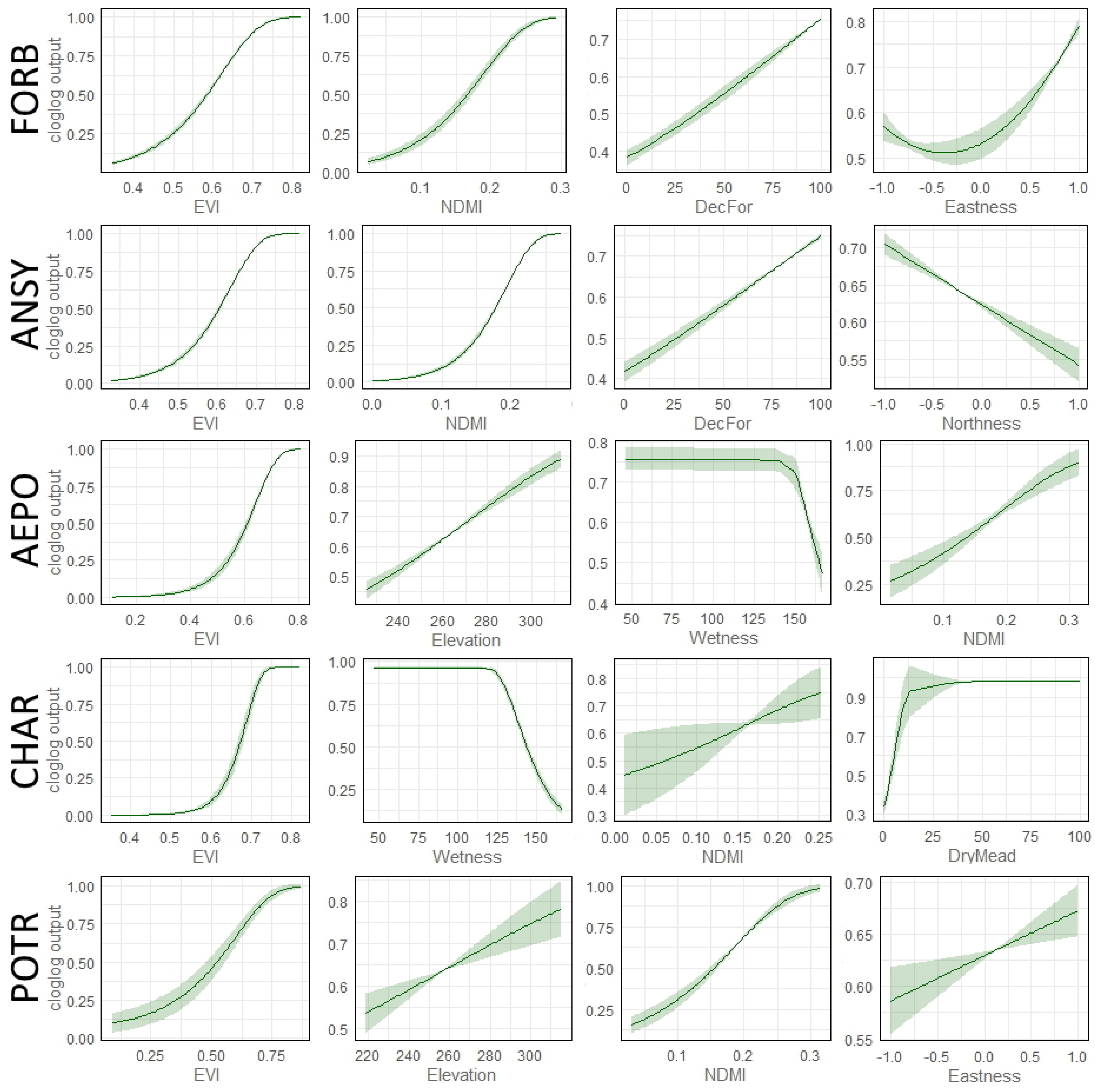
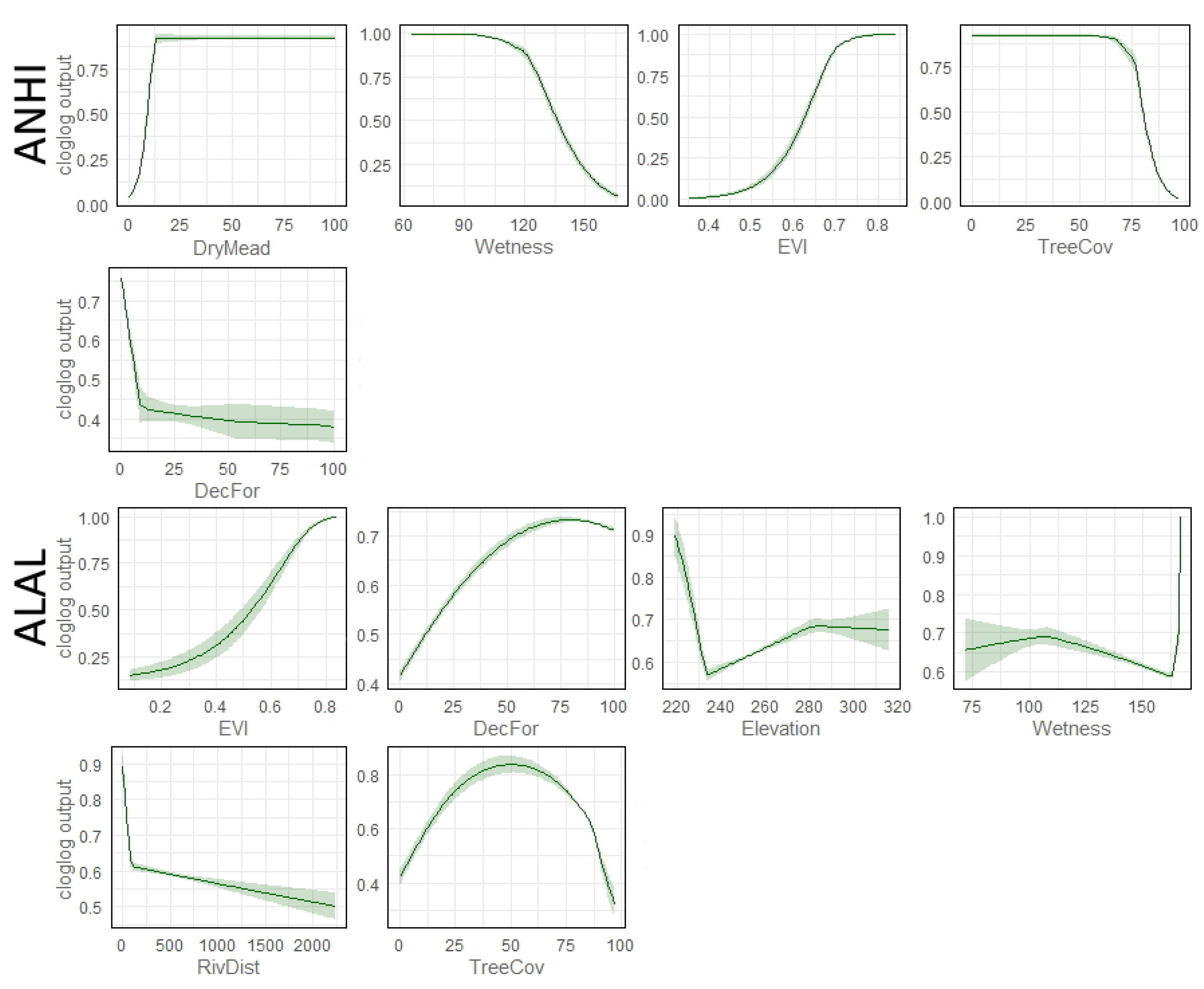
2.6. Model Evaluation (Testing) and Exploration
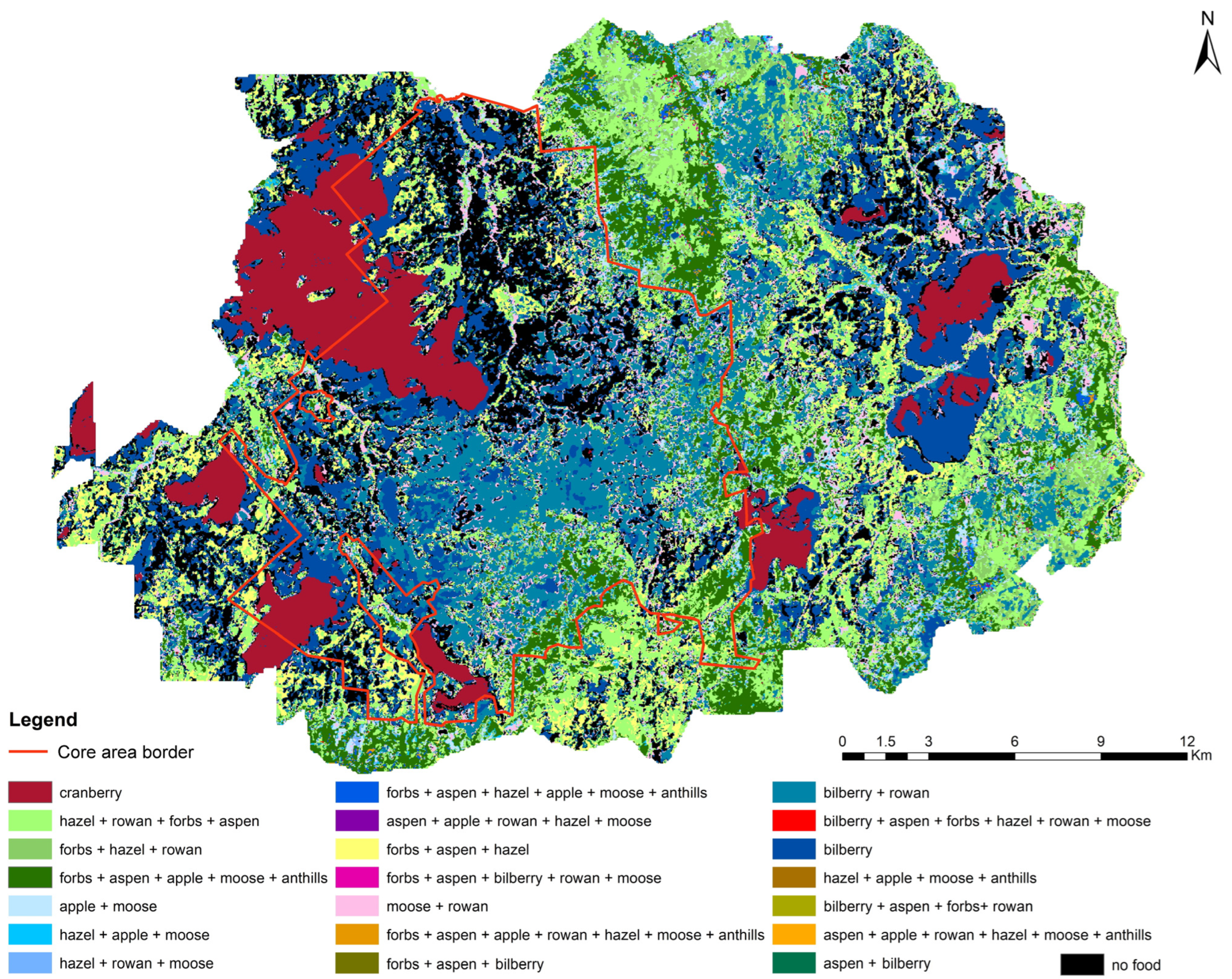
2.7. Mapping Spatial Distribution
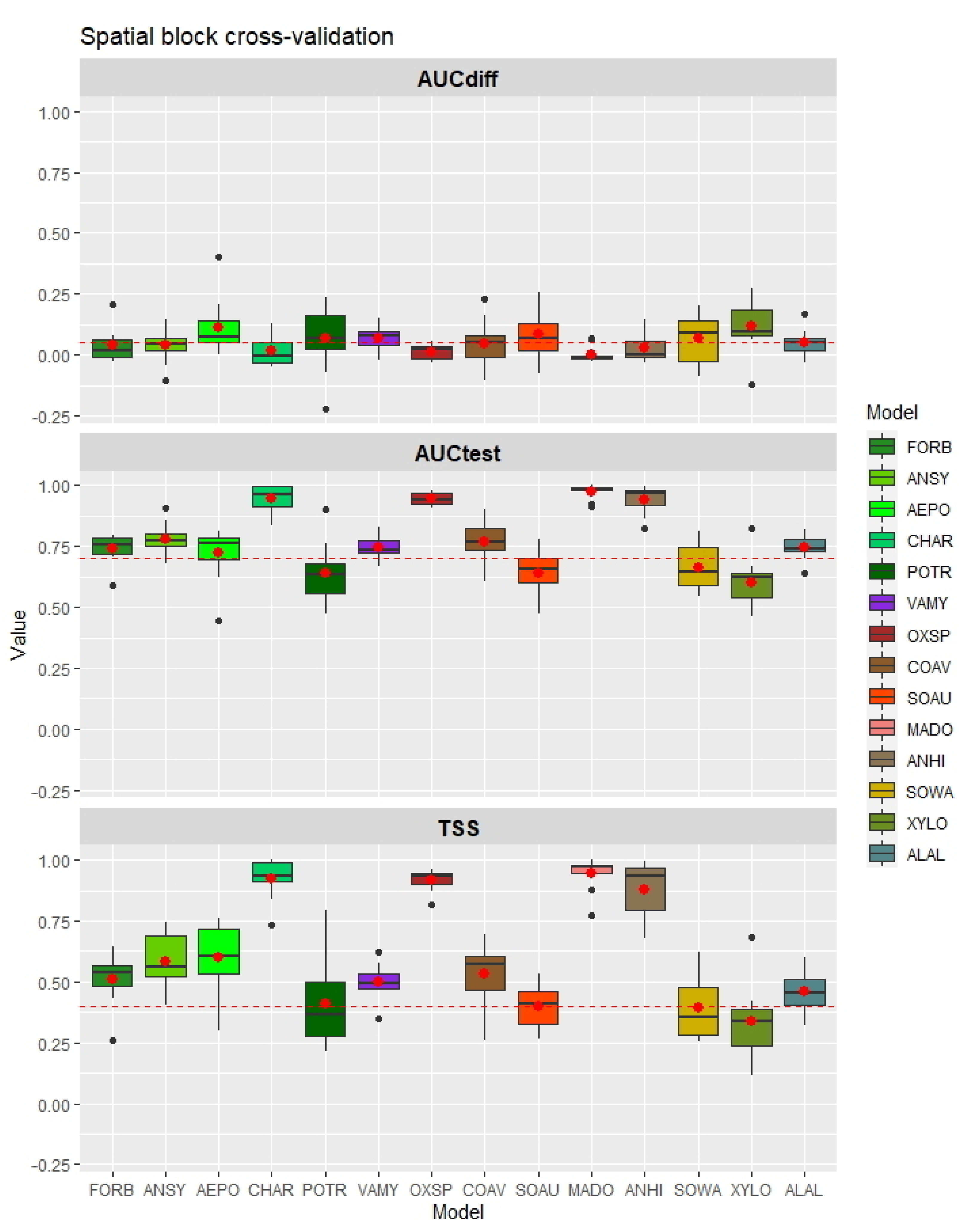
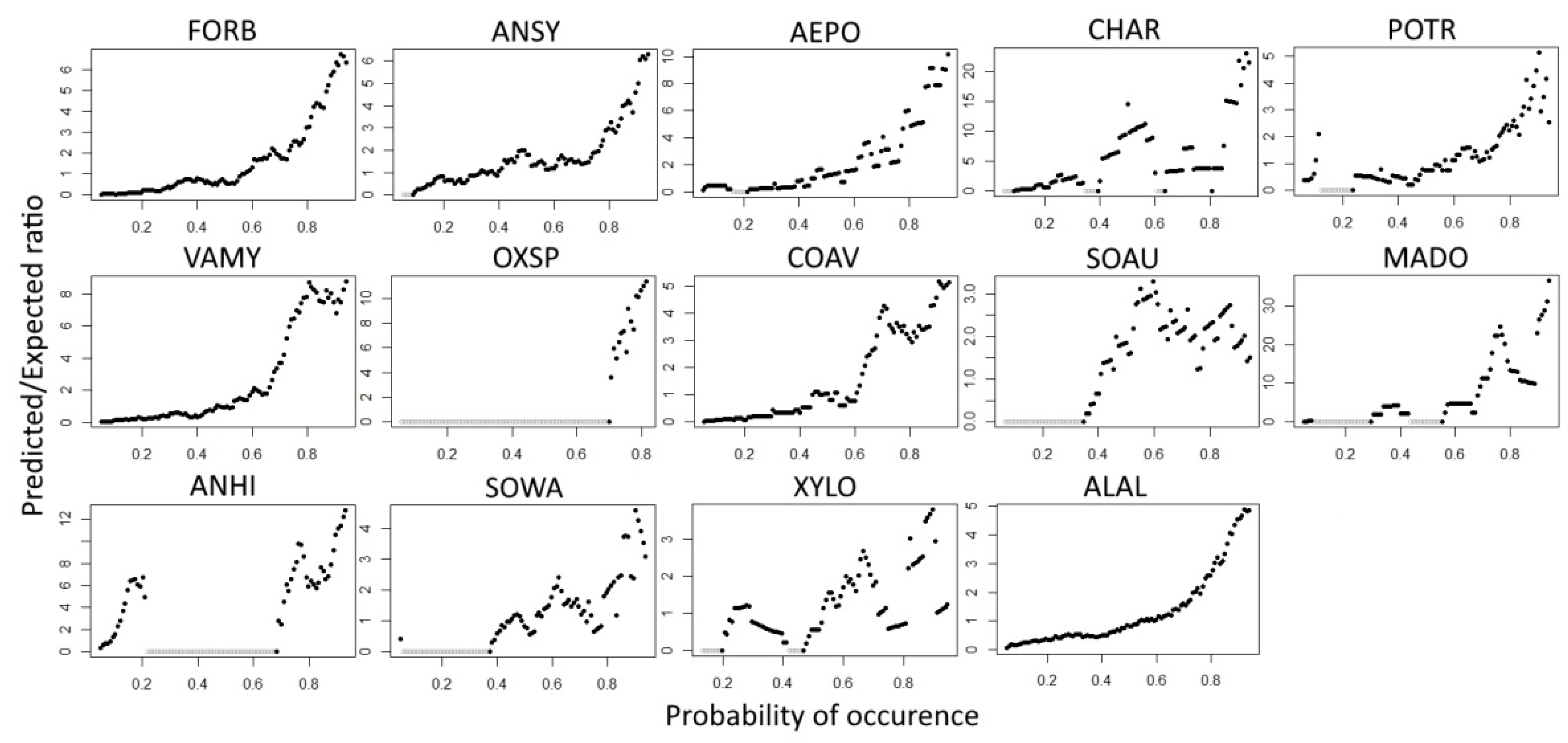
3. Results
3.1. Evaluation of Food-Probability Models
3.2. Variable Importance
3.3. Food-Probability Maps
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Model Name | Rarefy Distance (m) | NNI | z-Score | p-Value |
|---|---|---|---|---|
| FORB | 500 | 1.09 | 1.54 | 0.12 |
| ANSY | 450 | 1.10 | 1.57 | 0.12 |
| AEPO | 220 | 1.06 | 0.68 | 0.50 |
| CHAR | 270 | 1.27 | 2.18 | 0.03 |
| POTR | 30 | 1.17 | 2.27 | 0.02 |
| VAMY | 370 | 1.16 | 2.79 | 0.01 |
| OXSP | 650 | 0.88 | –1.09 | 0.28 |
| COAV | 350 | 1.07 | 1.16 | 0.25 |
| SOAU | 440 | 1.11 | 1.72 | 0.09 |
| MADO | 720 | 1.01 | 0.12 | 0.91 |
| ANHI | 560 | 1.04 | 0.53 | 0.60 |
| SOWA | 490 | 1.09 | 1.30 | 0.19 |
| XYLO | 140 | 1.07 | 1.14 | 0.25 |
| ALAL | 440 | 1.10 | 2.74 | 0.01 |
| Variable | FORB | ANSY | AEPO | CHAR | POTR | VAMY | OXSP | COAV | SOAU | MADO | ANHI | ALAL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EVI | 0.50 | 0.56 | 0.60 | 0.91 | 0.45 | 0.46 | 0.23 | 0.48 | 0.16 | 0.88 | 0.75 | 0.30 |
| NDMI | 0.33 | 0.44 | 0.39 | 0.50 | 0.36 | 0.35 | 0.51 | 0.36 | 0.31 | 0.02 | – | 0.15 |
| Wetness | 0.02 | 0.21 | 0.46 | 0.76 | 0.28 | 0.35 | 0.80 | 0.43 | 0.30 | 0.89 | 0.76 | 0.20 |
| Elevation | 0.08 | 0.17 | 0.49 | 0.42 | 0.38 | 0.30 | 0.69 | 0.39 | 0.38 | 0.00 | 0.02 | 0.21 |
| Slope | – | – | – | – | – | – | – | – | – | – | 0.37 | – |
| Northness | 0.22 | 0.27 | 0.28 | 0.01 | 0.25 | 0.08 | 0.02 | 0.15 | 0.19 | 0.48 | 0.24 | 0.06 |
| Eastness | 0.27 | 0.26 | 0.26 | 0.35 | 0.33 | 0.17 | 0.00 | 0.23 | 0.19 | 0.53 | 0.21 | 0.16 |
| CTI | 0.11 | 0.12 | 0.30 | 0.29 | 0.02 | 0.25 | 0.16 | 0.28 | 0.26 | 0.00 | – | – |
| SolRad | 0.22 | 0.24 | 0.29 | 0.00 | 0.01 | 0.03 | 0.02 | 0.23 | 0.02 | 0.12 | – | – |
| RivDist | – | – | – | – | – | – | – | – | – | – | – | 0.19 |
| BorSpr | 0.22 | 0.22 | 0.16 | 0.32 | 0.15 | 0.33 | 0.39 | 0.28 | 0.31 | 0.30 | 0.30 | 0.14 |
| NemSpr | 0.14 | 0.15 | 0.11 | 0.14 | 0.14 | 0.08 | 0.22 | 0.19 | 0.11 | 0.17 | 0.18 | 0.12 |
| SphFor | 0.14 | 0.14 | 0.11 | 0.16 | 0.13 | 0.22 | 0.00 | 0.08 | 0.10 | 0.13 | 0.15 | 0.09 |
| DecFor | 0.31 | 0.30 | 0.36 | 0.00 | 0.24 | 0.31 | 0.50 | 0.47 | 0.00 | 0.48 | 0.41 | 0.23 |
| YoungFor | 0.13 | 0.16 | 0.12 | 0.22 | 0.10 | 0.05 | 0.00 | 0.05 | 0.02 | 0.00 | 0.31 | 0.10 |
| RaisBog | 0.05 | 0.05 | 0.05 | 0.05 | 0.06 | 0.04 | 0.88 | 0.01 | 0.03 | 0.03 | 0.05 | 0.05 |
| DryMead | 0.09 | 0.11 | 0.14 | 0.47 | 0.00 | 0.05 | 0.00 | 0.02 | 0.05 | 0.79 | 0.83 | 0.07 |
| FloodMead | – | – | – | – | – | – | – | – | – | – | 0.07 | – |
| TreeCov | – | – | – | – | – | – | – | – | – | – | 0.67 | 0.18 |


References
- Nielsen, S.E.; Boyce, M.S.; Stenhouse, G.B.; Munro, R.H.M. Development and testing of phenologically driven grizzly bear habitat models. Écoscience 2003, 10, 1–10. [Google Scholar] [CrossRef]
- Milakovic, B.; Parker, K.L.; Gustine, D.D.; Lay, R.J.; Walker, A.B.D.; Gillingham, M.P. Seasonal habitat use and selection by grizzly bears in Northern British Columbia. J. Wildl. Manag. 2012, 76, 170–180. [Google Scholar] [CrossRef]
- Guisan, A.; Thuiller, W.; Zimmermann, N.E. Habitat Suitability and Distribution Models; Cambridge University Press: Cambridge, UK, 2017; 462p. [Google Scholar]
- Zurell, D.; Franklin, J.; König, C.; Bouchet, P.J.; Dormann, C.F.; Elith, J.; Fandos, G.; Feng, X.; Guillera-Arroita, G.; Guisan, A.; et al. A standard protocol for reporting species distribution models. Ecography 2020, 43, 1261–1277. [Google Scholar] [CrossRef]
- Boyce, M.S.; McDonald, L.L. Relating populations to habitats using resource selection functions. Trends Ecol. Evol. 1999, 14, 268–272. [Google Scholar] [CrossRef]
- Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction; Cambridge University Press: Cambridge, UK, 2010; 320p. [Google Scholar]
- Manly, B.F.J.; McDonald, L.L.; Thomas, D.L.; McDonald, T.L.; Erickson, W.P. Resource Selection by Animals. Statistical Design and Analysis for Field Studies, 2nd ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; 221p. [Google Scholar]
- Boyce, M.S.; Vernier, P.R.; Nielsen, S.E.; Schmiegelow, F.K.A. Evaluating resource selection functions. Ecol. Model. 2002, 157, 281–300. [Google Scholar] [CrossRef]
- Austin, M.P. Spatial prediction of species distribution: An interface between ecological theory and statistical modelling. Ecol. Model. 2002, 157, 101–118. [Google Scholar] [CrossRef]
- Nielsen, S.E.; McDermid, G.; Stenhouse, G.B.; Boyce, M.S. Dynamic wildlife habitat models: Seasonal foods and mortality risk predict occupancy-abundance and habitat selection in grizzly bears. Biol. Conserv. 2010, 143, 1623–1634. [Google Scholar] [CrossRef]
- Nielsen, S.E.; Larsen, T.A.; Stenhouse, G.B.; Coogan, S.C.P. Complementary food resources of carnivory and frugivory affect local abundance of an omnivorous carnivore. Oikos 2016, 126, 369–380. [Google Scholar] [CrossRef]
- Araújo, M.; Anderson, R.; Barbosa, A.M.; Beale, C.; Dormann, C.; Early, R.; Garcia, R.; Guisan, A.; Maiorano, L.; Naimi, B.; et al. Standards for distribution models in biodiversity assessments. Sci. Adv. 2019, 5, eaat4858. [Google Scholar] [CrossRef]
- Nielsen, S.E.; Boyce, M.S.; Stenhouse, G.B.; Munro, R.H.M. Modeling grizzly bear habitats in the Yellowhead ecosystem of Alberta: Taking autocorrelation seriously. Ursus 2002, 13, 45–56. [Google Scholar]
- Lyons, A.L.; Gaines, W.L.; Servheen, C. Black bear resource selection in the northeast Cascades, Washington. Biol. Conserv. 2003, 113, 55–62. [Google Scholar] [CrossRef]
- Hiller, T.L.; Belant, J.L.; Beringer, J.; Tyre, A.J. Resource selection by recolonizing american black bears in a fragmented forest landscape. Ursus 2015, 26, 116–128. [Google Scholar] [CrossRef]
- Lone, K.; Merkel, B.; Lydersen, C.; Kovacs, K.; Aars, J. Sea ice resource selection models for polar bears in the Barents Sea subpopulation. Ecography 2017, 41, 567–578. [Google Scholar] [CrossRef]
- Bai, W.; Huang, Q.; Zhang, J.; Stabach, J.; Huang, J.; Yang, H.; Songer, M.; Connor, T.; Liu, J.; Zhou, S.; et al. Microhabitat selection by giant pandas. Biol. Conserv. 2020, 247, 108615. [Google Scholar] [CrossRef]
- Ciarniello, L.M.; Boyce, M.S.; Beyer, H. Resource Selection Function for the Plateau Landscape of the Parsnip Grizzly Bear Project (An Update for 2003); Technical Report; British Columbia Ministry of Forests: Vancouver, BC, Canada, 2003; p. 18.
- Ciarniello, L.M.; Boyce, M.S.; Heard, D.C.; Seip, D.R. Components of grizzly bear habitat selection: Density; habitats; roads; and mortality risks. J. Wildl. Manag. 2007, 71, 1446–1457. [Google Scholar] [CrossRef]
- Wiegand, T.; Naves, J.; Garbulsky, M.F.; Fernandez, N. Animal habitat quality and ecosystem functioning: Exploring seasonal patterns using NDVI. Ecol. Monogr. 2008, 78, 87–103. [Google Scholar] [CrossRef]
- Chetkiewicz, C.L.B.; Boyce, M.S. Use of resource selection functions to identify conservation corridors. J. Appl. Ecol. 2009, 46, 1036–1047. [Google Scholar] [CrossRef]
- Graves, T.A.; Kendall, K.C.; Royle, J.A.; Stetz, J.B.; Macleod, A.C. Linking landscape characteristics to local grizzly bear abundance using multiple detection methods in a hierarchical model. Anim. Conserv. 2011, 14, 652–664. [Google Scholar] [CrossRef]
- Peters, W.; Hebblewhite, M.; Cavedon, M.; Pedrotti, L.; Mustoni, A.; Zibordi, F.; Cagnacci, F. Resource selection and connectivity reveal conservation challenges for reintroduced brown bears in the Italian Alps. Biol. Conserv. 2015, 186, 123–133. [Google Scholar] [CrossRef]
- Recio, M.R.; Knauer, F.; Molinari-Jobin, A.; Huber, D.; Filacorda, S.; Jerina, K. Context-dependent behaviour and connectivity of recolonizing brown bear populations identify transboundary conservation challenges in Central Europe. Anim. Conserv. 2020, 24, 73–83. [Google Scholar] [CrossRef]
- Roberts, D.R.; Nielsen, S.E.; Stenhouse, G.B. Idiosyncratic responses of grizzly bear habitat to climate change based on projected changes in their food resources. Ecol. Appl. 2014, 24, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Denny, C.K.; Stenhouse, G.B.; Nielsen, S.E. Scales of selection and perception: Landscape heterogeneity of an important food resource influences habitat use by a large omnivore. Wildl. Biol. 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- McClelland, C.J.R.; Coops, N.C.; Kearney, S.P.; Burton, A.C.; Nielsen, S.E.; Stenhouse, G.B. Variations in grizzly bear habitat selection in relation to the daily and seasonal availability of annual plant-food resources. Ecol. Inform. 2020, 58, 101116. [Google Scholar] [CrossRef]
- Twynham, K.; Ordiz, A.; Støen, O.-G.; Rauset, G.-R.; Kindberg, J.; Segerström, P.; Frank, J.; Uzal, A. Habitat Selection by Brown Bears with Varying Levels of Predation Rates on Ungulate Neonates. Diversity 2021, 13, 678. [Google Scholar] [CrossRef]
- Goldstein, M.I.; Poe, A.J.; Suring, L.H.; Nielson, R.M.; McDonald, T.L. Brown bear den habitat and winter recreation in South-Central Alaska. J. Wildl. Manag. 2010, 74, 35–42. [Google Scholar] [CrossRef]
- Hodder, D.P.; Johnson, C.J.; Rea, R.V.; Zedrosser, A. Application of a species distribution model to identify and manage bear den habitat in central British Columbia; Canada. Wildl. Biol. 2014, 20, 238–245. [Google Scholar] [CrossRef]
- Mateo-Sánchez, M.C.; Gastón, A.; Ciudad, C.; García-Viñas, J.I.; Cuevas, J.; López-Leiva, C.; Fernández-Landa, A.; Algeet-Abarquero, N.A.; Marchamalo, M.; Fortin, M.J.; et al. Seasonal and temporal changes in species use of the landscape: How do they impact the inferences from multiscale habitat modeling? Landsc. Ecol. 2016, 31, 1261–1276. [Google Scholar] [CrossRef]
- Berman, E.E.; Coops, N.C.; Kearney, S.P.; Stenhouse, G.B. Grizzly bear response to fine spatial and temporal scale spring snow cover in Western Alberta. PLoS ONE 2019, 14, e0215243. [Google Scholar] [CrossRef]
- Penteriani, V.; Zarzo-Arias, A.; Novo-Fernández, A.; Bombieri, G.; López-Sánchez, C.A. Responses of an endangered brown bear population to climate change based on predictable food resource and shelter alterations. Glob. Chang. Biol. 2019, 25, 1133–1151. [Google Scholar] [CrossRef]
- Munro, R.H.M.; Nielsen, S.E.; Price, M.H.; Stenhouse, G.B.; Boyce, M.S. Seasonal and diel patterns of grizzly bear diet and activity in West-Central Alberta. J. Mammal. 2006, 87, 1112–1121. [Google Scholar] [CrossRef]
- Nijland, W.; Nielsen, S.E.; Coops, N.C.; Wulder, M.A.; Stenhous, G.B. Fine-spatial scale predictions of understory species using climate- and LiDAR-derived terrain and canopy metrics. J. Appl. Remote Sens. 2014, 8, 083572. [Google Scholar] [CrossRef]
- Shores, C.R.; Mikle, N.; Graves, T.A. Mapping a keystone shrub species; huckleberry (Vaccinium membranaceum); using seasonal colour change in the Rocky Mountains. Int. J. Remote Sens. 2019, 40, 5695–5715. [Google Scholar] [CrossRef]
- Olchev, A.; Novenko, E.; Popov, V.; Pampura, T.; Meili, M. Evidence of temperature and precipitation change over the past 100 years in a high-resolution pollen record from the boreal forest of Central European Russia. Holocene 2017, 27, 740–751. [Google Scholar] [CrossRef]
- Puzachenko, Y.G.; Zheltukhin, A.S.; Kozlov, D.N.; Korablyov, N.P.; Fedyaeva, M.V.; Puzachenko, M.J.; Siunova, E.V. Central Forest State Nature Biosphere Reserve. Popular Science Essay, 2nd ed.; Pechatnya Press: Tver, Russia, 2016; 80p. [Google Scholar]
- Merow, C.; Maitner, B.S.; Owens, H.L.; Kass, J.M.; Enquist, B.J.; Jetz, W.; Guralnick, R. Species’ range model metadata standards: RMMS. Glob. Ecol. Biogeogr. 2019, 28, 1912–1924. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.R-project.org (accessed on 21 November 2020).
- Ogurtsov, S.S.; Khapugin, A.A.; Zheltukhin, A.S.; Fedoseeva, E.B.; Antropov, A.V.; Delgado, M.d.M.; Penteriani, V. Brown bear food habits in the natural and human-modified landscapes in the West-European Russia. Ursus, 2022; submitted. [Google Scholar]
- Frank, S.; Steyaert, S.; Swenson, J.; Storch, I.; Kindberg, J.; Barck, H.; Zedrosser, A. A “clearcut” case? Brown bear selection of coarse woody debris and carpenter ants on clearcuts. For. Ecol. Manag. 2015, 348, 164–173. [Google Scholar] [CrossRef]
- Dahle, B.; Wallin, K.; Cederlund, G.; Persson, I.; Selvaag, L.S.; Swenson, J.E. Predation on adult moose Alces alces by European brown bears Ursus arctos. Wildl. Biol. 2013, 19, 165–169. [Google Scholar]
- Coudun, C.; Gégout, J.-C. Quantitative prediction of the distribution and abundance of Vaccinium myrtillus with climatic and edaphic factors. J. Veg. Sci. 2007, 18, 517–524. [Google Scholar] [CrossRef]
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- Petrosyan, V.; Osipov, F.; Bobrov, V.; Dergunova, N.; Omelchenko, A.; Varshavskiy, A.; Danielyan, F.; Arakelyan, M. Species Distribution Models and Niche Partitioning among Unisexual Darevskia dahli and Its Parental Bisexual (D. portschinskii, D. mixta) Rock Lizards in the Caucasus. Mathematics 2020, 8, 1329. [Google Scholar] [CrossRef]
- Evans, J.S.; Murphy, M.A.; Ram, K. Package “spatialEco”. Spatial Analysis and Modelling Utilities, Version 1.3-7; 2021. Available online: https://cran.r-project.org/web/packages/spatialEco/spatialEco.pdf (accessed on 5 June 2022).
- Sillero, N.; Barbosa, A.M. Common mistakes in ecological niche models. Int. J. Geogr. Inf. Sci. 2021, 35, 213–226. [Google Scholar] [CrossRef]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.S.; Elith, J.; Guillera-Arroita, G.; Warton, D.I. Cross-validation strategies for data with temporal; spatial; hierarchical; or phylogenetic structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- Valavi, R.; Elith, J.; Lahoz-Monfort, J.J.; Guillera-Arroita, G. BlockCV: An R package for generating spatially or environmentally separated folds for k-fold cross-validation of species distribution models. Methods Ecol. Evol. 2019, 10, 225–232. [Google Scholar] [CrossRef]
- Mellert, K.H.; Fensterer, V.; Küchenhoff, H.; Reger, B.; Kölling, C.; Klemmt, H.J.; Ewald, J. Hypothesis-driven species distribution models for tree species in the Bavarian Alps. J. Veg. Sci. 2011, 22, 635–646. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Fernández, P.; Seymour, A.S.; Jordano, D. Habitat distribution models: Are mutualist distributions good predictors of their associates? Ecol. Appl. 2005, 15, 3–18. [Google Scholar] [CrossRef]
- Department of the Interior U.S. Geological Survey. Landsat 8 (L8) Data Users Handbook; Version 5.0; Department of the Interior U.S. Geological Survey: Sioux Falls, SD, USA, 2019; 106p.
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS Vegetation Indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band Enhanced Vegetation Index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the great plains with ERTS. In Proceedings of the Third ERTS Symposium NASA SP-351 1, NASA Goddard Space Flight Center, Greenbelt, MD, USA, 1 January 1974; Volume 1. [Google Scholar]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Wilson, E.H.; Sader, S.A. Detection of forest harvest type using multiple dates of Landsat TM imagery. Remote Sens. Environ. 2002, 80, 385–396. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.C.; Keydan, G.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30, 1248. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Kauth, R.J.; Thomas, G.S. The Tasseled Cap—A graphic description of the spectral-temporal development of agricultural crops as seen by Landsat. In Proceedings of the Symposium on Machine Processing of Remotely Sensed Data, Purdue University, West Lafayette, IN, USA, 29 June–1 July 1976. [Google Scholar]
- Baig, M.H.A.; Zhang, L.; Shuai, T.; Tong, Q. Derivation of a tasselled cap transformation based on Landsat 8 at-satellite reflectance. Remote Sens. Lett. 2014, 5, 423–431. [Google Scholar] [CrossRef]
- Riley, S.J.; Degloria, S.D.; Elliot, S.D. A Terrain Ruggedness Index that quantifies topographic heterogeneity. Intermt. J. Sci. 1999, 5, 23–27. [Google Scholar]
- Moore, I.D.; Grayson, R.B.; Ladson, A.R. Digital terrain modelling: A review of hydrological, geomorphological, and biological applications. Hydrol. Process. 1991, 5, 3–30. [Google Scholar] [CrossRef]
- Sørensen, R.; Zinko, U.; Seibert, J. On the calculation of the topographic wetness index: Evaluation of different methods based on field observations. Hydrol. Earth Syst. Sci. 2006, 10, 101–112. [Google Scholar] [CrossRef]
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Boehner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. An assessment of the effectiveness of decision tree methods for land cover classification. Remote Sens. Environ. 2003, 86, 554–565. [Google Scholar] [CrossRef]
- Potapov, P.; Turubanova, S.; Tyukavina, A.; Krylov, A.; McCarty, J.; Radeloff, V.; Hansen, M. Eastern Europe’s forest cover dynamics from 1985 to 2012 quantified from the full Landsat archive. Remote Sens. Environ. 2015, 159, 28–43. [Google Scholar] [CrossRef]
- Duque-Lazo, J.; van Gils, H.; Groen, T.A.; Navarro-Cerrillo, R.M. Transferability of species distribution models: The case of Phytophthora cinnamomi in Southwest Spain and Southwest Australia. Ecol. Model. 2016, 320, 62–70. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A., Jr. A practical guide to MaxEnt for modeling species’ distributions: What it does; and why inputs and settings matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black-box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Juiling, S.; Leon, S.K.; Jumian, J.; Tsen, S.; Lee, Y.L.; Khoo, E.; Sugau, J.B.; Nilus, R.; Pereira, J.T.; Damit, A.; et al. Conservation assessment and spatial distribution of endemic orchids in Sabah, Borneo. Nat. Conserv. Res. 2020, 5 (Suppl. 1), 136–144. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Schapire, R.E. Maxent Software for Modeling Species Niches and Distributions, Version 3.4.1. 2018. Available online: http://biodiversityinformatics.amnh.org/open_source/maxent/ (accessed on 12 October 2018).
- Syfert, M.M.; Smith, M.J.; Coomes, D.A. The effects of sampling bias and model complexity on the predictive performance of MaxEnt species distribution models. PLoS ONE 2013, 8, e55158. [Google Scholar] [CrossRef]
- Kramer-Schadt, S.; Niedballa, J.; Pilgrim, J.D.; Schröder, B.; Lindenborn, J.; Reinfelder, V.; Stillfried, M.; Heckmann, I.; Scharf, A.K.; Augeri, D.M.; et al. The importance of correcting for sampling bias in MaxEnt species distribution models. J. Conserv. Biogeogr. 2013, 19, 1366–1379. [Google Scholar] [CrossRef]
- Maiorano, L.; Boitani, L.; Monaco, A.; Tosoni, E.; Ciucci, P. Modeling the distribution of Apennine brown bears during hyperphagia to reduce the impact of wild boar hunting. Eur. J. Wildl. Res. 2015, 61, 241–253. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Vignali, S.; Barras, A.G.; Arlettaz, R.; Braunisch, V. SDMtune: An R package to tune and evaluate species distribution models. Ecol. Evol. 2020, 10, 11488–11506. [Google Scholar] [CrossRef]
- Phillips, S.J. A Brief Tutorial on Maxent. Lessons Conserv. 2010, 3, 108–135. [Google Scholar]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Vignali, S.; Barras, A.; Braunisch, V. Package “SDMtune”. Species Distribution Model Selection, Version 1.1.3; 2020. Available online: https://cran.r-project.org/web/packages/SDMtune/SDMtune.pdf (accessed on 5 June 2022).
- Hijmans, R.J.; Phillips, S.; Leathwick, J.; Elith, J. Package “Dismo”. Species Distribution Modeling, Version 1.1-4; 2017. Available online: https://cran.r-project.org/web/packages/dismo/dismo.pdf (accessed on 5 June 2022).
- Boria, R.A.; Olson, L.E.; Goodman, S.M.; Anderson, R.P. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol. Model. 2014, 275, 73–77. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Le Lay, G.; Helfer, V.; Randin, C.; Guisan, A. Evaluating the ability of habitat suitability models to predict species presences. Ecol. Model. 2006, 199, 142–152. [Google Scholar] [CrossRef]
- Zarzo-Arias, A.; Penteriani, V.; Delgado, M.d.M.; Torre, P.P.; García-González, R.; Mateo-Sánchez, M.C.; García, P.V.; Dalerum, F. Identifying potential areas of expansion for the endangered brown bear (Ursus arctos) population in the Cantabrian Mountains (NW Spain). PLoS ONE 2019, 14, e0209972. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.L.; Seifert, S.N. Ecological niche modeling in Maxent: The importance of model complexity and the performance of model selection criteria. Ecol. Appl. 2011, 21, 335–342. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence; kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Monserud, R.A.; Leemans, R. Comparing global vegetation maps with Kappa statistic. Ecol. Model. 1992, 62, 275–293. [Google Scholar] [CrossRef]
- Broennimann, O. Package “Ecospat”. Spatial Ecology Miscellaneous Methods, Version 3.0; 2018. Available online: https://cran.r-project.org/web/packages/ecospat/ecospat.pdf (accessed on 5 June 2022).
- Liu, C.; White, M.; Newell, G. Selecting thresholds for the prediction of species occurrence with presence-only data. J. Biogeogr. 2013, 40, 778–789. [Google Scholar] [CrossRef]
- Rodríguez, C.; Naves, J.; Fernández-Gil, A.; Obeso, J.R.; Delibes, M. Long-term trends in food habits of a relict brown bear population in northern Spain: The influence of climate and local factors. Environ. Conserv. 2007, 34, 36–44. [Google Scholar] [CrossRef]
- Rode, K.D.; Robbins, C.T.; Shipley, L.A. Constraints on herbivory by grizzly bears. Oecologia 2001, 128, 62–71. [Google Scholar] [CrossRef]
- Hertel, A.G.; Steyaert, S.M.J.G.; Zedrosser, A.; Mysterud, A.; Lodberg-Holm, H.K.; Gelink, H.W.; Kindberg, J.; Swenson, J.E. Bears and berries: Species-specific selective foraging on a patchily distributed food resource in a human-altered landscape. Behav. Ecol. Sociobiol. 2016, 70, 831–842. [Google Scholar] [CrossRef]
- Mangipane, L.S.; Belant, J.; Hiller, T.; Colvin, M.; Gustine, D.; Mangipane, B.; Hilderbrand, G. Influences of landscape heterogeneity on home-range sizes of brown bears. Mamm. Biol. 2018, 88, 1–7. [Google Scholar] [CrossRef]
- Robbins, C.T.; Schwartz, C.C.; Felicetti, L.A. Nutritional ecology of ursids: A review of newer methods and management implications. Ursus 2004, 15, 161–171. [Google Scholar] [CrossRef]
- Lopez-Alfaro, C.; Coogan, S.C.P.; Robbins, C.T.; Fortin, J.K.; Nielsen, S.E. Assessing nutritional parameters of brown bear diets among ecosystems gives insight into differences among populations. PLoS ONE 2015, 10, e82131. [Google Scholar] [CrossRef] [PubMed]
- Stenset, N.E.; Lutnæs, P.N.; Bjarnadóttir, V.; Dahle, B.; Fossum, K.H.; Jigsved, P.; Johansen, T.; Neumann, W.; Opseth, O.; Rønning, O.; et al. Seasonal and annual variation in the diet of brown bears (Ursus arctos) in the boreal forest of southcentral Sweden. Wildl. Biol. 2016, 22, 107–116. [Google Scholar] [CrossRef]
- Veloz, S.D. Spatially autocorrelated sampling falsely inflates measures of accuracy for presence-only niche models. J. Biogeogr. 2009, 36, 2290–2299. [Google Scholar] [CrossRef]
- Merckx, B.; Steyaert, M.; Vanreusel, A.; Vincx, M.; Vanaverbeke, J. Null models reveal preferential sampling, spatial autocorrelation and overfitting in habitat suitability modelling. Ecol. Model. 2011, 222, 588–597. [Google Scholar] [CrossRef]
- Halvorsen, R. A strict maximum likelihood explanation of MaxEnt, and some implications for distribution modelling. Sommerfeltia 2013, 36, 1–132. [Google Scholar] [CrossRef]
- Halvorsen, R.; Mazzoni, S.; Dirksen, J.W.; Næsset, E.; Gobakken, T.; Ohlson, M. How important are choice of model selection method and spatial autocorrelation of presence data for distribution modelling by MaxEnt? Ecol. Model. 2016, 328, 108–118. [Google Scholar] [CrossRef]
- Cannon, J.F.M. Angelica L. In Flora Europaea. Volume 2. Rosaceae to Umbelliferae, 3rd ed.; Tutin, T.G., Heywood, V.H., Burges, N.E., Moore, D.M., Valentine, D.H., Walters, S.M., Webb, D.A., Eds.; Cambridge University Press: Cambridge, UK, 1981; pp. 357–358. [Google Scholar]
- Minyaev, N.A.; Konechnaya, G.Y. Flora of the Central Forest State Nature Biosphere Reserve; Nauka: Leningrad, Russia, 1976; 104p. (In Russian) [Google Scholar]
- Petrova, S.E. Umbelliferae of Middle Russia: Biomorphological analysis. Bull. Mosc. Soc. Nat. 2015, 120, 46–56. (In Russian) [Google Scholar]
- Tsyganov, D.N. Phytoindication of Ecological Regimes in the Mixed Coniferous-Broad-Leaved Forest Subzone; Nauka: Moscow, Russia, 1983; 197p. (In Russian) [Google Scholar]
- D′Hertefeldt, T.; Eneström, J.M.; Pettersson, L.B. Geographic and Habitat Origin Influence Biomass Production and Storage Translocation in the Clonal Plant Aegopodium podagraria. PLoS ONE 2014, 9, e85407. [Google Scholar] [CrossRef]
- Semenishchenkov, Y.A. Phytocoenotic diversity of the gray alder forests in the South-West Nechernozemye of Russia. Rastit. Ross. 2014, 25, 71–88. [Google Scholar]
- Rogers, P.C.; Pinno, B.D.; Šebesta, J.; Albrectsen, B.R.; Li, G.; Ivanova, N.; Kusbach, A.; Kuuluvainen, T.; Landhäusser, S.M.; Liu, H.; et al. A global view of aspen: Conservation science for widespread keystone systems. Glob. Ecol. Conserv. 2020, 21, e00828. [Google Scholar] [CrossRef]
- Kivinen, S.; Koivisto, E.; Keski-Saari, S.; Poikolainen, L.; Tanhuanpää, T.; Kuzmin, A.; Viinikka, A.; Heikkinen, R.K.; Pykälä, J.; Virkkala, R.; et al. A keystone species, European aspen (Populus tremula L.), in boreal forests: Ecological role, knowledge needs and mapping using remote sensing. For. Ecol. Manag. 2020, 462, 118008. [Google Scholar] [CrossRef]
- Worrell, R. European aspen (Populus tremula L): A review with particular reference to Scotland I. Distribution, ecology and genetic variation. Forestry 1995, 68, 93–105. [Google Scholar] [CrossRef]
- Mäkipää, R. Response patterns of Vaccinium myrtillus and V. vitis-idaea along nutrient gradients in boreal forest. J. Veg. Sci. 1999, 10, 17–26. [Google Scholar] [CrossRef]
- Miina, J.; Hotanen, J.-P.; Salo, K. Modelling the abundance and temporal variation in the production of bilberry (Vaccinium myrtillus L.) in Finnish mineral soil forests. Silva Fenn. 2009, 43, 577–593. [Google Scholar] [CrossRef]
- Jacquemart, A.L. Vaccinium oxycoccos L. (Oxycoccus palustris Pers.) and Vaccinium microcarpum (Turcz. ex Rupr.) Schmalh. (Oxycoccus microcarpus Turcz. ex Rupr). J. Ecol. 1997, 85, 381–396. [Google Scholar] [CrossRef]
- Finsinger, W.; Tinner, W.; van der Knaap, W.O.; Ammann, B. The expansion of hazel (Corylus avellana L.) in the southern Alps: A key for understanding its early Holocene history in Europe? Quat. Sci. Rev. 2006, 25, 612–631. [Google Scholar] [CrossRef]
- Roellig, M.; Dorresteijn, I.; von Wehrden, H.; Hartel, T.; Fischer, J. Brown bear activity in traditional wood-pastures in Southern Transylvania, Romania. Ursus 2014, 25, 43–52. [Google Scholar] [CrossRef]
- Kaspari, M.; O’Donnell, S.; Kercher, J.R. Energy, Density, and Constraints to Species Richness: Ant Assemblages along a Productivity Gradient. Am. Nat. 2000, 155, 280–293. [Google Scholar] [CrossRef]
- Skuban, M.; Find’o, S.; Kajba, M. Human impacts on bear feeding habits and habitat selection in the Poľana Mountains, Slovakia. Eur. J. Wildl. Res. 2016, 62, 353–364. [Google Scholar] [CrossRef]
- Bjørneraas, K.; Solberg, E.J.; Herfindal, I.; Moorter, B.V.; Rolandsen, C.M.; Tremblay, J.; Astrup, R. Moose Alces alces habitat use at multiple temporal scales in a human-altered landscape. Wildl. Biol. 2011, 17, 44–54. [Google Scholar] [CrossRef]
- Swenson, J.E.; Ambarli, H.; Arnemo, J.M.; Baskin, L.; Ciucci, P.; Danilov, P.I.; Delibes, M.; Elfström, M.; Evans, A.L.; Groff, C.; et al. Brown Bear (Ursus arctos, Eurasia). In Bears of the World. Ecology, Conservation and Management; Penteriani, V., Melletti, M., Eds.; Cambridge University Press: Cambridge, UK, 2021; pp. 139–161. [Google Scholar]
- POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. 2021. Available online: http://www.plantsoftheworldonline.org/ (accessed on 19 March 2022).
- AntWeb. Version 8.68.7. California Academy of Science. Available online: https://www.antweb.org (accessed on 14 October 2021).
- Carpenter, J.M.; Kojima, J. Checklist of the species in the subfamily Vespinae (Insecta: Hymenoptera: Vespidae). Nat. Hist. Bull. Ibaraki Univ. 1997, 1, 51–92. [Google Scholar]
- Daglio, A. On the Taxonomy and Distribution of the Subfamily Vespinae (Insecta: Hymenoptera: Vespidae); LAP Lambert Academic Publishing: Beau Bassin, Mauritius, 2019; p. 49. [Google Scholar]
- Wilson, D.E.; Reeder, M.D.A. Mammal Species of the World. A Taxonomic and Geographic Reference, 3rd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; 142p. [Google Scholar]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21-st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef]
- Zuur, A.F.; Ieno, E.N.; Walker, N.J.; Saveliev, A.A.; Smith, G.M. Mixed Effects Models and Extensions in Ecology with R; Springer Science/Business Media, LLC: New York, NY, USA, 2009; 574p. [Google Scholar]


| Variable Name | Code in the Models | Organisms Included in the Models | Range |
|---|---|---|---|
| Vegetation Indices | |||
| EVI | P, M, I | 0.06–0.86 |
| NDMI | P, M | −0.02–0.33 |
| Wetness | P, M, I | 46.67–166.56 |
| Terrain | |||
| Elevation | P, M, I | 217–316 m a.s.l. |
| Slope | I | 0–15.85 degrees |
| Northness | P, M, I | −1–1 radians |
| Eastness | P, M, I | −1–1 radians |
| CTI | P | 0–250 |
| SolRad | P | 4721.23–5611.48 kWh/m2 |
| Proximity | |||
| RivDist | M | 0–3152 m |
| Landcover | |||
| BorSpr | P, M, I | 0%–100% |
| NemSpr | P, M, I | 0%–100% |
| SphFor | P, M, I | 0%–100% |
| DecFor | P, M, I | 0%–100% |
| YoungFor | P, M, I | 0%–100% |
| RaisBog | P, M, I | 0%–100% |
| DryMead | P, M, I | 0%–100% |
| FloodMead | I | 0%–100% |
| TreeCov | M, I | 0%–100% |
| Brown Bear Food Resource | Model Name | Raw Presence Points | Train Presence Points | Test Presence Points | RM | FT |
|---|---|---|---|---|---|---|
| Plant food resources | ||||||
| Apiaceae spp. | FORB | 365 | 87 | 159 | 4.0 | LQ |
| Angelica sylvestris | ANSY | 196 | 72 | 108 | 1.5 | L |
| Aegopodium podagraria | AEPO | 56 | 30 | 26 | 2.0 | LQH |
| Chaerophyllum aromaticum | CHAR | 113 | 18 | 24 | 2.5 | LQH |
| Aspen (Populus tremula) | POTR | 56 | 50 | 29 | 3.0 | LQ |
| Bilberry (Vaccinium myrtillus) | VAMY | 325 | 85 | 208 | 2.5 | LQHP |
| Cranberry (Oxycoccus spp.) | OXSP | 170 | 23 | 45 | 3.0 | H |
| Hazel (Corylus avellana) | COAV | 203 | 74 | 107 | 2.0 | LQH |
| Rowan (Sorbus aucuparia) | SOAU | 151 | 68 | 35 | 3 | LQ |
| Apple (Malus domestica) | MADO | 95 | 27 | 53 | 4.5 | LQH |
| Animal food resources | ||||||
| Ant hills | ANHI | 274 | 41 | 114 | 2.5 | LQHP |
| Social wasps | SOWA | 193 | 62 | 33 | 2.0 | LQ |
| Xylobiont insects | XYLO | 151 | 69 | 26 | 2.0 | H |
| Moose (Alces alces) | ALAL | 259 | 209 | 448 | 3.0 | LQH |
| No. | Model Name | AUCtrain | AUCtest | AUCdiff | TSS | CBI |
|---|---|---|---|---|---|---|
| Models for the plant food resources | ||||||
| 1 | FORB | 0.78 | 0.74 | 0.04 | 0.51 | 0.97 |
| 2 | ANSY | 0.82 | 0.78 | 0.04 | 0.59 | 0.92 |
| 4 | AEPO | 0.84 | 0.72 | 0.11 | 0.60 | 0.93 |
| 3 | CHAR | 0.96 | 0.95 | 0.01 | 0.92 | 0.71 |
| 5 | POTR | 0.70 | 0.64 | 0.07 | 0.41 | 0.87 |
| 6 | VAMY | 0.81 | 0.75 | 0.07 | 0.50 | 0.98 |
| 7 | OXSP | 0.96 | 0.94 | 0.01 | 0.92 | 0.95 |
| 9 | COAV | 0.82 | 0.77 | 0.05 | 0.53 | 0.97 |
| 8 | SOAU | 0.72 | 0.64 | 0.08 | 0.40 | 0.35 |
| 10 | MADO | 0.97 | 0.97 | 0.00 | 0.95 | 0.89 |
| Models for the animal food resources | ||||||
| 11 | ANHI | 0.97 | 0.94 | 0.03 | 0.88 | 0.82 |
| 12 | SOWA | 0.73 | 0.66 | 0.07 | 0.39 | 0.77 |
| 13 | XYLO | 0.72 | 0.60 | 0.12 | 0.34 | 0.54 |
| 14 | ALAL | 0.77 | 0.74 | 0.03 | 0.44 | 0.99 |
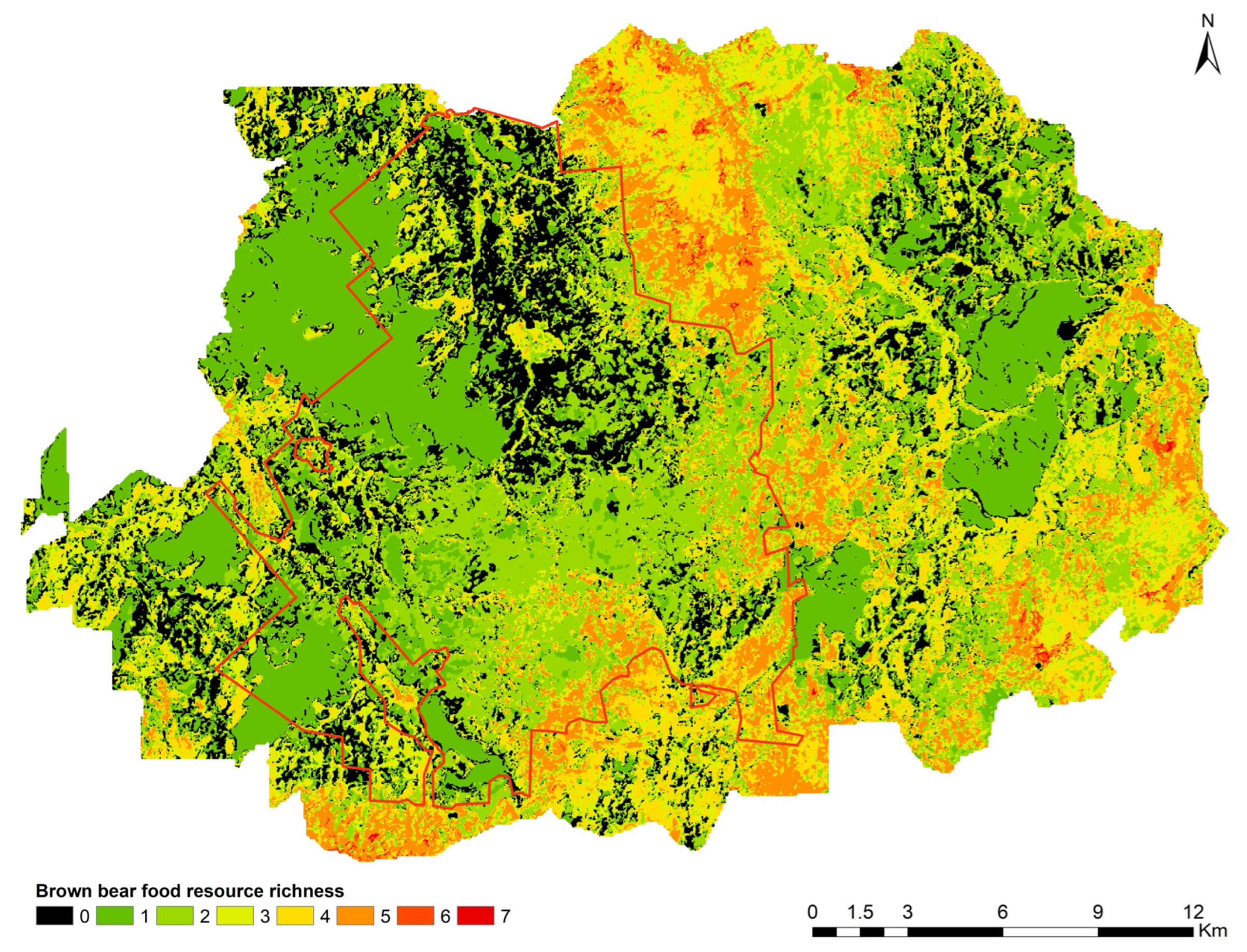
| Status of the Area and Proportion of Pixels | Number of Food Resources per Pixel in the Map of Figure 7 | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| CFNR core area (%) | 21.24 | 25.99 | 32.10 | 9.55 | 6.20 | 4.84 | 0.09 | 0 |
| CFNR buffer zone (%) | 11.47 | 22.80 | 20.18 | 16.22 | 16.49 | 11.87 | 0.81 | 0.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogurtsov, S.S.; Khapugin, A.A.; Zheltukhin, A.S.; Fedoseeva, E.B.; Antropov, A.V.; Delgado, M.d.M.; Penteriani, V. Brown Bear Food-Probability Models in West-European Russia: On the Way to the Real Resource Selection Function. Forests 2022, 13, 1247. https://doi.org/10.3390/f13081247
Ogurtsov SS, Khapugin AA, Zheltukhin AS, Fedoseeva EB, Antropov AV, Delgado MdM, Penteriani V. Brown Bear Food-Probability Models in West-European Russia: On the Way to the Real Resource Selection Function. Forests. 2022; 13(8):1247. https://doi.org/10.3390/f13081247
Chicago/Turabian StyleOgurtsov, Sergey S., Anatoliy A. Khapugin, Anatoliy S. Zheltukhin, Elena B. Fedoseeva, Alexander V. Antropov, María del Mar Delgado, and Vincenzo Penteriani. 2022. "Brown Bear Food-Probability Models in West-European Russia: On the Way to the Real Resource Selection Function" Forests 13, no. 8: 1247. https://doi.org/10.3390/f13081247
APA StyleOgurtsov, S. S., Khapugin, A. A., Zheltukhin, A. S., Fedoseeva, E. B., Antropov, A. V., Delgado, M. d. M., & Penteriani, V. (2022). Brown Bear Food-Probability Models in West-European Russia: On the Way to the Real Resource Selection Function. Forests, 13(8), 1247. https://doi.org/10.3390/f13081247









