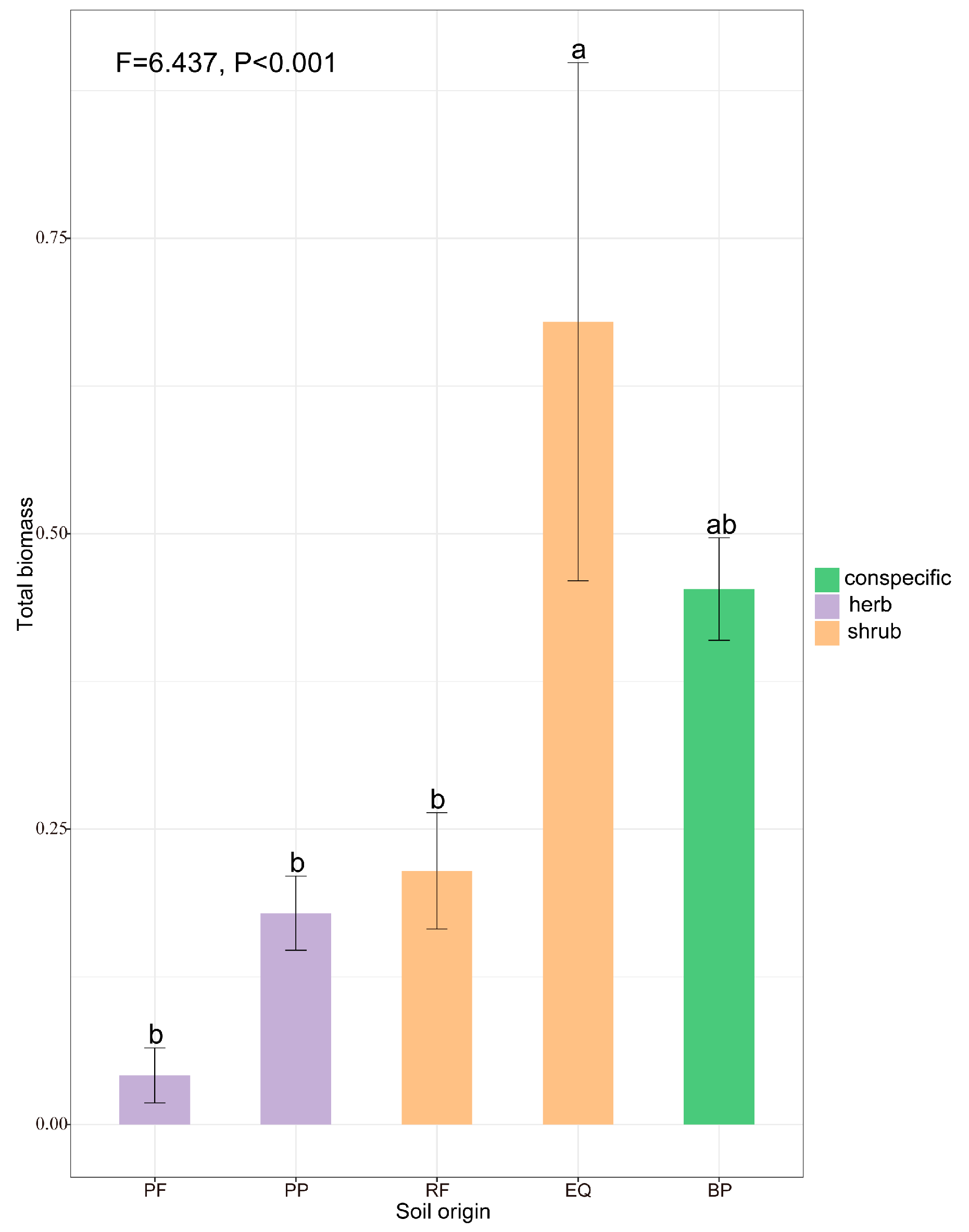Abstract
Fast-growing pioneer tree species play a crucial role in triggering late successional development in forests. Experimental evidence of the soil legacy effects of pre-existing plants on pioneer tree performance is lacking. We explored the legacy effects of soils conditioned by early successional herbs (Poa poophagorum Bor and Potentilla fragarioides L.) and mid-successional shrubs (Rhododendron fortunei Lindl. and Enkianthus quinqueflorus Lour.) on late-successional ectomycorrhizal (ECM) pioneer tree (Betula platyphylla Sukaczev) seedling growth. The soils were analyzed for soil nutrient status and fungal and bacterial compositions using ITS and 16S rRNA gene sequencing. B. platyphylla seedlings produced higher biomass in soils conditioned by shrubs. Soil organic carbon (SOC) and bacterial and fungal legacies most impacted pioneer tree seedling growth. Additionally, the partial least squares path model revealed that soil nutrients, especially SOC, indirectly affected seedling biomass by their direct effects on the bacterial and fungal communities. The changes in bacterial community composition had a stronger effect on seedling biomass than those of fungi because bacteria with shorter turnover times are generally considered to be more efficient than fungi in enhancing nutrient availability. Our study integrates soil microbial and nutrient legacies to explain the potential mechanisms of pioneer tree regeneration.
1. Introduction
The loss of natural forest is an unprecedented current challenge, and the remaining forests are fragmented and degraded [1,2,3]. Plant communities change from grassland and shrubland to forest during secondary forest succession [4]. A primary determinant of the recovery of forest community structure and composition is the successful establishment of pioneer tree seedlings, which can provide a template for further community assembly [5]. Pioneer tree species are generally light-demanding species [6], which can regenerate quickly, colonize large regions prosperously [7], and create canopy cover suitable for the establishment of mature forest trees [8]. Therefore, understanding the mechanisms regulating pioneer tree recruitment is of increasing importance for forest restoration [6,9].
The environmental conditions during forest succession change and influence the growth of trees over time [10]. Most studies on pioneer tree seedling establishment have focused on the microclimate (gap size, light, humidity, and temperature) [11,12], seed bank dynamics (seed dispersion and seeding predation) [13,14], and interspecific interactions (nutrient, water, light, and space competition) [15,16]. In addition, some reviews have suggested that durable beneficial legacy in soil supports the rapid colonization of pioneer species in secondary succession [17,18]. The allelochemical, microbial, and relevant effects can persist even if the initial plants are no longer present; therefore, such plants can have legacy effects on subsequent plant recruitment, reproduction, and growth, producing a selective advantage of one plant over another [19,20]. In forest ecosystems, the seedlings and saplings of tree species occur not only on bare soil but also in shrub and herb patches [21]. Thus, the legacies of pre-existing plants might also persist in the soils with pioneer tree colonization [22]. It is known that the abiotic tolerances and competitive ability of plants can be modified by soil legacies from pre-existing plants [20,23], which may potentially mediate pioneer tree establishment. However, experimental evidence for soil legacy effects on pioneer tree establishment is lacking.
Soil microbes have direct effects on plants via fungal mutualists (ectomycorrhizal, arbuscular, etc.), pathogens, or plant-associated bacteria. The beneficial interactions between plants and fungal mutualists could improve plant performance by increasing plant resistance to pathogens or stress and by enhancing nutrient acquisition [24]. Ectomycorrhizal (ECM) association is a major contributor to ecosystem functioning and a powerful regulator of forest productivity and composition [25]. Soil pathogens can decrease plant performance by causing a range of damping-off diseases [26]. Saprotrophic microbes are mainly involved in litter decomposition and carbon (C) cycling and influence plant growth by altering nutrient availability [27]. Some bacteria can promote the growth of plants by producing stimulative substances or providing protection against pathogens [28]. However, microbial activities can induce indirect effects on plants by changing the rates of nutrient acquisition and resource partitioning [29]. C and nitrogen (N) are the two most important soil nutrients in terrestrial ecosystems [30]. Both bacterial and fungal communities have been shown to be generally limited by soil C [31]. The availability of soil nutrients, including N and phosphorous (P), is an essential element that limits plant productivity and growth [32,33]. In addition, plants and microbes differ in terms of their threshold value for nutrient availability limitation [34], indicating that soil legacy effects may vary when nutrient availability changes [35]. However, the relative effects of the pre-existing fungi, bacteria, and nutrient availability left by plants in the soils on future pioneer tree seedling growth are still unclear.
A natural succession sequence of grassland–shrubland–secondary birch forest was formed due to damage to the original tree species in the southwestern subalpine forests [36]. Betula platyphylla is a representative ECM birch species, and it is the foundation tree species in this area [37]. Birches are widely distributed pioneer tree species in Asia, Europe, and America [38]. Recently, there has been increasing interest in reforestation with birch in post-agricultural lands [39]. Our goals were to determine whether early successional herbs and mid-successional shrubs contributed to legacy effects affecting B. platyphylla seedling growth. Here, we generated species-specific soil legacies by individually growing herbs (early successional species) or shrubs (mid-successional species) in a common starting soil. Then, we planted pioneer tree seedlings (B. platyphylla) in all soils to explore the pre-existing legacy effect affecting their performance. We characterized the bacterial and fungal compositions and nutrient availability and determined their relationships with pioneer tree seedling biomass. We hypothesized that i) the soil legacy effects on pioneer tree seedling performance could differ between herb and shrub soils because the soil microbial communities and nutrient availability depend on plant taxonomy and identity; ii) the fungal community will overwhelmingly promote B. platyphylla growth by increasing the resistance and nutrient acquisition associated with ECM fungi.
2. Materials and Methods
2.1. Soil and Seed Collection
The sampled sites were located in subalpine forests in Miyaluo, Lixian County, Sichuan, China (31°42′–31°51′ N, 102°41′–102°44′E). The altitude of the study area is 2200–5500 m above sea level, the mean annual temperature is 4.8–6.9 °C, and the mean annual precipitation is 700–800 mm. B. platyphylla appears frequently as a primary pioneer species in pure and mixed stands with Picea asperata and Abies fabri [37]. We used the typical pioneer tree B. platyphylla as the target species in the test phase of our experiment. Two early-successional herbs (Poa poophagorum and Potentilla fragarioides) and two mid-successional shrubs (Rhododendron fortunei and Enkianthus quinqueflorus) were selected as the pre-existing plants. In this region, the original coniferous tree species, such as P. asperata and A. fabri, were logged. A natural succession sequence of “grassland–shrubland–secondary birch forest” formed successively [36,40]. All species are typical in secondary succession after land abandonment in subalpine forests.
Common starting soil includes 90% sterilized bare soil and 10% specific inoculum collected from each species (bare soil and specific inoculum were collected from the sites with identical altitude and slope aspect) [41]. In June 2019, bare soil was collected from a bare area (unvegetated site) in this region, mixed 7:3 (v/v) with silica sand and sterilized by 25 kGy gamma irradiation [41]. To minimize the influence of different plant species’ overlapping roots, the specific soil inoculum was collected from monodominant plots of each species [42]. Soil inoculum was randomly collected (0–20 cm depth) from the rooting zones of each studied species (herbs, <0.2 m from the base of the stem; shrubs, <0.5 m from the base of the stem; pioneer tree, <1 m from the base of the stem) at five points (separated by >6 m) [43,44], homogenized, and then divided into two subsamples. One subsample was used for inoculating soil in the conditioning phase, and one was to analyze microbial communities. All soils were sieved using a mesh (<5 mm) to remove large stones and roots before experiments. Mature seeds of all species were obtained at their natural dispersal time from July to October in 2018 and 2019. Seeds of all species were collected from more than 20 plants. All seeds were surface-sterilized (70% ethanol, 2.625% NaOCl, ethanol, distilled water for 1 min, 3 min, 1 min, and 1 min, respectively) and sown on sterile sand and germinated in a cabinet under a regime of 16 h/21 °C (light):8 h/16 °C (dark).
2.2. Greenhouse Experiment
2.2.1. Conditioning Phase
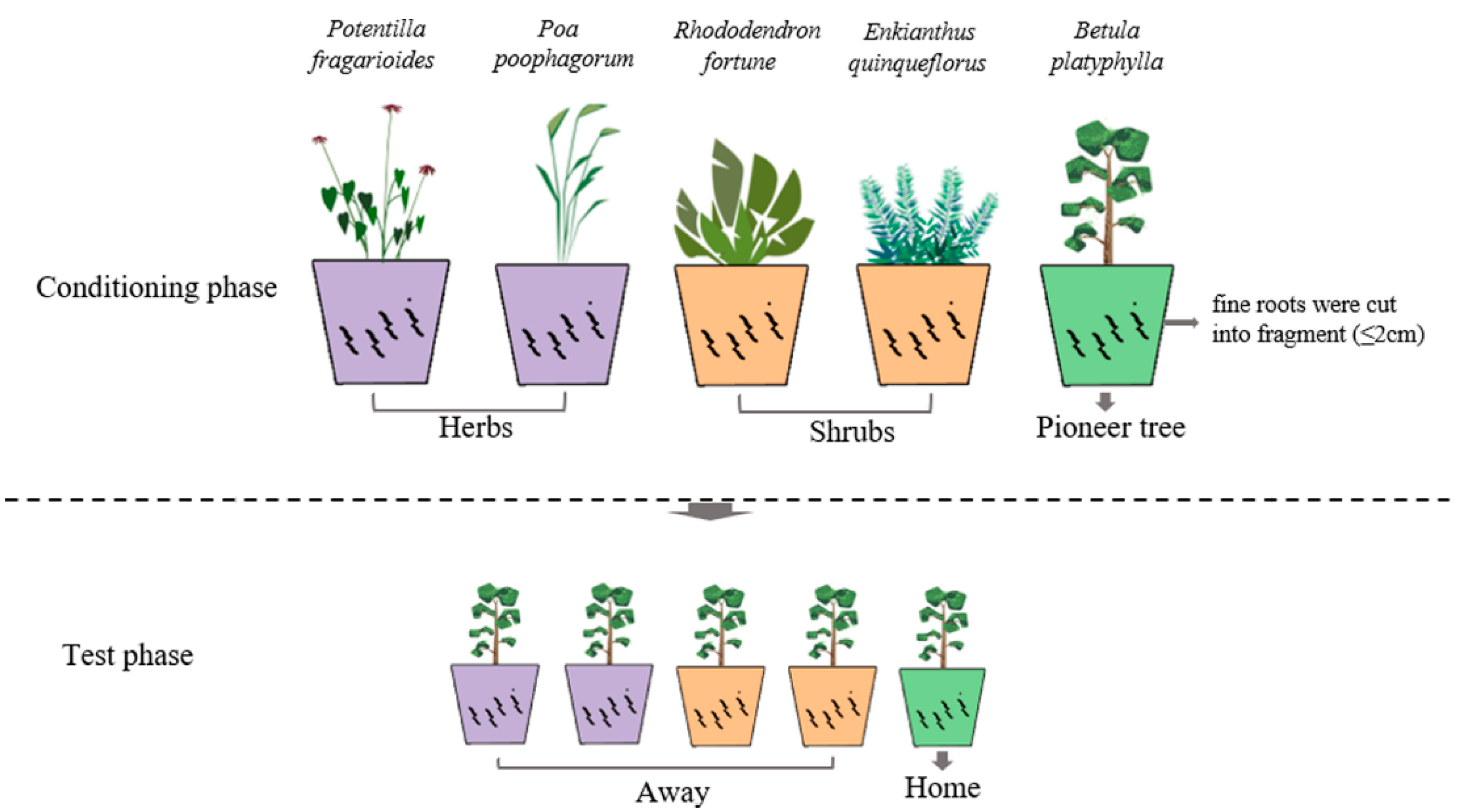
The major purpose of the conditioning phase was to obtain soils with specific soil communities of five species (two early-successional herbs, two mid-successional shrubs, and one target late-successional tree) (Figure 1, upper panel). In the conditioning phase, 25 pots (15 × 22 cm) were filled with 3.5 kg of common starting soil for each species. For shrubs and pioneer trees, saplings of comparable size were monocultured in each replicate soil for 5 and 8 months, respectively. For herbs, 20 one-week-old seedlings were transplanted into each pot for 3 months. Dead saplings and seedlings were replaced during the first week of the experiment. All pots were arranged in a completely randomized design in the greenhouse under a regime of 16 h/21 °C: 8 h/16 °C (light:dark) and 72% relative humidity. All pots were watered with demineralized water twice a week. At the end of the conditioning phase, shoots were clipped at the soil surface. The main roots were removed, and the fine roots of each conditioning species were cut into fragments (≤2 cm) and left in the soil so as to preserve the microbial communities and mined nutrients that had developed in the rhizosphere. Soils conditioned by each plant species were homogenized and pooled [45], and then divided into three subsamples. One subsample was used for determining nutrient concentration, one was stored at −20 °C in order to analyze microbial communities, and one was used in the following test phase.
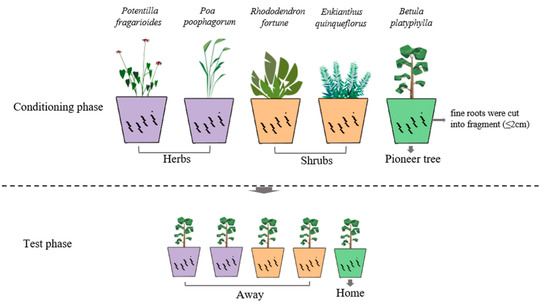
Figure 1.
Graphical illustration of the experimental design. In the conditioning phase, five different plant species, PF: Potentilla fragarioides L., PP: Poa poophagorum Bor, RF: Rhododendron fortune Lindl., EQ: Enkianthus quinqueflorus Lour., BP: Betula platyphylla Sukaczev, were monocultured in soils created by 90% sterilized bare soil and 10% specific inoculum. In the test phase, Betula platyphylla was monocultured in preconditioned soils.
2.2.2. Test Phase
In the test phase, the soils conditioned by different plant species were transferred to 1 L pots (7.5 × 9.8 cm) (Figure 1, lower panel). We had 50 1 L pots in the test phase (5 conditioned soil × 10 replicates = 50 pots). The recently germinated seedlings (at the second leaf stage) of B. platyphylla were replanted as single individuals in each pot (home vs. away) and grown for 5 months in a glasshouse. The seedlings that died were replaced during the first week of the experiment. The growth conditions and watering regime were consistent with those described for the conditioning phase. After 5 months of growth, all shoots were harvested, and roots were gently washed from the soil (the old root fragments in the conditioning phase were removed using 0.15 mm sieves). All shoots and roots were dried at 70 °C for 48 h, and the dry mass was weighed.
2.3. Soil DNA Extraction and Sequence Analyses
Soil DNA was extracted from 0.5 g soil samples using a PowerSoil DNA extraction kit (MoBio, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration and purity of the extracted DNA were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The V4 region of the 16S rRNA for bacteria and the first internal transcribed spacer (ITS1) region of the rRNA operon for fungi were amplified using the general primers 515F/806R [46] and ITS1F/ITS2 [47], respectively. The PCR volume was 50 μL, containing 25 μL 2 × Taq Master Mix, 20 μL ddH2O, 1 μL each primer, and 3 μL genomic DNA. The PCR amplification was conducted as follows: 5 min at 94 °C for initialization; 30 cycles of denaturation at 94 °C for 30 s, 30 s annealing at 52 °C, and 30 s extension at 72 °C; followed by 10 min final elongation at 72 °C. The PCR products from all samples were extracted and purified by 1% agarose gel electrophoresis and the E.Z.N.A. Gel Extraction Kit (Omega, Norcross, GA, USA). Sequencing was conducted on an Illumina NovaSeq 6000 Sequencing System (Guangdong Magigene Biotechnology Co., Ltd., Guangzhou, China). Trimming and filtering of raw sequencing data were performed with fastp [48]. Low-quality reads with lengths less than 200 bp and quality scores less than 20 were removed. Chimera sequences and singletons (with only one read) were removed during the procedure. Each sample was subsampled to the same number of reads as the lowest soil sample (31,468 and 50,251 reads for fungi and bacteria, respectively). Taxonomic annotation for fungi and bacteria was performed based on the UNITE [49] and SILVA [50] databases, respectively. The raw sequences have been uploaded to the NCBI Sequence Read Archive (SRA) database under accession number PRJNA843373.
2.4. Soil Properties
Soil inorganic N (ammonium (-N), nitrate (-N)) was extracted using 2 M KCL (1:5, w/v) to quantify the concentration on a calorimeter with a microplate reader (Varioskan LUX, Thermo Scientific, America) [51,52]. Soil organic carbon (SOC) was determined by the potassium dichromate oxidation method [52]. Soil available phosphorus (AP) was extracted using 0.5 M NaHCO3 (1:5, w/v) solution (pH = 8.5) and analyzed calorimetrically [52].
2.5. Statistical Analysis
We conducted one-way ANOVAs to analyze the effects of herbs and shrubs on soil chemical properties in the conditioning phase and biomass response in the test phase. We used plant species as explanatory variables, and pairwise differences between them were further evaluated using Tukey HSD tests. Log10 transform was used when necessary for homogeneity of variance and normality. A PERMANOVA model was used to investigate the effects of different successional plants on soil microbial communities using Bray–Curtis distances in the “vegan” package. Patterns of similarity among background (inoculum) and conditioning soils were visualized based on nonmetric multidimensional scaling (NMDS) ordination using the metaMDS function in the “vegan” package. Principal coordinates analysis (PCoA) based on Bray–Curtis distances was conducted to analyze the bacterial and fungal communities. We used the scores of the first and second PCoA axes as a variable to represent overall variation in bacterial and fungal community composition [53]. The Spearman correlation analyses between bacterial and fungal phyla and biomass are shown in a heatmap. The fungal putative functional guild was determined using the FUNGuild database [54]. The best subset model via stepwise multiple regression was conducted to assess the effects of biotic and abiotic soil legacies on pioneer tree biomass. Partial least squares path modeling (PLS-SM) was subsequently applied to assess the direct and indirect effects of soil nutrient availability and fungal and bacterial communities on seedling biomass. The model was conducted in the R package “plspm” using the “innerplot” function [55]. All statistical analyses were conducted in R v.4.0.4.
3. Results
3.1. Soil Physicochemical Properties and Microbial Community Composition
At the end of the soil conditioning stage, AP, -N, -N, and SOC were significantly affected by the conditioning species (p < 0.05) (Table 1). Soil nutrient availability generally decreased at the end of the conditioning phase, except for the inorganic nitrogen in P. fragarioides (PF) soil and the organic carbon in E. quinqueflorus (EQ) and R. fortunei (RF) soils. The lowest organic C was observed in soil conditioned by B. platyphylla (Table 1). The soils planted with grasses had the lowest -N. Soil conditioned by P. fragarioides had a higher -N content but a lower -N content than the sterile bare soil. Except for RF and PF, other species did not significantly affect available P (Table 1).

Table 1.
Soil physiochemical properties (mean ± SE) and statistical results at the end of the conditioning phase and in the sterile soil.
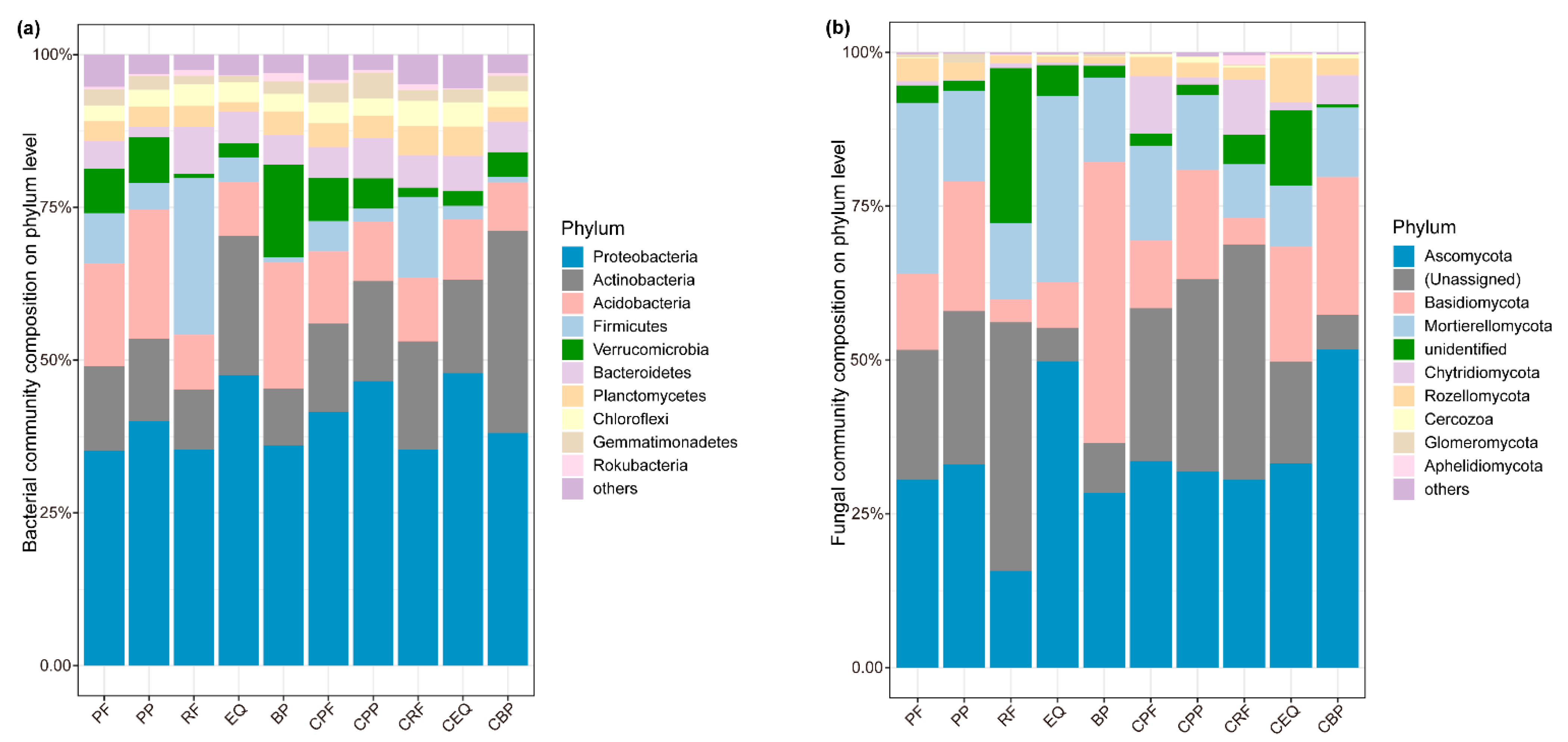
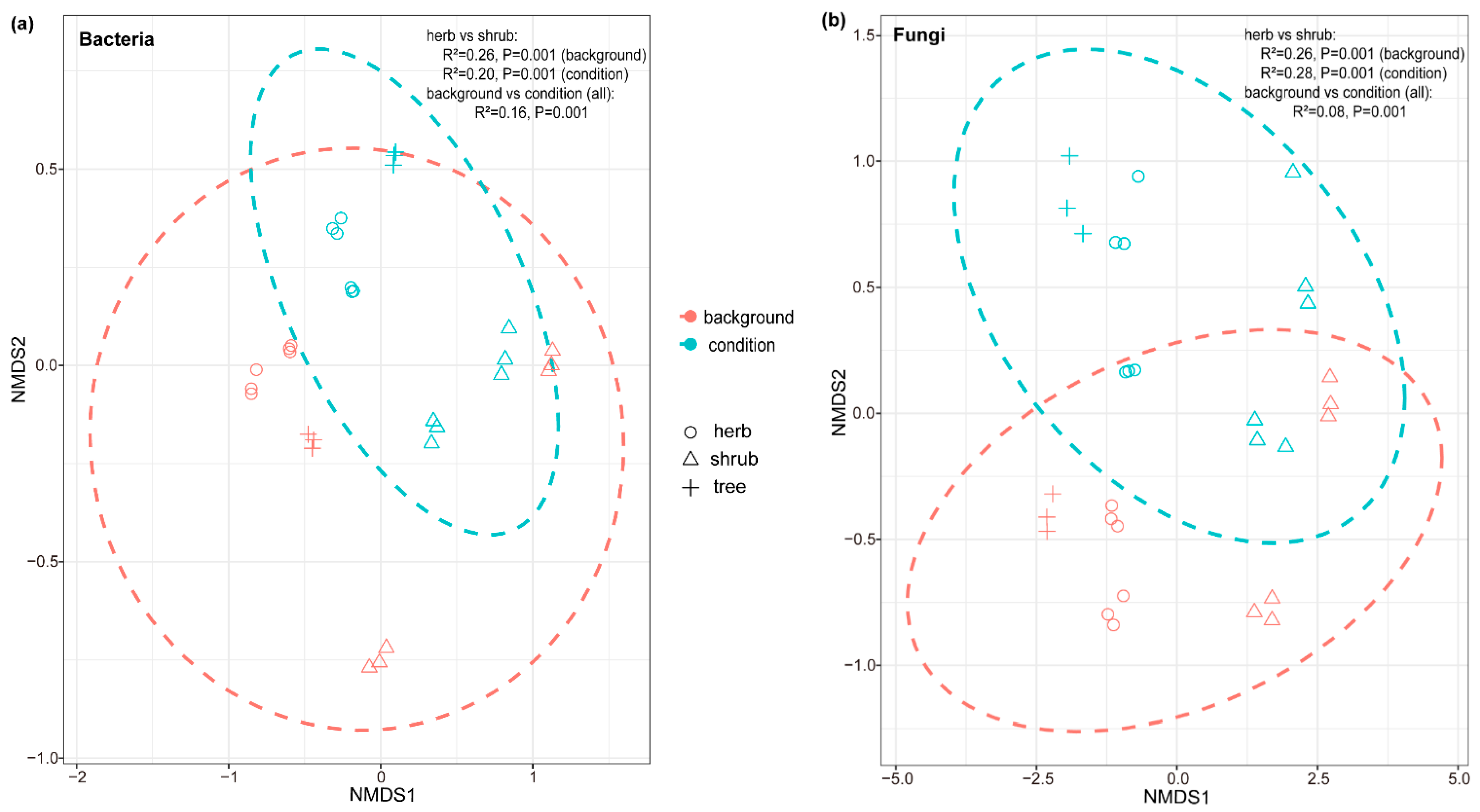
The fungal communities were dominated by Ascomycota, Basidiomycota, Mortierellomycota, Chytridiomycota, and Rozellomycota at the phylum level (Figure 2). Similarly, the dominant phyla of bacteria in soils conditioned by herbs were Proteobacteria, Actinobacteria, and Acidobacteria (Figure 2). The dominant fungal genera of fungi were Solicoccozyma, Mortierella, and Coniochaeta, while the dominant bacterial genera were Pseudarthrobacter, Massilia, and Candidatus Udaeobacter (Figure S1). PERMANOVA tests suggested that the species were significantly different in all background (inoculum) and conditioning soils (Figure 3). The first PCoA axis based on Bray–Curtis distances of bacterial communities explained 37.6% of the total variation, whereas the second axis explained 22.2% of the total variation (Figure S3a). In fungal communities, the first axis explained 32.8%, and the second axis explained 27.3%, of the total variation (Figure S3b). For fungi, the potential functional profiles were predicted using the FUNGuild database, including three main types of trophic modes: symbiotroph (ectomycorrhizal, endophyte, epiphyte), pathotroph (fungal parasite, insect parasite, plant pathogen), and saprotroph (wood/dung/plant saprotroph). No significant relationship was observed between the predicted fungal functional guilds and seedling biomass (Figure S2).
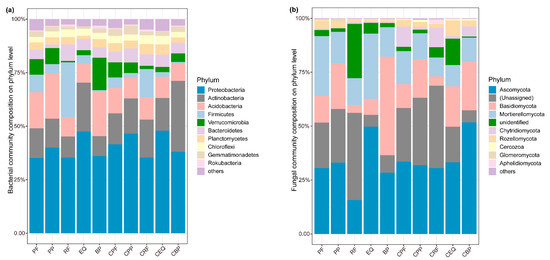
Figure 2.
Relative abundance of (a) bacterial and (b) fungal compositions at the phylum level. PP, PF, RF, EQ, and BP refer to the background (inoculum) soils of Poa poophagorum, Potentilla fragarioides, Rhododendron fortunei, Enkianthus quinqueflorus, and Betula platyphylla, respectively. CPP, CPF, CRF, CEQ, and CBP refer to the soil conditioned by the above species.
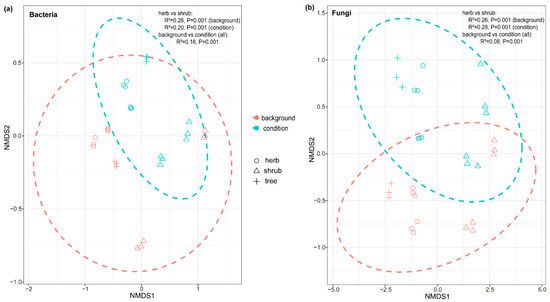
Figure 3.
(a) Bacterial (stress = 0.081) and (b) fungal (stress = 0.079) communities of background (inoculum) and conditioned soils. NMDS using Bray–Curtis distance of background (inoculum) soils and conditioning plants (colors: blue depicts conditioned soils, red depicts background soils) and differential successional stages (shapes: circles represent herbs, triangles represent shrubs, and crosses represent trees). PERMANOVA results, including R2 and significance p for the model, are given in the figure.
3.2. Linking Soil Biotic and Abiotic Legacies with Pioneer Tree Seedling Performance
The seedling biomass of B. platyphylla significantly responded to the conditioning plant guild (herbs vs. shrubs). Specifically, we found significantly lower total biomass in the soil conditioned by herbs than in the soil conditioned by conspecifics (Figure 4). The seedlings growing in E. uinqueflorus (EQ) soils exhibited a higher seedling biomass than those growing in conspecific soil and produced significantly higher biomass than those growing in herb soils (Figure 4).
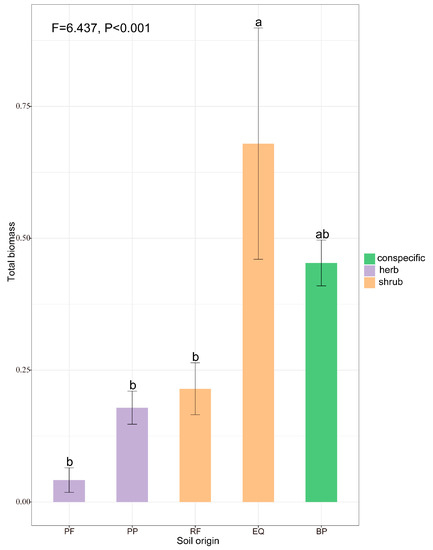
Figure 4.
The total biomass (g) of Betula platyphylla in soil conditioned by herbs (PP, PF), shrubs (EQ, RF), and conspecifics (BP). PP, Poa poophagorum; PF, Potentilla fragarioides; RF, Rhododendron fortunei; EQ, Enkianthus quinqueflorus; BP, Betula platyphylla. Values shown in the figure are mean values ± SE (n = 10). Different lowercase letters refer to significant differences at p < 0.05.
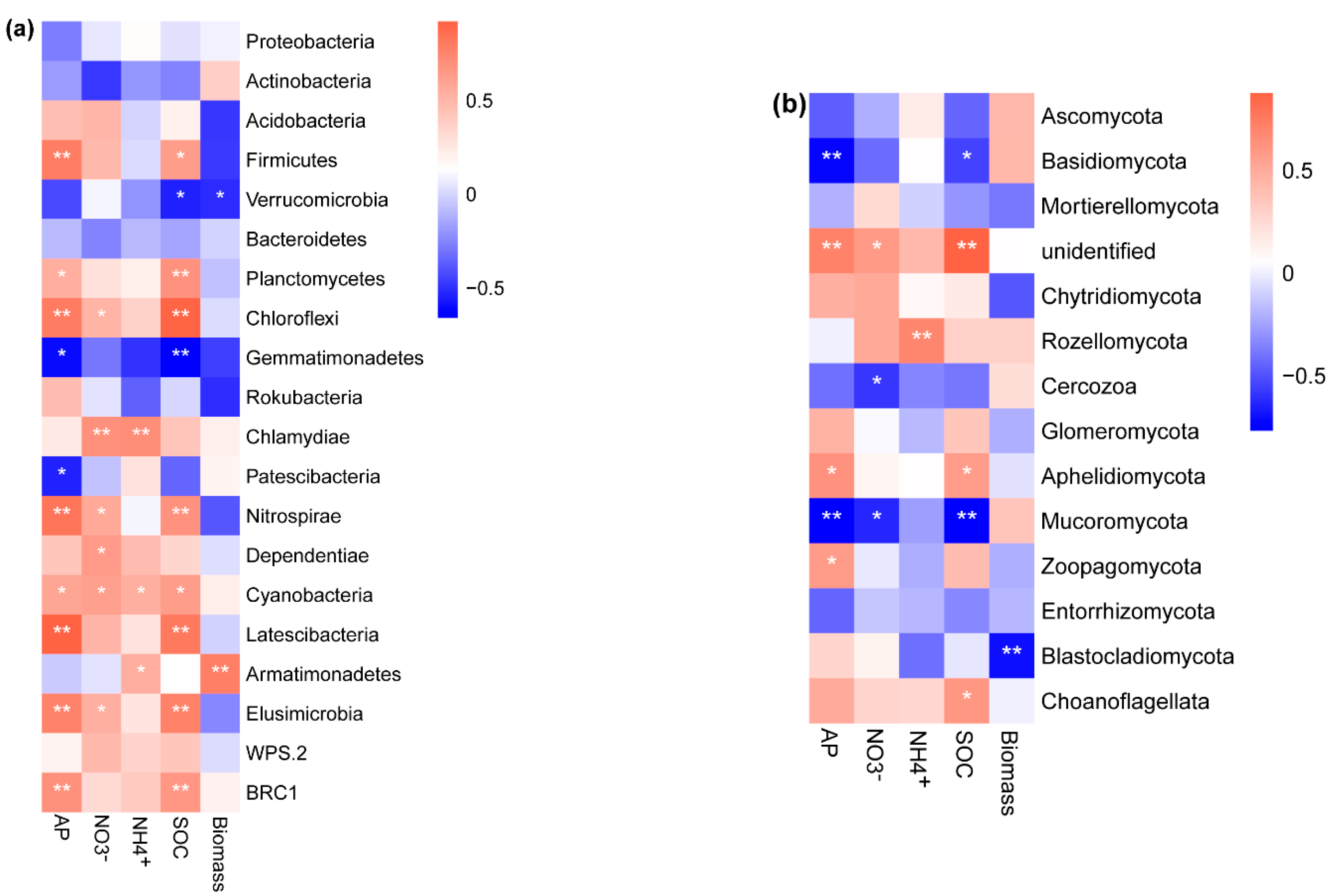
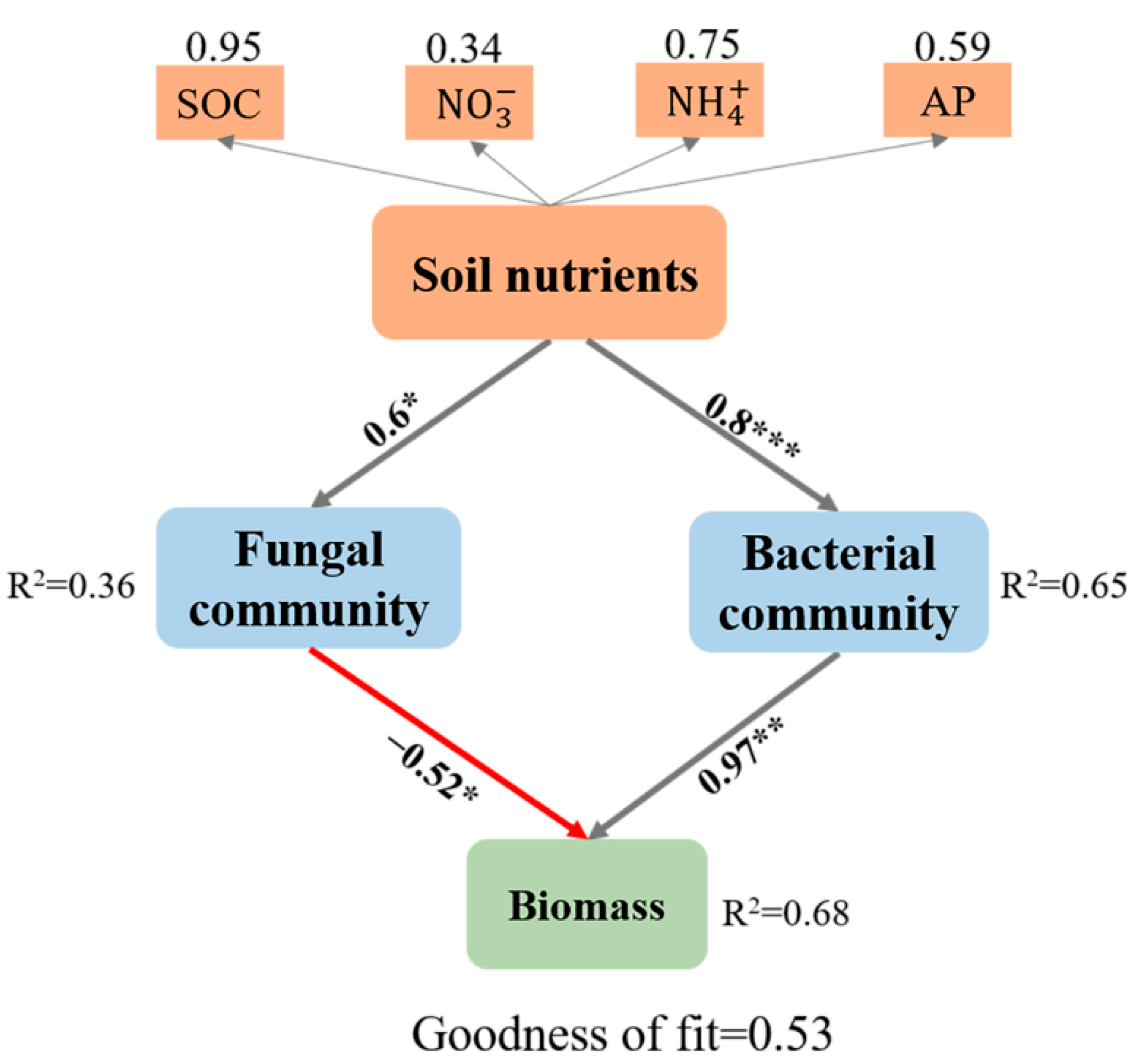
The relative abundance of the bacterial phylum Armatimonadetes (r = 0.78, p < 0.001) was significantly and positively correlated with the seedling biomass (Figure 5a). The relative abundances of the bacterial phylum Verrucomicrobia (Figure 5a, Spearman: r = −0.51, p = 0.05) and the fungal phylum Blastocladiomycota (Figure 5b, Spearman: r = −0.7, p = 0.004) were negatively correlated with seedling biomass. The best subset model via stepwise multiple regression showed that -N, SOC, bacterial community, and fungal community significantly explained the seedling biomass (Table 2). The SOC contents (27.8%), fungal community composition (29.7%), and bacterial community composition (34.8%) were the dominant determinants of the variation in seedling biomass (Table 2). Partial least squares path modeling indicated that the coefficients quantifying the directions and strengths between total biomass and fungi and bacteria were −0.52 and 0.97, respectively (Figure 6). B. platyphylla biomass was more significantly determined by soil bacterial communities than by fungal communities (Figure 6). In addition, the indirect effect of SOC (coefficient: 0.95) on seedling biomass via bacterial and fungal communities was stronger than that of -N (0.75), -N (0.34), and AP (0.59).
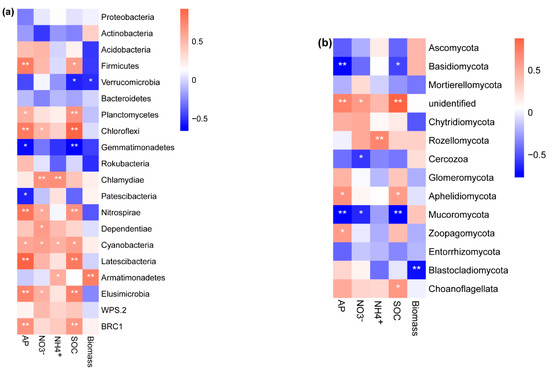
Figure 5.
Spearman correlation analysis between (a) bacterial phyla, (b) fungal phyla, soil nutrients, and seedling biomass. AP, soil available phosphorus; , soil nitrate nitrogen; , soil ammonium nitrogen; SOC, soil organic carbon; Biomass, total seedling biomass. Asterisks represent significant effects (*, p < 0.05; **, p < 0.01).

Table 2.
Results of the stepwise multiple regression using the best subset model for the effect of different soil nutrient contents and biotic (bacteria and fungi) factors on the biomass.
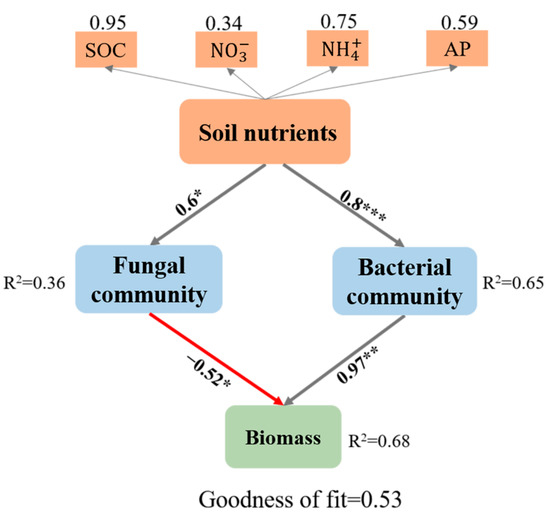
Figure 6.
Partial least squares path modeling disentangling the direct and indirect pathways of the soil nutrients, fungal communities, and bacterial communities in Betula platyphylla biomass. Red and gray arrows indicate negative and positive effects, respectively. Asterisks represent significant effects (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Only significant pathways are shown. Numbers on the arrows indicate standardized path coefficients.
4. Discussion
In this study, the legacies derived from herbs and shrubs significantly impacted the growth of pioneer tree seedlings. Consistent with hypothesis (i), the pioneer tree seedling biomasses in soils conditioned by herbs and shrubs were significantly different. Tree seedlings achieved more biomass when growing in the shrub (E. quinqueflorus) legacy compared to herb and conspecific legacy (Figure 4). The observed facilitative effect of shrub legacy on tree seedlings was consistent with previous observations showing that shrubs (e.g., Baccharis vernalis) had apparent facilitative effects on tree establishment [56]. Macek et al. (2018) suggested that shrubs provide situations (such as light, humidity, and temperature) suitable for tree growth. Here, our results suggested that soil nutrient content and bacterial and fungal community compositions changed by conditioning plants dominantly explained seedling biomass (Table 2). The content of -N being significantly greater in sterilized soil compared to soils that were conditioned by different plant species could be due to a release of nutrients from dead microbes in sterilized soil [57]. All conditioning plant species significantly decreased soil , resulting from plant absorption [58]. The contents of SOC were the highest in soils conditioned by two shrubs (R. fortunei and E. quinqueflorus). The SOC content of sterile bare soil was almost similar to those under R. fortunei, possibly because of recalcitrant debris of R. fortunei leading to lower C input [59] or higher microbial activity leading to greater C output in R. fortunei soils [60]. The primary sources of organic C entering the soil were root exudates, root debris, and border cells [61]. Significant changes in soil nutrient availability are usually associated with the accumulation of organic matter and increased abundance and activity of soil microbes [62]. B. platyphylla seedlings may largely profit from increased organic carbon in the conditioned soil of shrubs. In addition, the relative abundance of the bacterial phylum Armatimonadetes was positively correlated with the target seedling biomass (Figure 5a). This result is supported by previous studies showing that Armatimonadetes was positively correlated with plant growth [63] by protecting against fungal pathogens [64] and decomposing plant materials and photosynthetic substances [65]. Furthermore, pioneer tree seedlings possessed the highest relative biomass in E. quinqueflorus soils with the highest relative abundance of the fungal genus Acremonium (Figure S1). This result was consistent with previous studies showing that the species of Acremonium are not host-specific and could promote the development of different host plants by protecting against root herbivores [66]. The observed negative effects of herb legacy on tree seedlings were consistent with previous observations. Graves et al. (2006) suggested that there are negative effects of herbs on tree seedlings as they competitively inhibited tree seedlings via faster growth and consequent pre-emption of light or nutrients [15,67]. However, the decreased tree seedling growth were eventually ascribed to the changes in soil nutrient content and microbial communities in our study. For instance, the relative abundance of the fungal phylum Blastocladiomycota was negatively correlated with tree seedling biomass (Figure 5b). Some fungal species in Blastocladiomycota are considered pathogens of vascular plants and obligate parasites of plants [68,69], which may infect tree seedlings by having a pathogenic relationship with plants and thus inhibit their growth. Fast-growing B. platyphylla seedlings were more vulnerable to the pathogens because of a higher investment in advanced fine roots as a strategy to promote nutrient absorption [70,71] rather than the resistance to soil-borne pathogens [72]. Additionally, the contents of SOC were the lowest in soils conditioned by two herbs (P. poophagorum and P. fragarioides). This result might be due to higher microbial activity in herbs soils, which result in greater carbon mineralization and soil respiration intensity [60]. However, a higher carbon mineralization might inhibit plant performance due to nutrient starvation as a result of a rapid increment in microbial biomass [73,74]. Collectively, our results agreed with previous studies showing that fast-growing plants produced soil legacies that negatively influenced later plants, whereas slower-growing plants produced a more positive soil legacy effect [75,76]. However, we detected a higher biomass of pioneer tree seedlings when growing in shrub soil for only one plant species. This result indicated that the soil legacy effects of slower-growing shrubs were species-specific, and different soil legacies of pre-existing plant species may differentially influence subsequent plant performance [77].
Inconsistent with hypothesis (ii), our results showed that soil fungal community did not exhibit the overwhelming effects on B. platyphylla growth as expected. Instead, our results showed that variation in pioneer tree seedling biomass was driven more significantly by bacterial community composition than by fungal community composition (Figure 6). The different effects of bacterial and fungal communities on pioneer tree seedling growth in our study are partly in line with previous findings which suggested that the soil legacy effects were more strongly reflected in bacterial communities than fungal communities [78]. Bacterial communities can be responsible for direct growth promotion by enhancing nutrient acquisition, preventing the activity or growth of pathogens, and producing phytohormones [79,80]. For example, previous studies indicated that some species in the bacterial phylum Verrucomicrobia play important roles in organic carbon degradation and have negative impacts on the other microbial groups [81,82]. A probable explanation is that bacteria are generally considered to be more sensitive than fungi to the availability of C substrates due to their shorter turnover period [83,84]. Most of the easily available organic matter is first trapped and metabolized by bacteria [85,86]. Accelerating soil organic matter decomposition and available nutrient release is important for promoting plant growth [87,88]. Mycorrhization also plays a very important role in mutualistic interactions by obtaining nutrients and increasing resistance to biotic and abiotic stresses; however, hyphal-growing fungi have a relatively longer generation time [80,89]. Furthermore, fungal communities tend to have more host-specific relationships with plant species [89]. For example, our results showed that the most abundant sequence numbers of ECM were classified into Sphaerosporella after the conditioning phase (Table S1). Previous studies have reported that Sphaerosporella formed mycorrhizae with Pinus banksiana, Populus, Picea, and Larix [90] and may not effectively and specifically benefit B. platyphylla seedlings. Thus, our results implied that B. platyphylla, an ECM birch species, might be less influenced by pre-existing fungal legacies in this study.
Soil biotic (fungal and bacterial community compositions) and abiotic legacies (especially SOC) synergistically influenced tree seedling growth (Table 2). Soil nutrients (SOC, , , and AP) can indirectly influence plant growth by stimulating the soil microbial community, especially the bacterial community composition [91,92]. The strong association between soil nutrients and microbial communities (both bacteria and fungi) indicated that soil nutrients (especially SOC) were a potential factor influencing pioneer tree seedling recruitment. The interactions between plants and soil microbial communities are modified by variations in soil abiotic properties [93]. Therefore, our results suggested that soil nutrients (SOC, , , and AP) are potential and critical components of soil legacy effects. Furthermore, the results imply that the ability to predict soil legacy effects requires a better understanding of interactions between diverse soil microbes and nutrient availability rather than a focus on singular biotic legacies.
5. Conclusions
In conclusion, we found that early- and mid-successional plant species created distinct soil microbial and abiotic legacies, which in turn differentially influenced the subsequent pioneer tree seedlings that established in these soils. Our results suggested that the legacies of shrubs had a facilitative effect on pioneer tree seedling growth and that the bacterial and fungal communities associated with SOC were the crucial explanatory factors. Specifically, the seedling biomass of B. platyphylla was positively correlated with the relative abundance of bacterial phylum Armatimonadetes, and negatively correlated with the relative abundance of fungal phylum Blastocladiomycota. The changes in bacterial community composition had a stronger effect on seedling biomass than those of fungi. Soil nutrients (especially SOC) were inextricably connected with soil legacy effects on pioneer tree seedling growth. Overall, the results illustrated that soil microbial community composition and soil nutrients together modulated pioneer tree seedling growth. These mechanisms provide evidence for optimizing forest management practices. Further research that focuses on combined experimental studies between plant community legacy effects and individual legacy effects is essential for obtaining a better understanding of the mechanisms that govern pioneer tree species establishment.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13071110/s1, Figure S1: Relative abundance of dominant (a) bacterial and (b) fungal genera; Figure S2: Spearman correlation analysis between the relative abundance of soil fungal functional groups and seedling biomass; Figure S3: Principal coordinate analysis of (a) bacterial and (b) fungal communities based on Bray–Curtis distances; Table S1: Sequence numbers of the dominant genera of ECM under different treatments.
Author Contributions
T.L.: data curation, formal analysis, investigation, writing—original draft, writing—review and editing. W.Z.: conceptualization, methodology, supervision, writing—original draft, writing—review and editing. Y.K.: methodology, writing—review and editing. J.L.: data curation, writing—review and editing. Q.L.: conceptualization, funding acquisition, methodology, project administration, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (41930645, 31971637, 31870607), Youth Innovation Promotion Association of the Chinese Academy of Sciences (2019363, 2021371).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Dataset and associated codes used in the main results are available upon reasonable request to the corresponding author.
Acknowledgments
We thank Heliang He, Renjiao Lu, Xiaohu Wang, Jing Chen, Jiajia Wang, Yang Liu, and Xiaoying Zhang for their assistance during soil collecting, plant cultivation, and harvesting.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gomez-Aparicio, L.; Zamora, R.; Gomez, J.M.; Hodar, J.A.; Castro, J.; Baraza, E. Applying plant facilitation to forest restoration: A meta-analysis of the use of shrubs as nurse plants. Ecol. Appl. 2004, 14, 1128–1138. [Google Scholar] [CrossRef] [Green Version]
- Lamb, D.; Erskine, P.D.; Parrotta, J.A. Restoration of degraded tropical forest landscapes. Science 2005, 310, 1628–1632. [Google Scholar] [CrossRef] [Green Version]
- Mansourian, S.; Parrotta, J. From addressing symptoms to tackling the illness: Reversing forest loss and degradation. Environ. Sci. Policy 2019, 101, 262–265. [Google Scholar] [CrossRef]
- Harantová, L.; Mudrák, O.; Kohout, P.; Elhottová, D.; Frouz, J.; Baldrian, P. Development of microbial community during primary succession in areas degraded by mining activities. Land Degrad. Dev. 2017, 28, 2574–2584. [Google Scholar] [CrossRef]
- Gradel, A.; Haensch, C.; Ganbaatar, B.; Dovdondemberel, B.; Nadaldorj, O.; Günther, B. Response of white birch (Betula platyphylla Sukaczev) to temperature and precipitation in the mountain forest steppe and taiga of northern Mongolia. Dendrochronologia 2017, 41, 24–33. [Google Scholar] [CrossRef]
- Tiebel, K.; Huth, F.; Wagner, S. Soil seed banks of pioneer tree species in European temperate forests: A review. iForest 2018, 11, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Perala, D.A.; Alm, A.A. Reproductive ecology of birch: A review. Forest Ecol. Manag. 1990, 32, 1–38. [Google Scholar] [CrossRef]
- Ganade, G.; Miriti, M.N.; Mazzochini, G.G.; Paz, C.P. Pioneer effects on exotic and native tree colonizers: Insights for Araucaria forest restoration. Basic Appl. Ecol. 2011, 12, 733–742. [Google Scholar] [CrossRef]
- Pinard, M.; Howlett, B.; Davidson, D. Site conditions limit pioneer tree recruitment after logging of dipterocarp forests in Sabah, Malaysia. Biotropica 1996, 28, 2–12. [Google Scholar] [CrossRef]
- MacKenzie, W.H.; Mahony, C.R. An ecological approach to climate change-informed tree species selection for reforestation. Forest Ecol. Manag. 2021, 481, 118705. [Google Scholar] [CrossRef]
- Goodale, U.M.; Berlyn, G.P.; Gregoire, T.G.; Tennakoon, K.U.; Ashton, M.S. Differences in Survival and Growth Among Tropical Rain Forest Pioneer Tree Seedlings in Relation to Canopy Openness and Herbivory. Biotropica 2014, 46, 183–193. [Google Scholar] [CrossRef]
- Gómez-Aparicio, L. The role of plant interactions in the restoration of degraded ecosystems: A meta-analysis across life-forms and ecosystems. J. Ecol. 2009, 97, 1202–1214. [Google Scholar] [CrossRef] [Green Version]
- Dalling, J.W.; Muller-Landau, H.C.; Wright, S.J.; Hubbell, S.P. Role of dispersal in the recruitment limitation of neotropical pioneer species. J. Ecol. 2002, 90, 714–727. [Google Scholar] [CrossRef]
- Xia, Q.; Ando, M.; Seiwa, K.; Kitajima, K. Interaction of seed size with light quality and temperature regimes as germination cues in 10 temperate pioneer tree species. Funct. Ecol. 2015, 30, 866–874. [Google Scholar] [CrossRef]
- Graves, J.H.; Peet, R.K.; White, P.S. The influence of carbon-nutrient balance on herb and woody plant abundance in temperate forest understories. J. Veg. Sci. 2006, 17, 217–226. [Google Scholar] [CrossRef]
- Skinner, A.K.; Lunt, I.D.; McIntyre, S.; Spooner, P.G.; Lavorel, S. Eucalyptus recruitment in degraded woodlands: No benefit from elevated soil fertility. Plant Ecol. 2009, 208, 359–370. [Google Scholar] [CrossRef]
- Molina, R.; Amaranthus, M. Rhizosphere biology: Ecological linkages between soil processes, plant-growth, and community dynamics. In Proceedings of the Conf on Management and Productivity of Western-Montane Forest Soils, Boise, ID, USA, 10–12 April 1990; pp. 51–58. [Google Scholar]
- Cuddington, K. Legacy Effects: The Persistent Impact of Ecological Interactions. Biol. Theor. 2012, 6, 203–210. [Google Scholar] [CrossRef]
- Peña, E.; Baeten, L.; Steel, H.; Viaene, N.; De Sutter, N.; De Schrijver, A.; Verheyen, K.; Bailey, J. Beyond plant-soil feedbacks: Mechanisms driving plant community shifts due to land-use legacies in post-agricultural forests. Funct. Ecol. 2016, 30, 1073–1085. [Google Scholar] [CrossRef] [Green Version]
- Heinen, R.; Hannula, S.E.; De Long, J.R.; Huberty, M.; Jongen, R.; Kielak, A.; Steinauer, K.; Zhu, F.; Bezemer, T.M. Plant community composition steers grassland vegetation via soil legacy effects. Ecol. Lett. 2020, 23, 973–982. [Google Scholar] [CrossRef]
- Endo, M.; Yamamura, Y.; Tanaka, A.; Nakano, T.; Yasuda, T. Nurse-Plant Effects of a Dwarf Shrub on the Establishment of Tree Seedlings in a Volcanic Desert on Mt. Fuji, Central Japan. Arct. Antarct. Alp. Res. 2008, 40, 335–342. [Google Scholar] [CrossRef] [Green Version]
- Hannula, S.E.; Heinen, R.; Huberty, M.; Steinauer, K.; De Long, J.R.; Jongen, R.; Bezemer, T.M. Persistence of plant-mediated microbial soil legacy effects in soil and inside roots. Nat. Commun. 2021, 12, 5686. [Google Scholar] [CrossRef] [PubMed]
- Lekberg, Y.; Bever, J.D.; Bunn, R.A.; Callaway, R.M.; Hart, M.M.; Kivlin, S.N.; Klironomos, J.; Larkin, B.G.; Maron, J.L.; Reinhart, K.O.; et al. Relative importance of competition and plant-soil feedback, their synergy, context dependency and implications for coexistence. Ecol. Lett. 2018, 21, 1268–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting plant-microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, L.M.; Nilsson, M.C.; Wardle, D.A.; Zackrisson, O. Context dependent effects of ectomycorrhizal species richness on tree seedling productivity. Oikos 2001, 93, 353–364. [Google Scholar] [CrossRef]
- Garcia-Guzman, G.; Heil, M. Life histories of hosts and pathogens predict patterns in tropical fungal plant diseases. New Phytol. 2014, 201, 1106–1120. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, B.D.; Tunlid, A. Ectomycorrhizal fungi-potential organic matter decomposers, yet not saprotrophs. New Phytol. 2015, 205, 1443–1447. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Heijden, M.G.A.; Verkade, S.; De Bruin, S.J. Mycorrhizal fungi reduce the negative effects of nitrogen enrichment on plant community structure in dune grassland. Global Chang. Biol. 2008, 14, 2626–2635. [Google Scholar] [CrossRef]
- Wei, G.; Li, M.; Shi, W.; Tian, R.; Chang, C.; Wang, Z.; Wang, N.; Zhao, G.; Gao, Z. Similar drivers but different effects lead to distinct ecological patterns of soil bacterial and archaeal communities. Soil Biol. Biochem. 2020, 144, 107759. [Google Scholar] [CrossRef]
- Hogberg, M.N.; Baath, E.; Nordgren, A.; Arnebrant, K.; Hogberg, P. Contrasting effects of nitrogen availability on plant carbon supply to mycorrhizal fungi and saprotrophs-a hypothesis based on field observations in boreal forest. New Phytol. 2003, 160, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Lambers, H.; Raven, J.A.; Shaver, G.R.; Smith, S.E. Plant nutrient-acquisition strategies change with soil age. Trends Ecol. Evol. 2008, 23, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Amoo, A.E.; Babalola, O.O. Ammonia-oxidizing microorganisms: Key players in the promotion of plant growth. J. Soil Sci. Plant Nut. 2017, 17, 935–947. [Google Scholar] [CrossRef] [Green Version]
- Treseder, K.K. A meta-an.alysis of mycorrhizal responses to nitrogen, phosphorus, and atmospheric CO2 in field studies. New Phytol. 2004, 164, 347–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- in ‘t Zandt, D.; Hoekstra, N.J.; Wagemaker, C.A.M.; Caluwe, H.; Smit-Tiekstra, A.E.; Visser, E.J.W.; Kroon, H.; Pineda, A. Local soil legacy effects in a multispecies grassland community are underlain by root foraging and soil nutrient availability. J. Ecol. 2020, 108, 2243–2255. [Google Scholar] [CrossRef]
- Liu, Q. Ecological Research on Subalpine Coniferous Forests in China; Sichuan University Press: Chengdu, China, 2002; pp. 6–13. [Google Scholar]
- Hu, Z.; Liu, S.; Liu, X.; Hu, J.; Luo, M.; Li, Y.; Shi, S.; Wu, D.; Xiao, J. Soil and soil microbial biomass contents and C:N:P stoichiometry at different succession stages of natural secondary forest in sub-alpine area of western Sichuan, China. Acta Ecol. Sin. 2021, 41, 4900–4912. [Google Scholar]
- Beck, P.; Caudullo, G.; de Rigo, D.; Tinner, W. Betula pendula, Betula pubescens and other birches in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species, 2nd ed.; San-Miguel-Ayanz, J., de Rigo, D., Eds.; Publication Office of the European Union: Luxembourg, 2016; pp. 70–73. [Google Scholar]
- Jonczak, J.; Jankiewicz, U.; Kondras, M.; Kruczkowska, B.; Oktaba, L.; Oktaba, J.; Olejniczak, I.; Pawłowicz, E.; Polláková, N.; Raab, T.; et al. The influence of birch trees (Betula spp.) on soil environment-A review. Forest Ecol. Manag. 2020, 477, 118486. [Google Scholar] [CrossRef]
- Qiang, W.; He, L.; Zhang, Y.; Liu, B.; Liu, Y.; Liu, Q.; Pang, X. Aboveground vegetation and soil physicochemical properties jointly drive the shift of soil microbial community during subalpine secondary succession in southwest China. Catena 2021, 202, 105251. [Google Scholar] [CrossRef]
- Zhang, N.; Van der Putten, W.H.; Veen, G.F. Effects of root decomposition on plant-soil feedback of early- and mid-successional plant species. New Phytol. 2016, 212, 220–231. [Google Scholar] [CrossRef] [Green Version]
- Kivlin, S.N.; Bedoya, R.; Hawkes, C.V. Heterogeneity in arbuscular mycorrhizal fungal communities may contribute to inconsistent plant-soil feedback in a Neotropical forest. Plant Soil 2018, 432, 29–44. [Google Scholar] [CrossRef]
- He, H.; Yu, L.; Yang, X.; Luo, L.; Liu, J.; Chen, J.; Kou, Y.; Zhao, W.; Liu, Q. Effects of Different Soils on the Biomass and Photosynthesis of Rumex nepalensis in Subalpine Region of Southwestern China. Forests 2022, 13, 1–14. [Google Scholar] [CrossRef]
- Schittko, C.; Wurst, S. Above- and belowground effects of plant-soil feedback from exotic Solidago canadensis on native Tanacetum vulgare. Biol. Invasions 2013, 16, 1465–1479. [Google Scholar] [CrossRef]
- Kempel, A.; Rindisbacher, A.; Fischer, M.; Allan, E. Plant soil feedback strength in relation to large-scale plant rarity and phylogenetic relatedness. Ecology 2018, 99, 597–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachelot, B.; Uriarte, M.; Muscarella, R.; Forero-Montana, J.; Thompson, J.; McGuire, K.; Zimmerman, J.; Swenson, N.G.; Clark, J.S. Associations among arbuscular mycorrhizal fungi and seedlings are predicted to change with tree successional status. Ecology 2018, 99, 607–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glockner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, 259–264. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, W.; He, H.; Kou, Y.; Liu, Q. Variations in the community patterns of soil nematodes at different soil depths across successional stages of subalpine forests. Ecol. Indic. 2022, 136, 108624. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis, 3rd ed.; China Agriculture Press: Beijing, China, 2000; pp. 25–308. [Google Scholar]
- Lozano, Y.M.; Armas, C.; Hortal, S.; Casanoves, F.; Pugnaire, F.I. Disentangling above- and below-ground facilitation drivers in arid environments: The role of soil microorganisms, soil properties and microhabitat. New Phytol. 2017, 216, 1236–1246. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Sanchez, G.; Trinchera, L.; Russolillo, G. Plspm: Tools for Partial Least Squares Path Modeling (PLS-PM). 2015. Available online: https://github.com/gastonstat/plspm (accessed on 20 May 2022).
- Macek, P.; Schöb, C.; Núñez-Ávila, M.; Hernández Gentina, I.R.; Pugnaire, F.I.; Armesto, J.J.; Ohlemuller, R. Shrub facilitation drives tree establishment in a semiarid fog-dependent ecosystem. Appl. Veg. Sci. 2018, 21, 113–120. [Google Scholar] [CrossRef]
- Semchenko, M.; Leff, J.W.; Lozano, Y.M.; Saar, S.; Davison, J.; Wilkinson, A.; Jackson, B.G.; Pritchard, W.J.; De Long, J.R.; Oakley, S.; et al. Fungal diversity regulates plant-soil feedbacks in temperate grassland. Sci. Adv. 2018, 4, eaau4578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolan, N.S.; Hedley, M.J.; White, R.E. Processes of soil acidification during nitrogen cycling with emphasis on legume based pastures. Plant Soil 1991, 134, 53–63. [Google Scholar] [CrossRef]
- Osburn, E.D.; Elliottt, K.J.; Knoepp, J.D.; Miniat, C.F.; Barrett, J.E. Soil microbial response to Rhododendron understory removal in southern Appalachian forests: Effects on extracellular enzymes. Soil Biol. Biochem. 2018, 127, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, L.; Hu, J.; Zhang, W.; Fu, X.; Le, Y.; Jin, F. Organic carbon accumulation capability of two typical tidal wetland soils in Chongming Dongtan, China. J. Environ. Sci. 2011, 23, 87–94. [Google Scholar] [CrossRef]
- Marando, G.; Jiménez, P.; Hereter, A.; Julià, M.; Ginovart, M.; Bonmatí, M. Effects of thermally dried and composted sewage sludges on the fertility of residual soils from limestone quarries. Appl. Soil Ecol. 2011, 49, 234–241. [Google Scholar] [CrossRef]
- Tscherko, D.; Rustemeier, J.; Richter, A.; Wanek, W.; Kandeler, E. Functional diversity of the soil microflora in primary succession across two glacier forelands in the Central Alps. Eur. J. Soil Sci. 2003, 54, 685–696. [Google Scholar] [CrossRef]
- Ma, H.-K.; Pineda, A.; Hannula, S.E.; Kielak, A.M.; Setyarini, S.N.; Bezemer, T.M. Steering root microbiomes of a commercial horticultural crop with plant-soil feedbacks. Appl. Soil Ecol. 2020, 150, 103468. [Google Scholar] [CrossRef]
- Bejarano-Bolívar, A.A.; Lamelas, A.; Aguirre von Wobeser, E.; Sánchez-Rangel, D.; Méndez-Bravo, A.; Eskalen, A.; Reverchon, F. Shifts in the structure of rhizosphere bacterial communities of avocado after Fusarium dieback. Rhizosphere 2021, 18, 100333. [Google Scholar] [CrossRef]
- Lee, K.C.Y.; Dunfield, P.F.; Stott, M.B. The Phylum Armatimonadetes. In The Prokaryotes, 4th ed.; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 447–458. [Google Scholar]
- Han, L.; Zhou, X.; Zhao, Y.; Zhu, S.; Wu, L.; He, Y.; Ping, X.; Lu, X.; Huang, W.; Qian, J.; et al. Colonization of endophyte Acremonium sp. D212 in Panax notoginseng and rice mediated by auxin and jasmonic acid. J. Integr. Plant Biol. 2020, 62, 1433–1451. [Google Scholar] [CrossRef]
- Gafta, D.; Peet, R.K.; Morgan, J. Interaction of herbs and tree saplings is mediated by soil fertility and stand evergreenness in southern Appalachian forests. J. Veg. Sci. 2019, 31, 95–106. [Google Scholar] [CrossRef]
- Hoffman, Y.; Aflalo, C.; Zarka, A.; Gutman, J.; James, T.Y.; Boussiba, S. Isolation and characterization of a novel chytrid species (phylum Blastocladiomycota), parasitic on the green alga Haematococcus. Mycol. Res. 2008, 112, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Porter, T.M.; Martin, W.; James, T.Y.; Longcore, J.E.; Gleason, F.H.; Adler, P.H.; Letcher, P.M.; Vilgalys, R. Molecular phylogeny of the Blastocladiomycota (Fungi) based on nuclear ribosomal DNA. Fungal Biol. 2011, 115, 381–392. [Google Scholar] [CrossRef]
- Meier, I.C.; Leuschner, C. Nutrient dynamics along a precipitation gradient in European beech forests. Biogeochemistry 2014, 120, 51–69. [Google Scholar] [CrossRef]
- Sawada, Y.; Aiba, S.-I.; Takyu, M.; Repin, R.; Nais, J.; Kitayama, K. Community dynamics over 14 years along gradients of geological substrate and topography in tropical montane forests on Mount Kinabalu, Borneo. J. Trop. Ecol. 2015, 31, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Velmala, S.M.; Rajala, T.; Smolander, A.; Petäistö, R.L.; Lilja, A.; Pennanen, T. Infection with foliar pathogenic fungi does not alter the receptivity of Norway spruce seedlings to ectomycorrhizal fungi. Plant Soil 2014, 385, 329–342. [Google Scholar] [CrossRef]
- Liu, E.; Takahashi, T.; Hitomi, T. Effect of pruning material compost on the nitrogen dynamic, soil microbial biomass, and plant biomass in different soil types. Landsc. Ecol. Eng. 2019, 15, 413–419. [Google Scholar] [CrossRef]
- Thiessen, S.; Gleixner, G.; Wutzler, T.; Reichstein, M. Both priming and temperature sensitivity of soil organic matter decomposition depend on microbial biomass-An incubation study. Soil Biol. Biochem. 2013, 57, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Lemmermeyer, S.; Lorcher, L.; van Kleunen, M.; Dawson, W. Testing the Plant Growth-Defense Hypothesis Belowground: Do Faster-Growing Herbaceous Plant Species Suffer More Negative Effects from Soil Biota than Slower-Growing Ones? Am. Nat. 2015, 186, 264–271. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, J.; Verbruggen, E.; Heinze, J.; Xiang, D.; Chen, B.; Joshi, J.; Rillig, M.C. The interplay between soil structure, roots, and microbiota as a determinant of plant-soil feedback. Ecol. Evol. 2016, 6, 7633–7644. [Google Scholar] [CrossRef]
- Hassan, K.; Carrillo, Y.; Nielsen, U.N.; Pugnaire, F. The effect of prolonged drought legacies on plant–soil feedbacks. J. Veg Sci. 2021, 32, e13100. [Google Scholar] [CrossRef]
- Schmid, M.W.; van Moorsel, S.J.; Hahl, T.; De Luca, E.; De Deyn, G.B.; Wagg, C.; Niklaus, P.A.; Schmid, B. Effects of plant community history, soil legacy and plant diversity on soil microbial communities. J. Ecol. 2021, 109, 3007–3023. [Google Scholar] [CrossRef]
- Blom, D.; Fabbri, C.; Connor, E.C.; Schiestl, F.P.; Klauser, D.R.; Boller, T.; Eberl, L.; Weisskopf, L. Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ. Microbiol. 2011, 13, 3047–3058. [Google Scholar] [CrossRef]
- Chamkhi, I.; El Omari, N.; Balahbib, A.; El Menyiy, N.; Benali, T.; Ghoulam, C. Is the rhizosphere a source of applicable multi-beneficial microorganisms for plant enhancement? Saudi J. Biol. Sci. 2022, 29, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Nixon, S.L.; Daly, R.A.; Borton, M.A.; Solden, L.M.; Welch, S.A.; Cole, D.R.; Mouser, P.J.; Wilkins, M.J.; Wrighton, K.C. Genome-Resolved Metagenomics Extends the Environmental Distribution of the Verrucomicrobia Phylum to the Deep Terrestrial Subsurface. mSphere 2019, 4, e00613-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altshuler, I.; Hamel, J.; Turney, S.; Magnuson, E.; Levesque, R.; Greer, C.W.; Whyte, L.G. Species interactions and distinct microbial communities in high Arctic permafrost affected cryosols are associated with the CH4 and CO2 gas fluxes. Environ. Microbiol. 2019, 21, 3711–3727. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y. Priming effects: Interactions between living and dead organic matter. Soil Biol. Biochem. 2010, 42, 1363–1371. [Google Scholar] [CrossRef]
- Ren, J.; Liu, X.; Yang, W.; Yang, X.; Li, W.; Xia, Q.; Li, J.; Gao, Z.; Yang, Z. Rhizosphere soil properties, microbial community, and enzyme activities: Short-term responses to partial substitution of chemical fertilizer with organic manure. J. Environ. Manag. 2021, 299, 113650. [Google Scholar] [CrossRef]
- Paterson, E.; Gebbing, T.; Abel, C.; Sim, A.; Telfer, G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2007, 173, 600–610. [Google Scholar] [CrossRef]
- Moore-Kucera, J.; Dick, R.P. Application of 13C-labeled litter and root materials for in situ decomposition studies using phospholipid fatty acids. Soil Biol. Biochem. 2008, 40, 2485–2493. [Google Scholar] [CrossRef]
- Blagodatskaya, E.V.; Blagodatsky, S.A.; Anderson, T.H.; Kuzyakov, Y. Contrasting effects of glucose, living roots and maize straw on microbial growth kinetics and substrate availability in soil. Eur. J. Soil Sci. 2009, 60, 186–197. [Google Scholar] [CrossRef]
- Cheng, W. Rhizosphere priming effect: Its functional relationships with microbial turnover, evapotranspiration, and C-N budgets. Soil Biol. Biochem. 2009, 41, 1795–1801. [Google Scholar] [CrossRef]
- Sun, S.; Li, S.; Avera, B.N.; Strahm, B.D.; Badgley, B.D. Soil Bacterial and Fungal Communities Show Distinct Recovery Patterns during Forest Ecosystem Restoration. Appl. Environ. Microbiol. 2017, 83, e00966-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danielson, R.M. Ectomycorrhiza formation by the operculate discomycete Sphaerosporella brunnea (Pezizales). Mycologia 1984, 76, 454–461. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fert. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Liang, B.; Yang, X.; Murphy, D.V.; He, X.; Zhou, J. Fate of 15 N-labeled fertilizer in soils under dryland agriculture after 19 years of different fertilizations. Biol. Fert. Soils 2013, 49, 977–986. [Google Scholar] [CrossRef]
- Nash, J.; Laushman, R.; Schadt, C. Ectomycorrhizal fungal diversity interacts with soil nutrients to predict plant growth despite weak plant-soil feedbacks. Plant Soil 2020, 453, 445–458. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).