Abstract
Woodlands are pivotal to carbon stocks, but the process of cycling C is slow and may be most effective in the biodiverse root zone. How the root zone impacts plants has been widely examined over the past few decades, but the role of the root zone in decomposition is understudied. Here, we examined how mycorrhizal association and macroinvertebrate activity influences wood decomposition across diverse tree species. Within the root zone of six predominantly arbuscular mycorrhizal (AM) (Acer negundo, Acer saccharum, Prunus serotina, Juglans nigra, Sassafras albidum, and Liriodendron tulipfera) and seven predominantly ectomycorrhizal (EM) tree species (Carya glabra, Quercus alba, Quercus rubra, Betula alleghaniensis, Picea rubens, Pinus virginiana, and Pinus strobus), woody litter was buried for 13 months. Macroinvertebrate access to woody substrate was either prevented or not using 0.22 mm mesh in a common garden site in central Pennsylvania. Decomposition was assessed as proportionate mass loss, as explained by root diameter, phylogenetic signal, mycorrhizal type, canopy tree trait, or macroinvertebrate exclusion. Macroinvertebrate exclusion significantly increased wood decomposition by 5.9%, while mycorrhizal type did not affect wood decomposition, nor did canopy traits (i.e., broad leaves versus pine needles). Interestingly, there was a phylogenetic signal for wood decomposition. Local indicators for phylogenetic associations (LIPA) determined high values of sensitivity value in Pinus and Picea genera, while Carya, Juglans, Betula, and Prunus yielded low values of sensitivity. Phylogenetic signals went undetected for tree root morphology. Despite this, roots greater than 0.35 mm significantly increased woody litter decomposition by 8%. In conclusion, the findings of this study suggest trees with larger root diameters can accelerate C cycling, as can trees associated with certain phylogenetic clades. In addition, root zone macroinvertebrates can potentially limit woody C cycling, while mycorrhizal type does not play a significant role.
1. Introduction
Woody plants are quintessential to carbon storage and net primary productivity [1,2]. These rigid organisms inhabit 30% of Earth’s landmass and contain 50% of the carbon that makes up the aboveground terrestrial biosphere [3,4]. In addition to their impact on C flux, woody plants can shape the pedosphere [5] through root recruitment of mycorrhizal fungi, recycling of organic material, and litter deposition [6,7]. Particularly, plant material can be fragmented by natural events (e.g., freeze–thaw events, windstorms, and animal activity), as well as deposited into soil communities from plant standing mass. Coarse and fine wood debris can make up anywhere from 1 to 25% of the forest floor [8,9,10], as the brown food web is supplemented by rotten logs, snags and stumps, which can provide resources for seed germination [11], as well as habitats for invertebrates and microbes. Select soil microbes have a primary role in wood degradation and are key components of SOM formation and decomposition [12]. Yet, soil macroinvertebrates (>2 mm) may have an equally important role as microbial decomposers [13], but macroinvertebrate function is often context dependent and can at times be disruptive (e.g., burrowing, predation, and ecosystem engineering) to microbial function.
Determining ways in which diverse root zones may impact soil fauna is essential to understanding brown food web processes. Many organisms are involved in mineralizing wood: ~20–30 percent of forest arthropods, including insects, are either wood dependent (i.e., saproxylic) or opportunistically utilize wood [14,15]. Wood can be broken down through modes of tunneling and nesting [16], while some invertebrates rely on hind-gut microbes to metabolize wood [17,18]. Lumbricida (i.e., earthworms) and collembola (i.e., springtails) harbor Bacillus spp. in their intestinal systems, which plays a role in the degradation of chitin and lignocellulose [17]. Radiolabeling studies have found invertebrates to consume both mycorrhizae and saprotrophic fungi [19], perhaps suggesting that macroinvertebrates can harness microorganisms involved in C exchange and cycling. Invertebrate activity, including the communition of wood debris and burrowing activity, can increase wood surface area, potentially leading to increased decomposition through mycelium contact with wood surfaces. Invertebrates may also slow down decomposition, as collembola (i.e., Folsomia candida) activity has been reported to sever and disrupt mycelial cords from connecting with wood substrate [20], which can slow C cycling. This may be pertinent to recent studies that suggest hyphal extension to be an important predictor of wood decomposition [21].
Wood is decomposed by a limited number of invertebrate and fungal classes [15,22,23]. Decomposer efficacy may also be influenced by foliar litter input [24,25,26], as minerals can differentially accumulate in foliar tissue prior to being introduced into soils [27]. Differences in pine versus broad leaf tissue may differentially alter soil chemistry (i.e., pH) [28], as aging pine needles can lead to an increase in Mn2+ oxidation state (i.e., Mn3+ and Mn4+), which negatively corresponds with C: N [29]. Such changes in litter chemistry may directly affect decomposers [30], as white, brown, and soft rot presence in sapwood corresponds to wood pH [31]. White and brown rot fungi can decompose wood through enzymatic activity or Fenton redox chemistry [32,33,34,35].
Aside from free-living decomposers, root-associated fungi may also play a role in decomposition, especially in the context of the root zone. Some root-partnering fungi, termed ectomycorrhizal (EM) fungi, have been reported to degrade cellulose through Fenton redox chemistry and hydroxyl radical attacks [36,37]. This suggests that the life history of EM fungi spans a biotroph–saprotroph continuum [38], but this characterization may depend on life history, evolutionary divergence, and the retention/expression of lignocellulolytic genes that are localized in mycelial networks [39]. In contrast, arbuscular mycorrhizal (AM) fungi are not known for lignocellulolytic capabilities, but still, AM fungi can increase water-stable aggregates [40,41] and make the soil environment conducive toward decomposers. Interestingly, fine roots can reduce soil moisture (i.e., drying effect), which may be antagonistic toward decomposers [42]. Thus, root morphology, or variation in root diameter, may help shape root zone decomposition, as previous studies found correlations with root diameter and soil organic matter respiration [43,44]. Moreover, root-zone-mediated decomposition may also be autocorrelated with phylogenetic signals or links between phylogeny and continuous trait values [45]. For example, decomposition in hardwood tree soils (e.g., black cherry) may differ from softwoods (e.g., pines), but in a manner that is not to be confused with phylogenetic conservatism [46].
To date, a myriad of studies has increased our understanding of wood decomposition [47,48,49,50,51], but the effect of the soil-mediated root zone is unclear. In addition to that, it is unclear how macroinvertebrates are differentially influenced by the root zone of EM- versus AM-associated trees, as well as trees of varying root diameter and phylogenetic distance. In this study, we sought to unravel how soils conditioned by diverse tree species can affect the decomposition of woody litter. Here, mycorrhizal type, phylogenetic distance, root diameter, litter quality (canopy trait), and macroinvertebrates were assessed. To address this knowledge gap, we sought to determine how mycorrhizal type interacts with macroinvertebrates to influence decomposition. To limit the influence of environmental heterogeneity, we made use of a common garden forest by manipulating the presence (+) or absence (−) of macroinvertebrates > 0.22 mm on added woody material in the root zone of six AM and seven EM trees. Under these conditions, wood was allowed to decompose for 13 months in the root zone of 13 tree species. We hypothesized that: (1) the exclusion of macroinvertebrates (>0.22-mm) would enhance decomposition due to less negative interactions on microbes; (2) root diameter would be a good predictor of decomposition, and there might be a phylogenetic signal for this trait. Additionally, (3) differences in mycorrhizal association would lead to differences in wood decomposition rates.
2. Materials and Methods
2.1. Study Site
This study was conducted within a common garden forest in central Pennsylvania, USA (40°42′ N, 77°57′ W). From 1981 to 2010, annual rainfall in this area has been ~1006 mm, and the average annual high temperature is recorded as 15 °C and average annual low temperature is recorded as 5 °C (National Climatic Data Center [52]). The research site rests on continuously flat semi-active mesic Typic Hapludalfs soil composed of silt loam (Hagerstown series, 0–3 percent slope), with an approximate pH of 5.48 [53]. The previous land use at this site was grassy hay-field (Cheng et al., 2016), prior to becoming a research site in the mid-1990s. This site features tree species occurring in conspecific monocultures (i.e., conspecific blocks), which is a stand of 6 trees of the same species separated belowground from other species by plastic barriers to approximately a 1 m depth. The trees are over 20 years old, and prior research at this site has described root morphology and mycorrhizal root colonization levels [54,55].
2.2. Study System
Macroinvertebrates can be characterized on a size-class basis, as size-class delineation does not take away from the essential role of invertebrates involved in brown food webs, resource redistribution, and decomposition. Soil macroinvertebrates, ranging from earthworms, soil-dwelling insects, myriapods, isopods, and Staphylinids, are of a size class larger than 2 mm [56,57] and differ from the meso fauna that are between 0.1 mm and 0.2 mm [13,58,59]. The manipulation of invertebrate presence, absence, or density can require laborious sterilization or inoculation in a controlled setting [60], but as it relates to this study, polyester mesh (0.22 mm) was used to restrict macroinvertebrates from interacting with decomposing wood substrate placed in the root zone of 6 AM and 7 EM trees.
The impact of the root zone of 6 AM (Acer negundo, Acer saccharum, Prunus serotina, Juglans nigra, Sassafras albidum, and Liriodendron tulipfera) and 7 EM trees (Carya glabra, Quercus alba, Quercus rubra, Betula alleghaniensis, Picea rubens, Pinus virginiana, and Pinus strobus) (Table 1) on wood decomposition was compared. AM and EM trees are known to differentially impact underlying soil and microorganisms, and at our site, AM-associated trees generally had increased soil pH and available N [61].

Table 1.
Root diameter, mycorrhizal type, and foliar litter trait from 13 temperate trees at a plantation in Pennsylvania, USA.
2.3. Experimental Design
The experiment tested the interactive effect of mycorrhizal type, tree canopy (i.e., foliar traits), root morphology, and macroinvertebrates on decomposition (Figure 1). In addition to selecting a variety of AM and EM trees for this study, the 13 trees were chosen because of their broad root diameter range (Table 1), which was previously determined by assessing roots of the first three orders [54]. As a caveat, DNA and microscopic evidence were not used to verify the mycorrhizal root colonization rate or rare bimodal associations, as this study was designed to only include well-defined mycorrhizal types and tree-specific associations [54,62,63]. The experimental design included 2 conspecific blocks (i.e., 6 trees per conspecific block) × 13 tree species (6 AM + 7 EM) × 2 macroinvertebrates levels for a total of 52 observations. Specifically, this allowed 26 observations that included macroinvertebrates (+M) and 26 observations that excluded macroinvertebrates (−M). In addition, each observation was contrasted by mycorrhizal type, canopy litter trait (needle or broadleaf), and root morphology (roots greater than 0.35 mm (i.e., coarse) versus roots less than 0.35 mm (i.e., fine). The 0.35 mm cut off was chosen because past studies have shown bifurcations in root behavior at this measurement. This includes differences in root length proliferation, root dry mass proliferation, arbuscule mycorrhizal colonization, and extramatrical hyphae length [64].
Figure 1.
Testing the effects of mycorrhizal type, foliar litter input, root morphology, and macroinvertebrates on wood decomposition in the root zone of 13 temperate trees.
2.4. Wood Substrate
Assessing the rate of wood decomposition for a single species (Acer rubrum) was ideal for this study, especially since woody litter can vary in the ratio of labile to recalcitrant tissue [65]. Focusing on a common wood type across diverse tree species also allowed us to examine the influence of tree type and phylogenetic distance on decomposition. Commercial wood cubes with a volume of 1.905 cm3 (Woodpeckers Inc., Lakewood, OH, USA), derived from Acer rubrum, were oven-dried for 20 h at 40 °C to remove residual moisture in the woody tissue. Wood substrate was handled in pairs (Figure A1), as fine polyester mesh with 0.22 mm openings enclosed one wood substrate (−macroinvertebrates), while another was void of polyester mesh (+macroinvertebrates). Wood pairs were deposited into mesh tube cylindrical cores (10 cm long × 5 cm diameter) with 50 mm × 50 mm openings (Figure A1). These cylindrical cores were found to be unrestrictive to macroinvertebrates and salamanders in previous field studies (i.e., unpublished observations [53,66]).
2.5. Field Burial and Incubation
Cores were filled and buried vertically (Figure A2) with root-zone soil collected at the burial site. Similar to Malik, Trexler [66], cores were buried ~37.5 cm from the trunk of each specified EM or AM tree. Excavations for core burials were made to about 10 cm, because the decomposition of woody debris frequently occurs at shallow depths [67]. Additionally, cylindrical cores’ subsurface placement enabled wood cubes to stay at a constant depth, as soil surface placement could have potentially led to stochastically uneven burials. Field incubation occurred from October 2017 to November 2018, after which, cores were removed from the field and brought to the laboratory for analysis.
2.6. Analysis: Mycorrhizal Type, Foliar Litter, and Macro Invertebrate Exclusion
Wood was oven-dried for 5 days at 40 °C. Decomposition was quantified as proportionate mass loss, because mass loss corresponds to C and N mineralization and lignocellulose solubilization [68,69]. Relative wood decomposition or mass loss was evaluated as the difference in the initial and final mass divided by the initial mass (Delta mass/initial mass). On average, the initial mass was 3 g, and due to the nature of decomposition, the final mass was always less than the initial. This always made relative wood decomposition a positive number between 0 and 1 when plugged into the described formula. Data were analyzed with R version 4.0.2. The Shapiro–Wilk’s test and Levene’s test were used to assess normality and the equality of variance, respectively. Macroinvertebrates, mycorrhizal type, root morphology, and foliar litter trait were held as explanatory variables, while relative wood decomposition was set as the response variable. A four-way ANOVA was performed to determine which explanatory variables were significant predictors at an alpha of 0.05. Statistical outliers were examined with diagnostic plots of standardized residuals and the ‘boxplot () $out’ command. StepAICc was used to evaluate model selection with the ‘drop1()’ command. An association between root diameter and average root zone decomposition was assessed using Pearson product moment correlation through the ‘Hmisc’ package [70], using the ‘rcorr’ command. Correlations between root diameter and average root zone decomposition were further examined with 95% CI ellipses using ggplot2.
2.7. Analysis: Phylogenetic Signals
Using R version 4.1.3, phylogenetic distance was captured with respect to the diverse plant species used in this study. Particularly, ‘V. Phylomaker’, an R package with pre-determined vascular plant relatedness, was employed [71], and the ‘phytools’ library [72] was used to help generate a phylogenetic construct. Phylosignal for decomposition, root diameter, and Brownian motion (e.g., random effect) was plotted, measured, and tested using the ‘phylosignal’ package [45]. Signal depth was assessed using the ‘phylocorrelogram’ command. Localized signals were then identified using Local indices of phylogenetic association (LIPA), or the ‘lipaMoran’ command, which determined sensitivities and local significance by comparing Local Moran’s index values. The behaviors of general phylogenetic signals were assessed with all methods (Cmean, I, Blomberg’s K, K.star, and Pagel’s λ), as seen in Figure A3 and Figure A4, and final statistics were reported with Blomberg’s K and Pagel’s λ.
3. Results
3.1. Phylosignal Associations
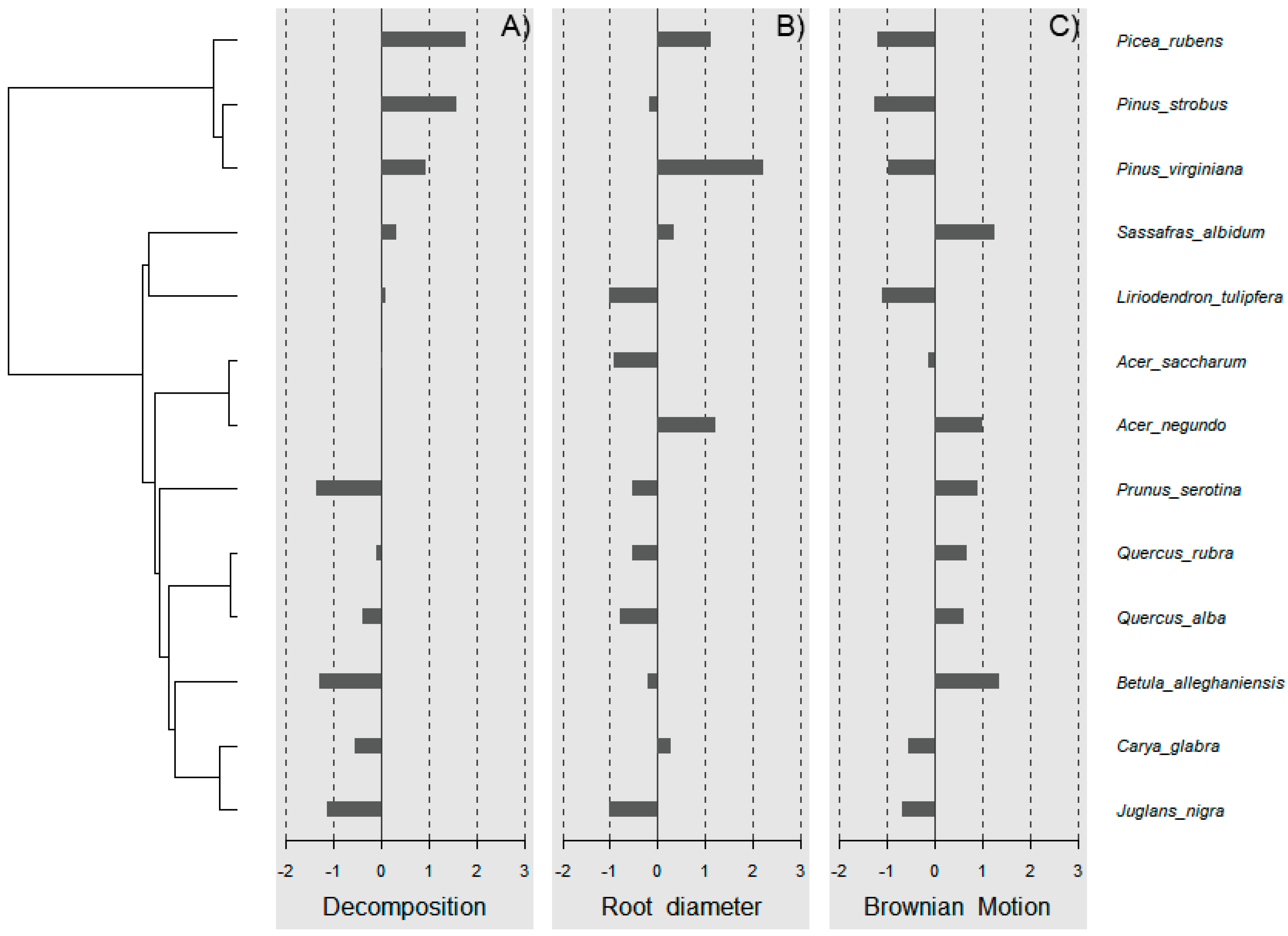
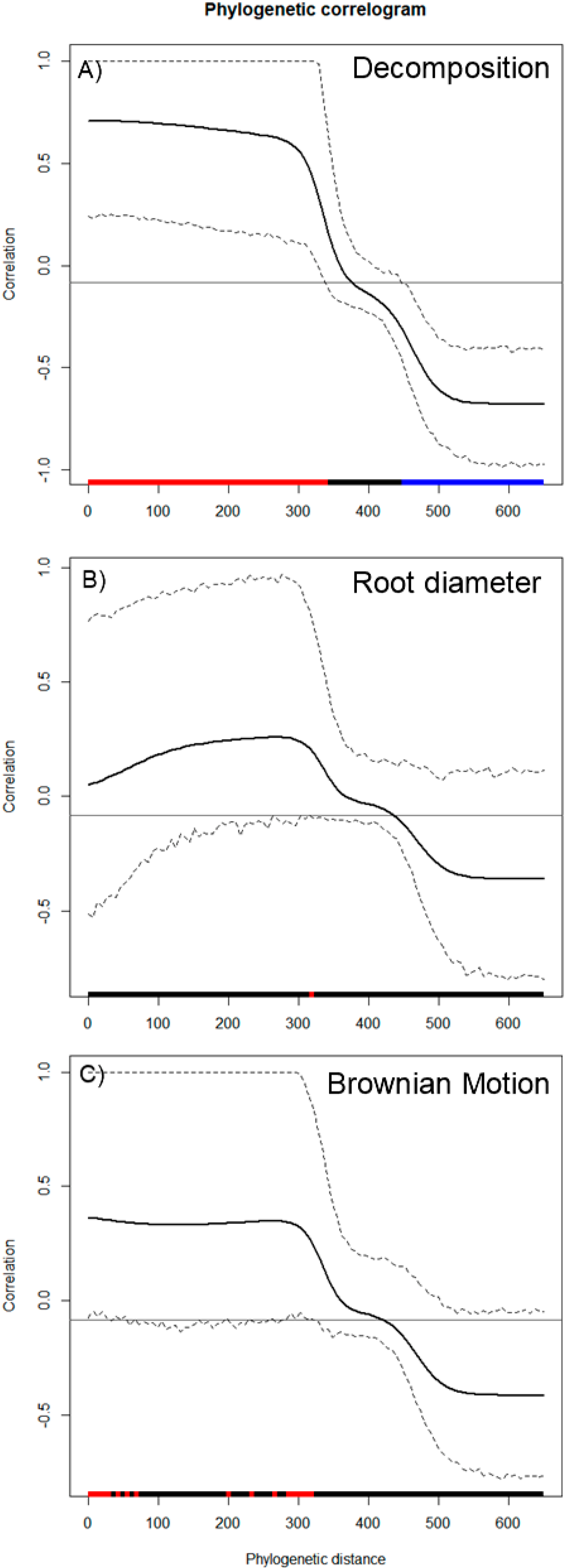
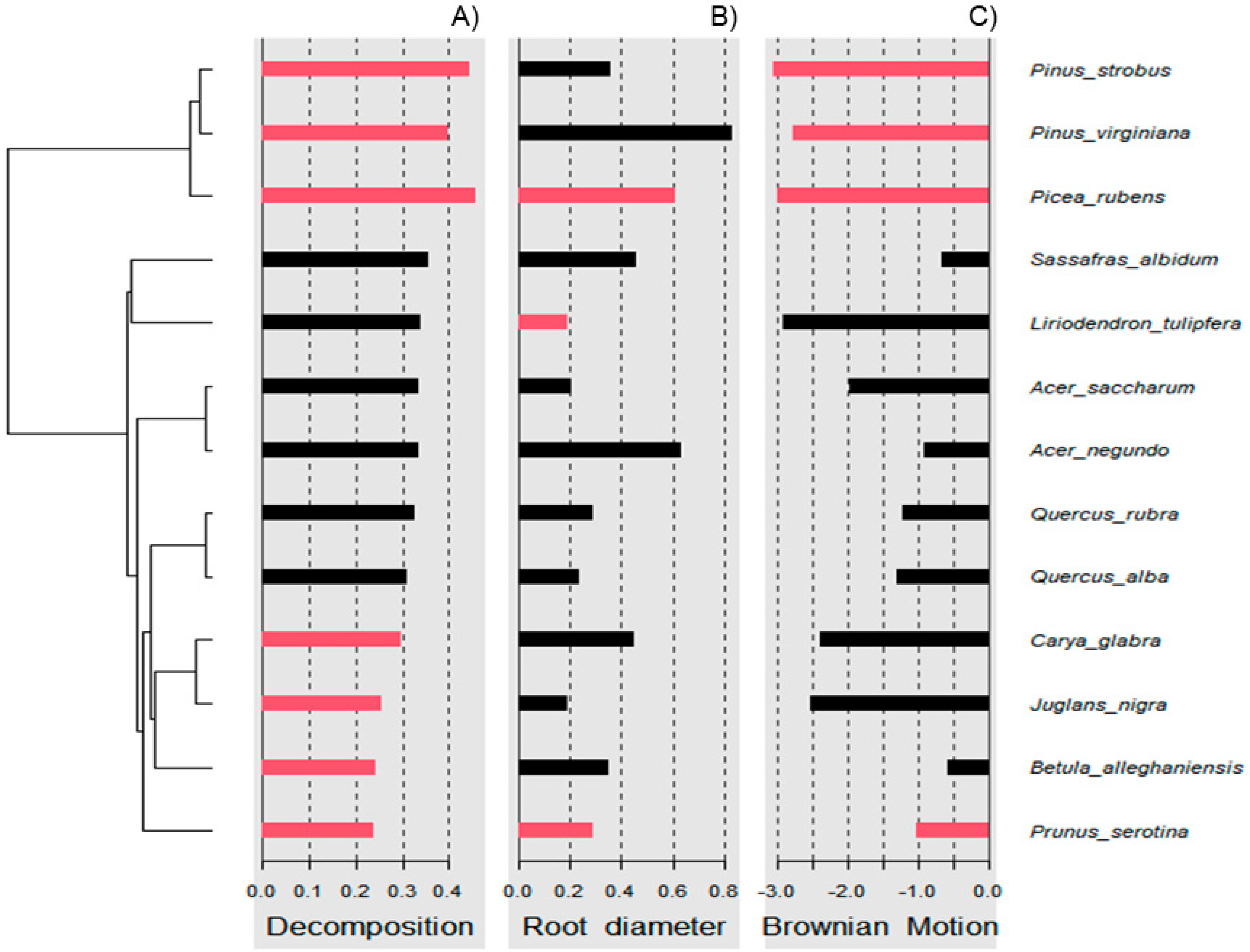
Wood decomposition outcomes were associated with a phylogenetic signal (Figure 2A, General tests for phylogenetic signals, Blomberg’s K = 1.30, Pagel’s λ = 1.00, p = 0.001). The assessment of phylogenetic signal depth via correlograms revealed significant long ranges of positive autocorrelations, and significant long ranges of negative autocorrelation (Figure 3A) for decomposition. However, this was not the case for root diameter and Brownian motion model (Figure 2B,C and Figure 3B,C), as general phylogenetic signals were insignificant for root diameter (Blomberg’s K = 0.16, Pagel’s λ = 0.20, p = 0.46) and the Brownian motion (Blomberg’s K = 0.55, Pagel’s λ = 0.89, p = 0.07). With respect to decomposition, Local indices of phylogenetic association (LIPA) revealed bimodal clustering for local Moran’s index values. These clusters were on opposing ends of the phylogenetic spectrum. On one hand, P. strobus, P. virginiana, and P. rubens had high values of sensitivity (Figure 4A). On the other hand, Carya, Juglans, Betula, and Prunus species had low values of sensitivity (Figure 4A). Local Moran’s index values were sparse for root diameter (Figure 4B).
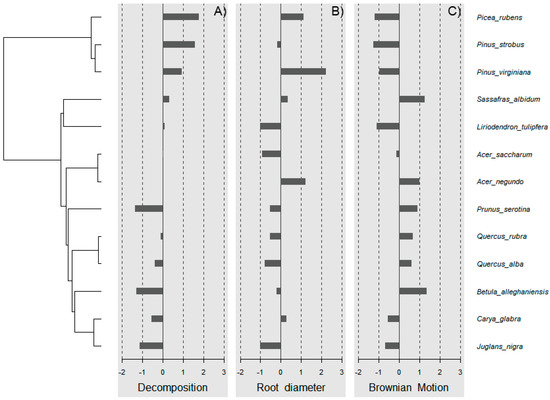
Figure 2.
Testing for general phylogenetic signal. A phylogenetic signal detected for tree-soil-mediated wood decomposition (A) (Blomberg’s K = 1.30, Pagel’s λ = 1.00, p = 0.001). The signal was undetected or insignificant for root diameter (B) and Brownian motion model (C).
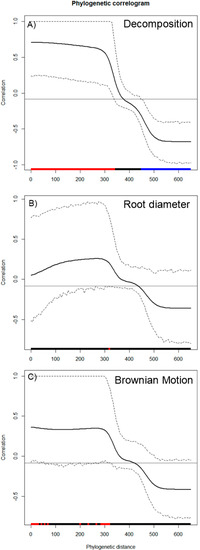
Figure 3.
Phylogenetic correlograms for 3 traits: (A) decomposition, (B) root diameter, and (C) Brownian motion model. Here, correlations (y-axis) are assessed with respect to phylogenetic distance (x-axis). Red bars along the x-axis indicate significant positive autocorrelation, while blue bars along the x-axis indicate significant negative autocorrelation. With respect to the plotted lines, the solid black line represents Moran’s I index of autocorrelation. Hashed lines represent the upper and lower bounds of 95% CI.
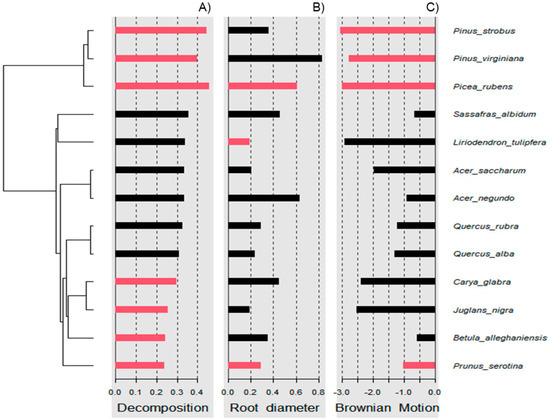
Figure 4.
Local indices of phylogenetic association (LIPA) for (A) decomposition, (B) root diameter, and (C) Brownian motion. Red bars indicate significance with respect to Local Moran’s index. As it relates to decomposition, bimodal clustering was observed. Particularly, high sensitivity values were observed for pine species. Meanwhile, low sensitivity values were observed for species distant to pines (i.e., black cherry). With respect to root diameter, sensitivity values were sparse.
3.2. Root Diameter, Macroinvertebrates or Mycorrhizae as Drivers
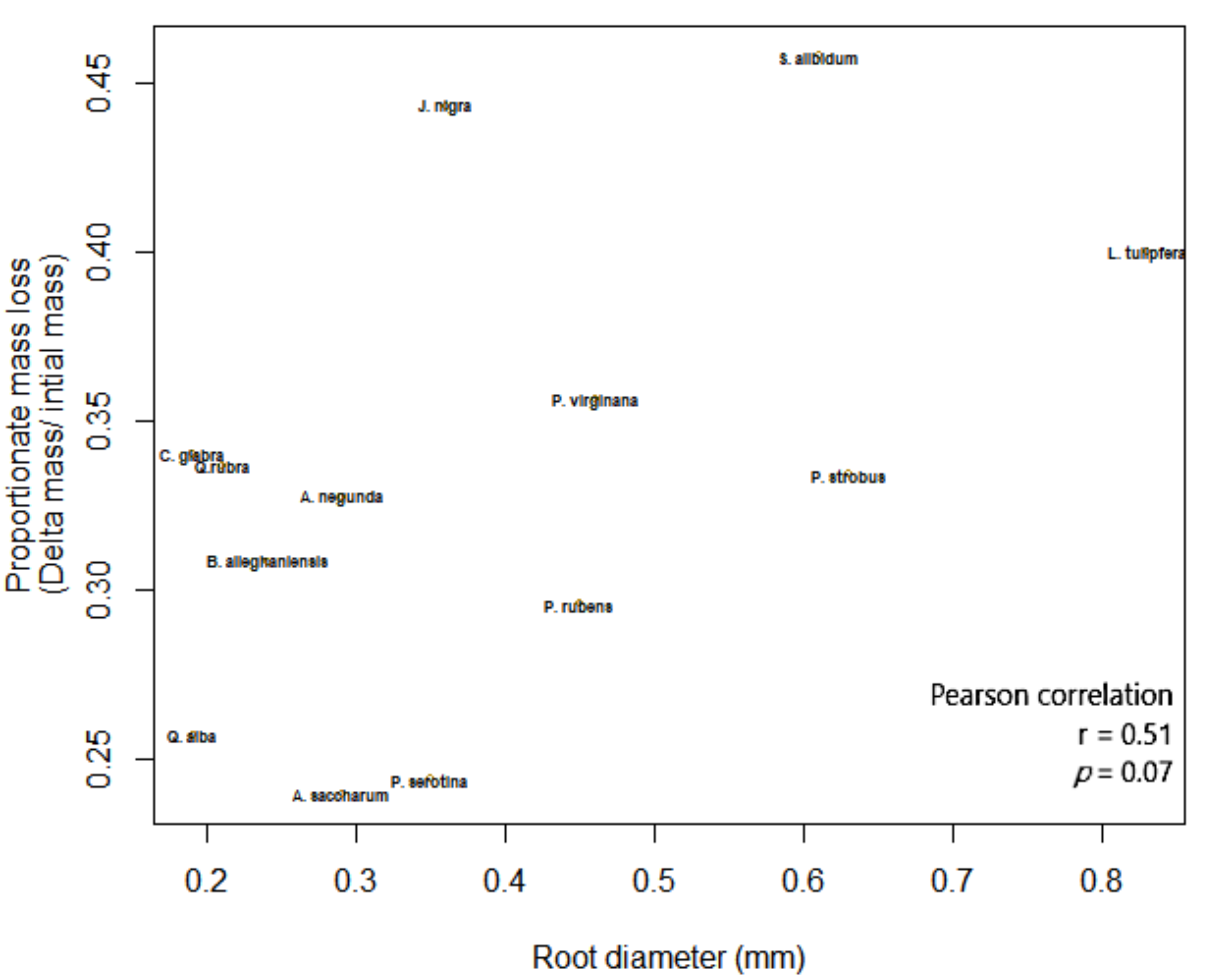
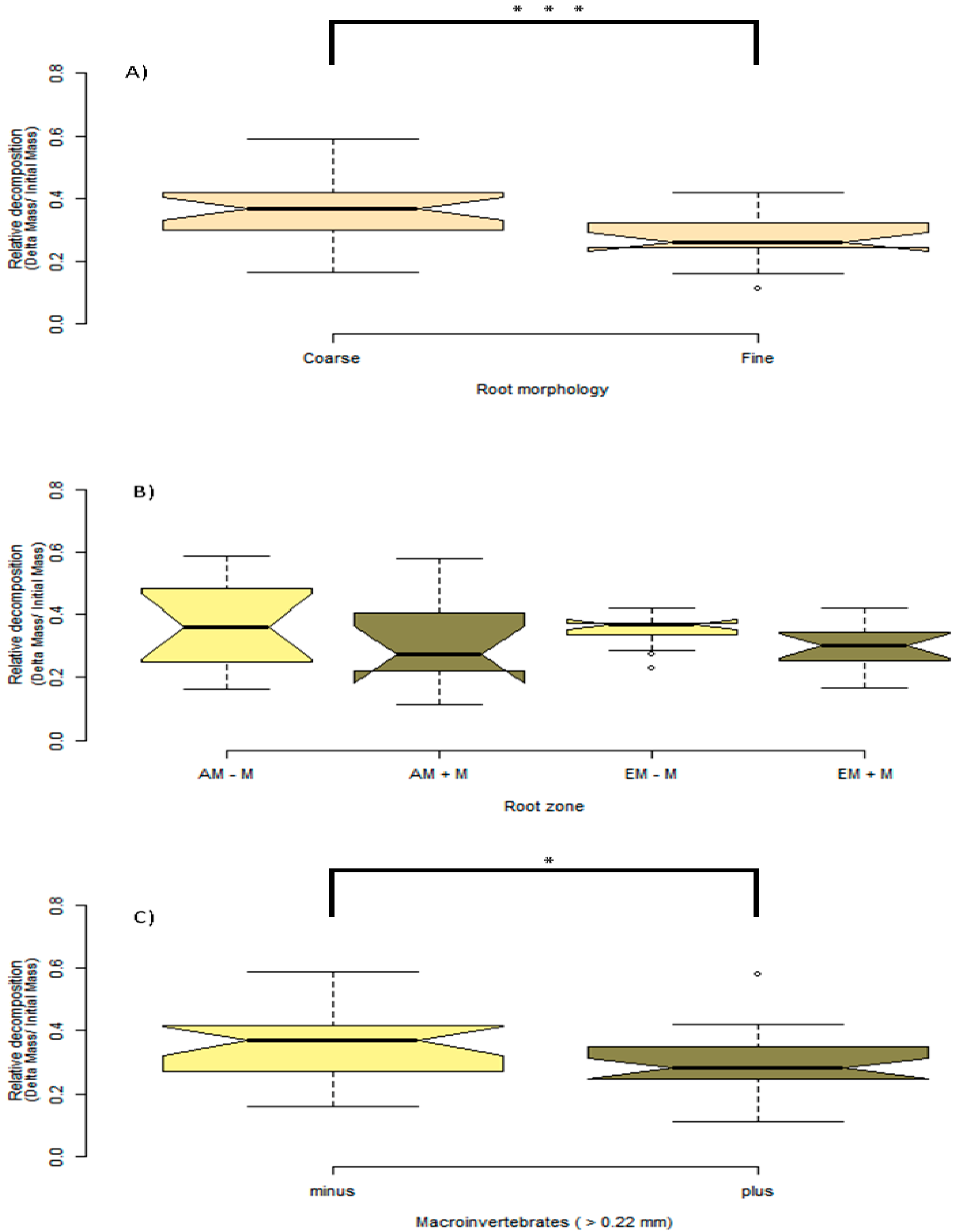
The outcome of macroinvertebrate, mycorrhizal type, and foliar litter trait on wood decomposition was assessed. Wood decomposition was lowest in the root zone of two AM trees, A. saccharum and P. serotina, and highest in the root zone of another pair of AM trees, J. nigra and S. albidum (Figure 5). Foliar litter legacy (broad leaf versus pine needle) did not affect wood decomposition (Table 2), as Step AIC found foliar litter weakened the model. Irrespective of mycorrhizal type, the relationship between root diameter and decomposition was moderately correlated (Figure 5). Specifically, the association between root diameter and wood decomposition was positive and marginally significant (Pearson correlation r = 0.51, n = 13, p = 0.07). Coarse-rooted species were found to increase decomposition by 8% (Figure 6A, four-way ANOVA, F1,38 = 0.348, p = 0.0004). Statistical overlap/non-overlap between EM and AM root zone outcomes were determined with 95% CI ellipses. With the exception of the AM trees, J. nigra, S. albidum, and L. tulipfera, there was a great deal of overlap between AM and EM root diameter effects on decomposition (Figure A5). The AM ellipse suggests a positive relationship between root diameter and decomposition, and the EM ellipse suggests a mildly positive relationship (Figure A5). Mycorrhizal type did not significantly influence decomposition (four-way ANOVA, F1,38 = 0.465, p = 0.499), and the difference in decomposition when in AM versus EM root zone was relatively small (i.e., 1.8%). Interestingly, the absence of macroinvertebrates (−macroinvertebrates) increased wood decomposition by about 5.9% (Figure 6C; four-way ANOVA, F1,38 = 5.10, p = 0.029). When only considering AM root zone, −macroinvertebrate was greater than +macroinvertebrate. Similarly, in the EM root zone, −macroinvertebrate was greater than +macroinvertebrate (Figure 6B). No interaction between mycorrhizal type and macroinvertebrates was detected (four-way ANOVA, F1,44 = 0. 001, p = 0.974), but a trend toward significance was observed when examining the interaction between root morphology and mycorrhizal type (four-way ANOVA, F1,38 = 3.168, p = 0.083).
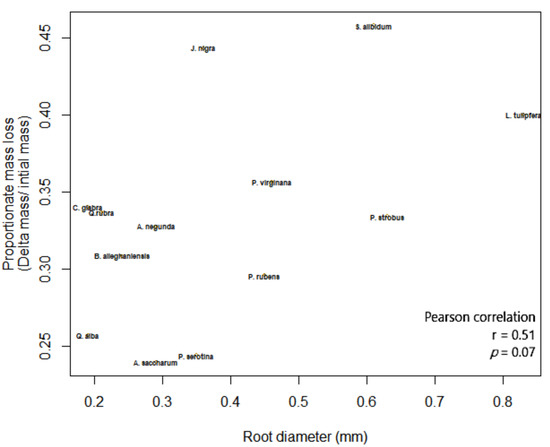
Figure 5.
The association between root diameter (mm) and wood decomposition.

Table 2.
Four-way ANOVA.
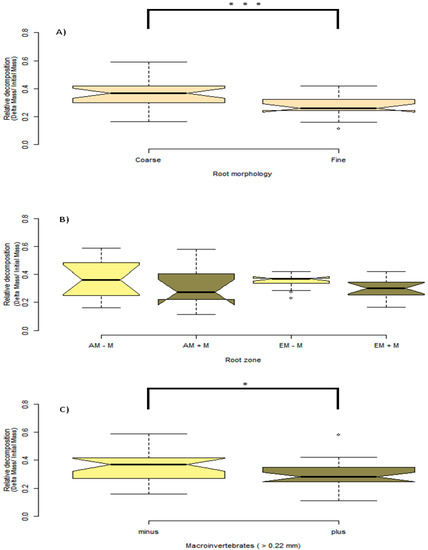
Figure 6.
Provided here are boxplots showing the outcome of experimental factors on wood decomposition. The notches in each boxplot represent 95% CI around the mean. The upper edge and lower edge of each boxplot is the 25% and 75% quartile, or the lower and upper median, which are the boundaries of the interquartile range (IQR). In addition, the boxplot whiskers represent 1.5 × IQR, and points beyond the whiskers are outliers. (A) Seen here are coarse versus fine root species, and coarse roots are defined as roots > 0.35-mm, and fine roots are defined as < 0.35-mm. (B) Root zone and macroinvertebrate effect on decomposition. Light-khaki-colored boxplots represent macroinvertebrate exclusion (−M); dark-khaki-colored boxplots represent non-macroinvertebrate exclusion (+M). (C) The main effect of macroinvertebrate exclusion. Significance: * p < 0.05; *** p < 0.001.
4. Discussion
Wood decay provides a sink for N2O and a source for CO2 and CH4 [73], as well as a source for soil phosphorus [65]. In addition, it is widely accepted that wood decomposes faster in soil communities than in suspension [65,73]. The novelty of the present study is the interactive role of root morphology, mycorrhizal type, and macroinvertebrates on wood degradation in rhizosphere-adjacent soils. Soils are differentially influenced by plant-specific traits (i.e., rhizodeposition and foliage), which in turn helps structure belowground communities [74]. To our knowledge, this is the first study to show an association between phylogenetic distance and wood decomposition. Species that were more closely related to pines had highly sensitive values for local positive auto correlations. Tree mycorrhizal type also has the potential to impact root zone dynamics. Past studies have predicted differences in AM and EM trees in biogeochemical cycles [63,75,76], which may be pertinent to shifts in forest demography [77], which can impact predominant mycorrhizal type [78]. As it relates to this study, mycorrhizal type did not impact wood decomposition (Table 2), which suggests a mixed effect of mycorrhizal type on C cycling that may depend on the species of fungi colonizing the host tree species. To our knowledge, we are the first to examine wood decomposition in root zones of contrasting mycorrhizal association and root morphology. Mild interaction was observed between mycorrhizal type and root diameter (Table 2). Irrespective of mycorrhizal type, root diameter was positively correlated with wood decomposition at a marginal level of significance (Figure 5), as coarse roots significantly increased decomposition (Figure 6A). While coarse roots promoted decomposition, a phylogenetic signal was not detected for this trait. Interestingly, excluding macroinvertebrates from the soil environment led to increased wood degradation (Figure 6C), which corroborates the findings of Wood, Tordoff [20], which suggested that macroinvertebrates can disrupt microbial decomposer involvement in wood decomposition. Taken together, these findings provide insight into wood decomposition as influenced by biodiverse root zones.
4.1. Foliar Trait Legacy and Wood Decomposition
Phylogenetic signals were observed in tree-specific soils (Figure 2). These signals were likely due to root exudates and litter input from the canopy trees. Litter inputs can fuel soil food webs through rhizodeposits, foliage, and surface accumulation [79]. Specifically, plant identity can structure communities of arachnida, nematoda, collembola, enchytraeidae, and mycorrhizae [74]. Together, these communities may help facilitate the outcome of wood decomposition. Lignin is an important component of wood and makes up 20–32% of lignocellulosic biomass and is dramatically resistant to chemical degradation [33,80]. According to our ANOVA model, the decomposition rate beneath pine versus broad leaf canopies was insignificant (Table 2), but these differences may be gradual and amplified over phylogenetic distance (Figure 2A and Figure 3A). This may also explain why local positive autocorrelation was detected on opposite ends of the phylogenetic spectrum (Figure 4A). Sensitivity values waned in species that were phylogenetically distant from Pinus and Picea species, including Carya and Juglans spp. Ironically, Carya glabra and Juglans nigra were previously shown to differ from Pinus strobus and Pinus virginiana in stem wood density [81] by about 45 percent. The tree species used in this study have been structuring the soil community for over 20 years at this particular site, yielding notable changes in some soil characteristics [61]. The effect of tree litter deposits may amplify with an increase in stand age [82], as tree-specific litter has been reported to impact invertebrate richness, diversity, and assemblage [24]. The phylogenetic signal observed in the present study suggests tree species can condition the soil environment in way that may be predictable across species of closely related genera.
4.2. Macroinvertebrate Exclusion Improved Wood Decomposition
Macroinvertebrate exclusion increased wood decomposition (Table 2). This may suggest that macroinvertebrates can disrupt saprotroph mycelial cords and reduce C cycling. On the contrary, there may be a specific context in which macroinvertebrates and fungal decomposers can have an additive effect on wood decomposition, and this may depend on the abundance of macroinvertebrates that specialize in wood (i.e., termites, carpenter ants, etc.). For example, +macroinvertebrate communities that are overrepresented with termites and carpenter ants are likely to increase wood decomposition, as this select class of arthropods can specialize in recalcitrant forms of carbon. However, this experiment was conducted in the root zone, which is rich in labile carbon (i.e., active photosynthates, soluble C, etc.) and root-derived C [83], thereby providing resources to a broad array of organisms. Macroinvertebrate exclusion improved subsurface wood decomposition (Figure 6C), and similar effects have been found for surface wood decomposition, as macroinvertebrate exclusion was reported to alter saprotroph communities [16]. In subtropical forests, wood contact with soil surface, combined with invertebrates, was found to be optimal for wood decomposition [84]. The role of macroinvertebrates may also depend on canopy tree cover (i.e., shading); an increase in ambient radiation has been shown to decrease invertebrate density [47].
4.3. Mycorrhizal Type and Wood Decomposition
Mycorrhizal type has important implications on ecosystem processes, including soil structure, C storage, and N and P cycling [85,86,87]. As it relates to global change biology, predictable demographic shifts in eastern USA deciduous forests [77] can directly lead to shifts in the dominant mycorrhizal type. Mycorrhizal fungi can increase the uptake of soluble nutrients, and in some cases, the mycorrhizal type (e.g., EM fungi) can even lead to the decomposition of organic matter [37]. However, this may depend on the forest system (i.e., boreal versus temperate forest), as EM’s lignocellulolytic enzyme capabilities may not necessarily apply to recalcitrant woody litter, although EM fungi have been found to modify SOM [88,89]. Mycorrhizal type did not influence wood decomposition in this study (Table 2), perhaps suggesting that models predicting the role of mycorrhizae in C and nutrient cycling may not apply to wood-derived C in temperate forests.
4.4. Moderate Correlation between Decomposition and Root Diameter
Our findings suggest wood decomposition may not be impacted by tree demographic shifts affecting mycorrhizal type, but instead, by tree-species shifts affecting absorptive root diameter. Across the root zone of 13 diverse tree species, we found a positive correlation between root diameter and wood decomposition (Figure 5), with most points falling within the 95% CI (Figure A5). Wood decomposition in coarse root zone soils was increased by ~8% (p < 0.001; Figure 6A). Perhaps this may be explained by the “drying effect hypothesis” [42], where fine roots are better able to remove soil residual moisture from soil and suppress decomposer activity. In grasses, root diameter was found to be positively correlated with decomposition and soil organic carbon respiration [43]. Interestingly, a similar trend was found among woody plants [44], perhaps suggesting that increased root diameter facilitates increased C and N cycling. Collectively, these findings show an additive effect of root diameter on decomposition. In addition, we did not observe interaction of root diameter with the macroinvertebrate exclusion barrier (0.22 mm mesh). This suggests that while the barrier may have inhibited a certain amount of root growth around the wood substrate, it did not limit the growth of coarse-root species more than fine-root species. To our knowledge, this is the first study to show a correlation of root diameter with the decomposition of wood debris.
5. Conclusions
Unraveling ways in which biodiverse trees can influence ecosystem processes may provide additional insight into C and nutrient cycling. With respect to the root zone, a phylogenetic signal was observed for decomposition. Pinus and Picea species were most sensitive to decomposition–phylogenetic distance autocorrelations. The presence of macroinvertebrates lessened recalcitrant litter decomposition, perhaps suggesting an antagonistic effect of macroinvertebrates on saprotrophs. Additionally, the finding that mycorrhizal type did not affect wood decomposition suggests neutral outcomes for forest demography shifts, specifically those that affect predominant mycorrhizal type (i.e., AM versus EM). Additionally, results of this study support the premise that woodlands overrepresented by trees of a large root diameter can potentially accelerate the cycling of recalcitrant C.
Author Contributions
R.J.M.: Conceptualization—Lead, Data Curation—Lead, Formal Analysis—Lead, Investigation—Lead, Methodology—Lead, Visualization—Lead, Writing—Original Draft—Lead, Writing—Review and Editing—Lead: M.A.V.B.: Investigation—Supporting; Writing—Review and Editing—Supporting: T.H.B.: Investigation—Supporting; Writing—Review and Editing—Supporting: D.M.E.: Conceptualization—Supporting, Investigation—Supporting, Methodology—Supporting, Resources—Supporting, Writing—Review and Editing—Supporting. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was provided Button Waller Fellowship, National Science Foundation PRFB Award #1907242; NIH-IRACDA (P.I. Dr. Roberto De Guzman, project #2K12GM063651-16A1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is to be made available upon request.
Acknowledgments
Special thanks to Charles David Ray and D. Christopher Rogers for technical support, and James D. Bever for helpful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Figure A1.
Seen here are cylindrical core mesh tubes with 50 mm × 50 mm window openings. At the base of the cylindrical core are a pair of wood cubes derived from Acer rubrum. The pair is composed of one cube enclosed in a polyester mesh (–M), while the adjacent pair is void of polyester mesh +M.
Figure A1.
Seen here are cylindrical core mesh tubes with 50 mm × 50 mm window openings. At the base of the cylindrical core are a pair of wood cubes derived from Acer rubrum. The pair is composed of one cube enclosed in a polyester mesh (–M), while the adjacent pair is void of polyester mesh +M.


Figure A2.
Depicted here is a burial site; cylindrical core was placed vertically into the burial site. The burial and cylindrical core was then filled in with native soil, and wood substrate was allowed to decompose.
Figure A2.
Depicted here is a burial site; cylindrical core was placed vertically into the burial site. The burial and cylindrical core was then filled in with native soil, and wood substrate was allowed to decompose.

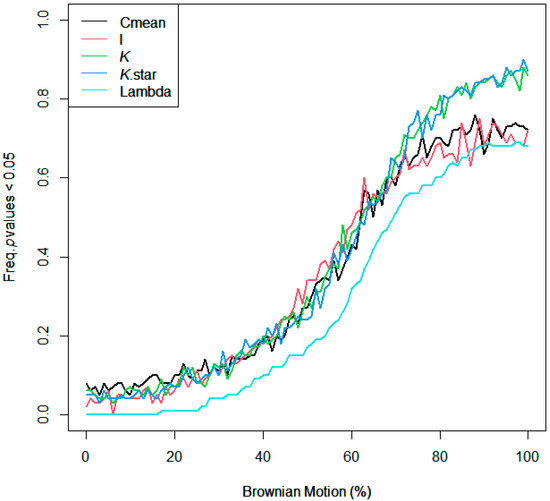
Figure A3.
Assessing the behavior of phylogenetic signal via Brownian motion influence gradient.
Figure A3.
Assessing the behavior of phylogenetic signal via Brownian motion influence gradient.
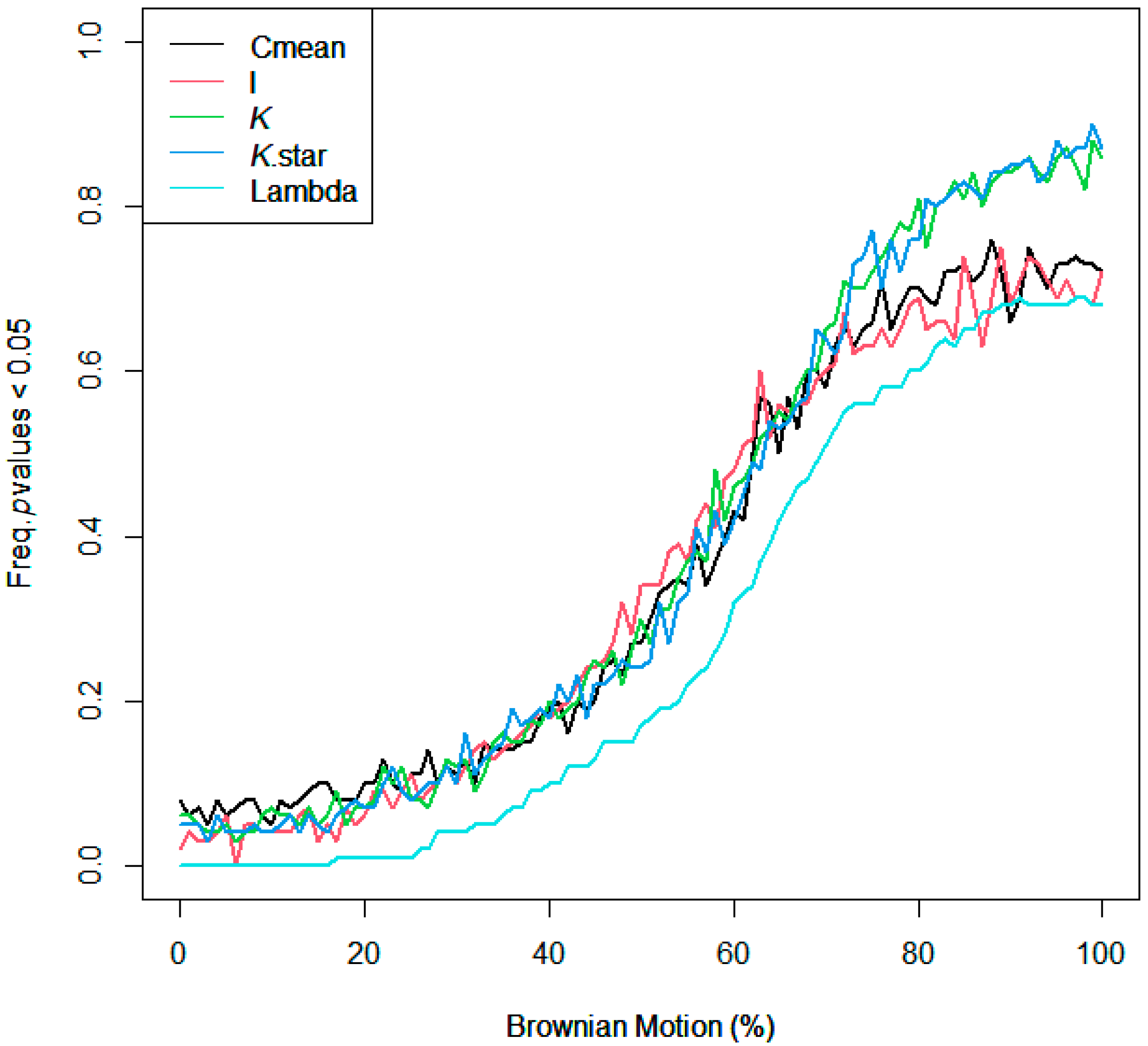
Figure A4.
Assessing the behavior of phylogenetic signal via Brownian motion influence gradient and 95% CI.
Figure A4.
Assessing the behavior of phylogenetic signal via Brownian motion influence gradient and 95% CI.
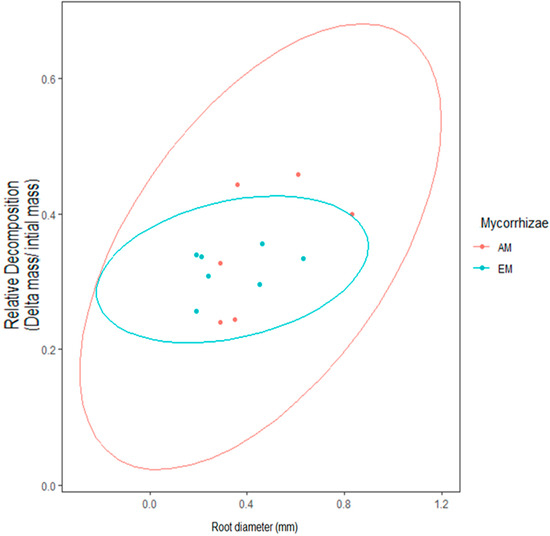
Figure A5.
Observed here are 95% CI ellipses in the context of mycorrhizal type. AM root zones had a broader range in decomposition outcomes in comparison to EM root zones.
Figure A5.
Observed here are 95% CI ellipses in the context of mycorrhizal type. AM root zones had a broader range in decomposition outcomes in comparison to EM root zones.
References
- Potter, C.S.; Randerson, J.T.; Field, C.B.; Matson, P.A.; Vitousek, P.M.; Mooney, H.A.; Klooster, S.A. Terrestrial ecosystem production: A process model based on global satellite and surface data. Glob. Biogeochem. Cycles 1993, 7, 811–841. [Google Scholar] [CrossRef]
- Šímová, I.; Sandel, B.; Enquist, B.J.; Michaletz, S.T.; Kattge, J.; Violle, C.; McGill, B.J.; Blonder, B.; Engemann, K.; Peet, R.K.; et al. The relationship of woody plant size and leaf nutrient content to large-scale productivity for forests across the Americas. J. Ecol. 2019, 107, 2278–2290. [Google Scholar] [CrossRef]
- Weedon, J.T.; Cornwell, W.K.; Cornelissen, J.H.; Zanne, A.E.; Wirth, C.; Coomes, D.A. Global meta-analysis of wood decomposition rates: A role for trait variation among tree species? Ecol. Lett. 2009, 12, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Malhi, Y. Carbon in the atmosphere and terrestrial biosphere in the 21st century. Philos. Trans. R. Soc. Lond. Ser. A Math. Phys. Eng. Sci. 2002, 360, 2925–2945. [Google Scholar] [CrossRef]
- Hu, B.; Flemetakis, E.; Rennenberg, H. Pedospheric Microbial Nitric Oxide Production Challenges Root Symbioses. Trends Plant Sci. 2020, 26, 104–107. [Google Scholar] [CrossRef]
- Poirier, V.; Roumet, C.; Munson, A.D. The root of the matter: Linking root traits and soil organic matter stabilization processes. Soil Biol. Biochem. 2018, 120, 246–259. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, H.; Li, S.; Fu, X.; Dai, X.; Yan, H.; Kou, L. Mycorrhizal and environmental controls over root trait–decomposition linkage of woody trees. New Phytol. 2021, 229, 284–295. [Google Scholar] [CrossRef]
- Harmon, M.E. Logs as Sites of Tree Regeneration in Picea Sitchensis-Tsuga Heterophylla Forests of Coastal Washington and Oregon. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, 1986. [Google Scholar]
- Christy, E.J.; Mack, R.N. Variation in demography of juvenile Tsuga heterophylla across the substratum mosaic. J. Ecol. 1984, 72, 75–91. [Google Scholar] [CrossRef]
- McKee, A.; LaRoi, G.; Franklin, J.F. Structure, composition, and reproductive behavior of terrace forests, South Fork Hoh River, Olympic National Park. Ecol. Res. Natl. Parks Pac. Northwest 1982, 22–29. [Google Scholar]
- Harmon, M.E. Effects of bark fragmentation on plant succession on conifer logs in the Picea-Tsuga forests of Olympic National Park, Washington. Am. Midl. Nat. 1989, 121, 112–124. [Google Scholar] [CrossRef]
- Cotrufo, M.F.; Wallenstein, M.D.; Boot, C.M.; Denef, K.; Paul, E. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Change Biol. 2013, 19, 988–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, D.C.; Callaham, M.A.; Crossley, D., Jr. Fundamentals of Soil Ecology; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Stokland, J.N.; Siitonen, J.; Jonsson, B.G. Biodiversity in Dead Wood; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Ulyshen, M.D. Wood decomposition as influenced by invertebrates. Biol. Rev. 2016, 91, 70–85. [Google Scholar] [CrossRef]
- Jacobsen, R.M.; Sverdrup-Thygeson, A.; Kauserud, H.; Mundra, S.; Birkemoe, T. Exclusion of invertebrates influences saprotrophic fungal community and wood decay rate in an experimental field study. Funct. Ecol. 2018, 32, 2571–2582. [Google Scholar] [CrossRef]
- König, H. Bacillus species in the intestine of termites and other soil invertebrates. J. Appl. Microbiol. 2006, 101, 620–627. [Google Scholar] [CrossRef]
- Griffiths, B.; Bracewell, J.; Robertson, G.; Bignell, D. Pyrolysis-mass spectrometry confirms enrichment of lignin in the faeces of a wood-feeding termite, Zootermopsis nevadensis and depletion of peptides in a soil-feeder, Cubitermes ugandensis. Soil Biol. Biochem. 2013, 57, 957–959. [Google Scholar] [CrossRef]
- Jonas, J.L.; Wilson, G.W.; White, P.M.; Joern, A. Consumption of mycorrhizal and saprophytic fungi by Collembola in grassland soils. Soil Biol. Biochem. 2007, 39, 2594–2602. [Google Scholar] [CrossRef]
- Wood, J.; Tordoff, G.M.; Jones, T.H.; Boddy, L. Reorganization of mycelial networks of Phanerochaete velutina in response to new woody resources and collembola (Folsomia candida) grazing. Mycol. Res. 2006, 110, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Lustenhouwer, N.; Maynard, D.S.; Bradford, M.A.; Lindner, D.L.; Oberle, B.; Zanne, A.E.; Crowther, T.W. A trait-based understanding of wood decomposition by fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 11551–11558. [Google Scholar] [CrossRef]
- Rubino, D.L.; McCarthy, B.C. Composition and ecology of macrofungal and myxomycete communities on oak woody debris in a mixed-oak forest of Ohio. Can. J. For. Res. 2003, 33, 2151–2163. [Google Scholar] [CrossRef]
- Yuan, J.; Zheng, X.; Cheng, F.; Zhu, X.; Hou, L.; Li, J.; Zhang, S. Fungal community structure of fallen pine and oak wood at different stages of decomposition in the Qinling Mountains, China. Sci. Rep. 2017, 7, 13866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanowski, C.K.; Horodecki, P.; Kamczyc, J.; Skorupski, M.; Jagodziński, A.M. Does litter decomposition affect mite communities (Acari, Mesostigmata)? A five-year litterbag experiment with 14 tree species in mixed forest stands growing on a post-industrial area. Geoderma 2021, 391, 114963. [Google Scholar] [CrossRef]
- Hansen, R.A. Red oak litter promotes a microarthropod functional group that accelerates its decomposition. Plant Soil 1999, 209, 37–45. [Google Scholar] [CrossRef]
- Hansen, R.A. Effects of habitat complexity and composition on a diverse litter microarthropod assemblage. Ecology 2000, 81, 1120–1132. [Google Scholar] [CrossRef]
- Buresova, A.; Tejnecky, V.; Kopecky, J.; Drabek, O.; Madrova, P.; Rerichova, N.; Omelka, M.; Krizova, P.; Nemecek, K.; Parr, T.B. Litter chemical quality and bacterial community structure influenced decomposition in acidic forest soil. Eur. J. Soil Biol. 2021, 103, 103271. [Google Scholar] [CrossRef]
- Cresser, M.; Killham, K.; Edwards, T. Soil Chemistry and Its Applications; Cambridge University Press: Cambridge, UK, 1993; Volume 5. [Google Scholar]
- Keiluweit, M.; Nico, P.; Harmon, M.E.; Mao, J.; Pett-Ridge, J.; Kleber, M. Long-term litter decomposition controlled by manganese redox cycling. Proc. Natl. Acad. Sci. USA 2015, 112, E5253–E5260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, N.C.; Weil, R.R. The Nature and Properties of Soils; Prentice Hall: Upper Saddle River, NJ, USA, 2008; Volume 13. [Google Scholar]
- Fukasawa, Y.; Takahashi, K.; Arikawa, T.; Hattori, T.; Maekawa, N. Fungal wood decomposer activities influence community structures of myxomycetes and bryophytes on coarse woody debris. Fungal Ecol. 2015, 14, 44–52. [Google Scholar] [CrossRef]
- Blanchette, R.A. Screening wood decayed by white rot fungi for preferential lignin degradation. Appl. Environ. Microbiol. 1984, 48, 647–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Zeng, G.; Tan, Z.; Jiang, M.; Li, H.; Liu, L.; Zhu, Y.; Yu, Z.; Wei, Z.; Liu, Y.; et al. Understanding Lignin-Degrading Reactions of Ligninolytic Enzymes: Binding Affinity and Interactional Profile. PLoS ONE 2011, 6, e25647. [Google Scholar] [CrossRef] [Green Version]
- Tien, M.; Kirk, T.K. Lignin-degrading enzyme from Phanerochaete chrysosporium: Purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc. Natl. Acad. Sci. USA 1984, 81, 2280–2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arantes, V.; Jellison, J.; Goodell, B. Peculiarities of brown-rot fungi and biochemical Fenton reaction with regard to their potential as a model for bioprocessing biomass. Appl. Microbiol. Biotechnol. 2012, 94, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Rineau, F.; Roth, D.; Shah, F.; Smits, M.; Johansson, T.; Canbäck, B.; Olsen, P.; Persson, P.; Grell, M.; Lindquist, E.; et al. The ectomycorrhizal fungus Paxillus involutus converts organic matter in plant litter using a trimmed brown-rot mechanism involving Fenton chemistry. Environ. Microbiol. 2012, 14, 1477–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindahl, B.D.; Tunlid, A. Ectomycorrhizal fungi—Potential organic matter decomposers, yet not saprotrophs. New Phytol. 2015, 205, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Koide, R.T.; Sharda, J.N.; Herr, J.R.; Malcolm, G.M. Ectomycorrhizal fungi and the biotrophy–saprotrophy continuum. New Phytol. 2008, 178, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Pellitier, P.T.; Zak, D.R. Ectomycorrhizal fungi and the enzymatic liberation of nitrogen from soil organic matter: Why evolutionary history matters. New Phytol. 2018, 217, 68–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leifheit, E.; Verbruggen, E.; Rillig, M. Arbuscular mycorrhizal fungi reduce decomposition of woody plant litter while increasing soil aggregation. Soil Biol. Biochem. 2015, 81, 323–328. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Zhao, L.; Zhang, J.; Xu, W. Dynamics of arbuscular mycorrhizal fungi and glomalin under Psammochloa villosa along a typical dune in desert, North China. Symbiosis 2017, 73, 145–153. [Google Scholar] [CrossRef]
- Cheng, W.; Kuzyakov, Y. Root effects on soil organic matter decomposition. In Roots and Soil Management: Interactions between Roots and the Soil; Wiley: Hoboken, NJ, USA, 2005; Volume 48, pp. 119–143. [Google Scholar]
- de Graaff, M.-A.; Six, J.; Jastrow, J.D.; Schadt, C.W.; Wullschleger, S.D. Variation in root architecture among switchgrass cultivars impacts root decomposition rates. Soil Biol. Biochem. 2013, 58, 198–206. [Google Scholar] [CrossRef]
- Mao, R.; Zeng, D.-H.; Li, L.-J. Fresh root decomposition pattern of two contrasting tree species from temperate agroforestry systems: Effects of root diameter and nitrogen enrichment of soil. Plant Soil 2011, 347, 115–123. [Google Scholar] [CrossRef]
- Keck, F.; Rimet, F.; Bouchez, A.; Franc, A. phylosignal: An R package to measure, test, and explore the phylogenetic signal. Ecol. Evol. 2016, 6, 2774–2780. [Google Scholar] [CrossRef] [PubMed]
- Losos, J.B. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 2008, 11, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Møller, A.; Mousseau, T. Reduced colonization by soil invertebrates to irradiated decomposing wood in Chernobyl. Sci. Total Environ. 2018, 645, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Cheesman, A.W.; Cernusak, L.A.; Zanne, A.E. Relative roles of termites and saprotrophic microbes as drivers of wood decay: A wood block test. Austral Ecol. 2018, 43, 257–267. [Google Scholar] [CrossRef]
- Ulyshen, M.D.; Müller, J.; Seibold, S. Bark coverage and insects influence wood decomposition: Direct and indirect effects. Appl. Soil Ecol. 2016, 105, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Torres, J.A. Wood decomposition of Cyrilla racemiflora in a tropical montane forest. Biotropica 1994, 26, 124–140. [Google Scholar] [CrossRef]
- Ausmus, B. Regulation of wood decomposition rates by arthropod and annelid populations. Ecol. Bull. 1977, 25, 180–192. [Google Scholar]
- National Climatic Data Center. NOAA’s 1981–2010 Climate Normals; National Climatic Data Center: Asheville, NC, USA, 2012. [Google Scholar]
- Malik, R.J. No “Gadgil effect”: Temperate tree roots and soil lithology are effective predictors of wood decomposition. For. Pathol. 2019, 49, e12506. [Google Scholar] [CrossRef]
- Chen, W.; Koide, R.T.; Adams, T.S.; DeForest, J.L.; Cheng, L.; Eissenstat, D.M. Root morphology and mycorrhizal symbioses together shape nutrient foraging strategies of temperate trees. Proc. Natl. Acad. Sci. USA 2016, 113, 8741–8746. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Chen, W.; Adams, T.S.; Wei, X.; Li, L.; McCormack, M.L.; DeForest, J.L.; Koide, R.T.; Eissenstat, D.M. Mycorrhizal fungi and roots are complementary in foraging within nutrient patches. Ecology 2016, 97, 2815–2823. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.; Kao-Kniffin, J.; Frey, S.D.; Fahey, T.; Wickings, K. Soil macroinvertebrate presence alters microbial community composition and activity in the rhizosphere. Front. Microbiol. 2019, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.; Tordoff, G.M.; Black, H.; Cook, R.; Eggers, T.; Garnett, M.; Grayston, S.; Hutcheson, K.; Ineson, P.; Newington, J. Carbon dynamics in a model grassland with functionally different soil communities. Funct. Ecol. 2007, 21, 690–697. [Google Scholar] [CrossRef]
- Swift, M.J.; Heal, O.W.; Anderson, J.M.; Anderson, J. Decomposition in Terrestrial Ecosystems; Blackwell Scientific Publications: Oxford, UK, 1979. [Google Scholar]
- Beare, M.; Coleman, D.; Crossley, D.; Hendrix, P.; Odum, E. A hierarchical approach to evaluating the significance of soil biodiversity to biogeochemical cycling. In The Significance and Regulation of Soil Biodiversity; Springer: Berlin/Heidelberg, Germany, 1995; pp. 5–22. [Google Scholar]
- Bonkowski, M.; Clarholm, M. Stimulation of plant growth through interactions of bacteria and protozoa: Testing the auxiliary microbial loop hypothesis. Acta Protozool. 2012, 51, 237–247. [Google Scholar]
- Yates, C.F.; Guo, J.; Bell, T.H.; Fleishman, S.M.; Bock, H.W.; Trexler, R.V.; Eissenstat, D.M.; Centinari, M. Tree-induced alterations to soil properties and rhizoplane-associated bacteria following 23 years in a common garden. Plant Soil 2021, 461, 591–602. [Google Scholar] [CrossRef]
- Bennett, J.A.; Maherali, H.; Reinhart, K.O.; Lekberg, Y.; Hart, M.M.; Klironomos, J. Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 2017, 355, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Craig, M.E.; Turner, B.L.; Liang, C.; Clay, K.; Johnson, D.J.; Phillips, R.P. Tree mycorrhizal type predicts within-site variability in the storage and distribution of soil organic matter. Glob. Change Biol. 2018, 24, 3317–3330. [Google Scholar] [CrossRef]
- Liu, B.; Li, H.; Zhu, B.; Koide, R.T.; Eissenstat, D.M.; Guo, D. Complementarity in nutrient foraging strategies of absorptive fine roots and arbuscular mycorrhizal fungi across 14 coexisting subtropical tree species. New Phytol. 2015, 208, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Romero, L.M.; Smith, T.J.; Fourqurean, J.W. Changes in mass and nutrient content of wood during decomposition in a south Florida mangrove forest. J. Ecol. 2005, 93, 618–631. [Google Scholar] [CrossRef]
- Malik, R.J.; Trexler, R.V.; Eissenstat, D.M.; Bell, T.H. Bark decomposition in white oak soil outperforms eastern hemlock soil, while bark type leads to consistent changes in soil microbial composition. Biogeochemistry 2020, 150, 329–343. [Google Scholar] [CrossRef]
- Posada, R.H.; Madriñan, S.; Rivera, E.-L. Relationships between the litter colonization by saprotrophic and arbuscular mycorrhizal fungi with depth in a tropical forest. Fungal Biol. 2012, 116, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Melillo, J.M.; Aber, J.D.; Muratore, J.F. Nitrogen and lignin control of hardwood leaf litter decomposition dynamics. Ecology 1982, 63, 621–626. [Google Scholar] [CrossRef]
- Pointing, S.B.; Parungao, M.M.; Hyde, K.D. Production of wood-decay enzymes, mass loss and lignin solubilization in wood by tropical Xylariaceae. Mycol. Res. 2003, 107, 231–235. [Google Scholar] [CrossRef]
- Harrell, F.E., Jr.; Harrell, M.F.E., Jr. Package ‘hmisc’. In CRAN2018; 2019; Volume 2019, pp. 235–236. Available online: https://hbiostat.org/R/Hmisc/ (accessed on 1 February 2021).
- Jin, Y.; Qian, H.V. PhyloMaker: An R package that can generate very large phylogenies for vascular plants. Ecography 2019, 42, 1353–1359. [Google Scholar] [CrossRef] [Green Version]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Oberle, B.; Covey, K.R.; Dunham, K.M.; Hernandez, E.J.; Walton, M.L.; Young, D.F.; Zanne, A.E. Dissecting the effects of diameter on wood decay emphasizes the importance of cross-stem conductivity in Fraxinus americana. Ecosystems 2018, 21, 85–97. [Google Scholar] [CrossRef]
- Bezemer, T.M.; Fountain, M.; Barea, J.; Christensen, S.; Dekker, S.; Duyts, H.; Van Hal, R.; Harvey, J.; Hedlund, K.; Maraun, M. Divergent composition but similar function of soil food webs of individual plants: Plant species and community effects. Ecology 2010, 91, 3027–3036. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Dijkstra, F.A.; Phillips, R.P.; Zhu, B.; Wang, P.; Cheng, W. Arbuscular mycorrhizal trees cause a higher carbon to nitrogen ratio of soil organic matter decomposition via rhizosphere priming than ectomycorrhizal trees. Soil Biol. Biochem. 2021, 157, 108246. [Google Scholar] [CrossRef]
- Phillips, R.P.; Brzostek, E.; Midgley, M.G. The mycorrhizal-associated nutrient economy: A new framework for predicting carbon–nutrient couplings in temperate forests. New Phytol. 2013, 199, 41–51. [Google Scholar] [CrossRef]
- Au, T.F.; Maxwell, J.T.; Novick, K.A.; Robeson, S.M.; Warner, S.M.; Lockwood, B.R.; Phillips, R.P.; Harley, G.L.; Telewski, F.W.; Therrell, M.D. Demographic shifts in eastern US forests increase the impact of late-season drought on forest growth. Ecography 2020, 43, 1475–1486. [Google Scholar] [CrossRef]
- Steidinger, B.S.; Bhatnagar, J.M.; Vilgalys, R.; Taylor, J.W.; Qin, C.; Zhu, K.; Bruns, T.D.; Peay, K.G. Ectomycorrhizal fungal diversity predicted to substantially decline due to climate changes in North American Pinaceae forests. J. Biogeogr. 2020, 47, 772–782. [Google Scholar] [CrossRef]
- Neher, D.A.; Barbercheck, M.E. Soil microarthropods and soil health: Intersection of decomposition and pest suppression in agroecosystems. Insects 2019, 10, 414. [Google Scholar] [CrossRef] [Green Version]
- Munk, L.; Sitarz, A.K.; Kalyani, D.C.; Mikkelsen, D.J.; Meyer, A.S. Can laccases catalyze bond cleavage in lignin? Biotechnol. Adv. 2015, 33, 13–24. [Google Scholar] [CrossRef]
- McCormack, M.L.; Adams, T.S.; Smithwick, E.A.; Eissenstat, D.M. Predicting fine root lifespan from plant functional traits in temperate trees. New Phytol. 2012, 195, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yesilonis, I.; Szlavecz, K. Microbial and environmental controls on wood decomposition in deciduous forests of different ages. Appl. Soil Ecol. 2021, 166, 103986. [Google Scholar] [CrossRef]
- Keller, A.B.; Brzostek, E.R.; Craig, M.E.; Fisher, J.B.; Phillips, R.P. Root-derived inputs are major contributors to soil carbon in temperate forests, but vary by mycorrhizal type. Ecol. Lett. 2021, 24, 626–635. [Google Scholar] [CrossRef]
- Wu, C.; Ulyshen, M.D.; Shu, C.; Zhang, Z.; Zhang, Y.; Liu, Y.; Geoff Wang, G. Stronger effects of termites than microbes on wood decomposition in a subtropical forest. For. Ecol. Manag. 2021, 493, 119263. [Google Scholar] [CrossRef]
- Johnson, N.C.; Gehring, C.; Jansa, J. Mycorrhizal Mediation of Soil: Fertility, Structure, and Carbon Storage; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Shen, Y.; Zhu, B. Arbuscular mycorrhizal fungi reduce soil nitrous oxide emission. Geoderma 2021, 402, 115179. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Hartley, I.P.; Dungait, J.A.; Wen, X.; Li, D.; Guo, Z.; Quine, T.A. Contrasting rhizosphere soil nutrient economy of plants associated with arbuscular mycorrhizal and ectomycorrhizal fungi in karst forests. Plant Soil 2021, 470, 81–93. [Google Scholar] [CrossRef]
- Shah, F.; Nicolás, C.; Bentzer, J.; Ellström, M.; Smits, M.; Rineau, F.; Canbäck, B.; Floudas, D.; Carleer, R.; Lackner, G. Ectomycorrhizal fungi decompose soil organic matter using oxidative mechanisms adapted from saprotrophic ancestors. New Phytol. 2016, 209, 1705–1719. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.D. Mycorrhizal fungi as mediators of soil organic matter dynamics. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 237–259. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).