Genome-Wide Identification and Expression Analysis of MYC Transcription Factor Family Genes in Rubber Tree (Hevea brasiliensis Muell. Arg.)
Abstract
1. Introduction
2. Methods
2.1. Plant Materials
2.2. Analyses of Tissue-Specific Expression
2.3. Coronatine (COR) Treatment and Cambium Tissue Isolation
2.4. Tapping Treatment and Latex Collection
2.5. Total RNA Isolation and cDNA Synthesis
2.6. Amplification of the Open Reading Frame (ORF)
2.7. Conserved Domain Analysis and Phylogenetic Tree
2.8. Real-Time PCR Analysis
2.9. Statistic Analysis of Real-Time PCR Analysis Data
3. Results
3.1. Identification of HblMYC Gene Family
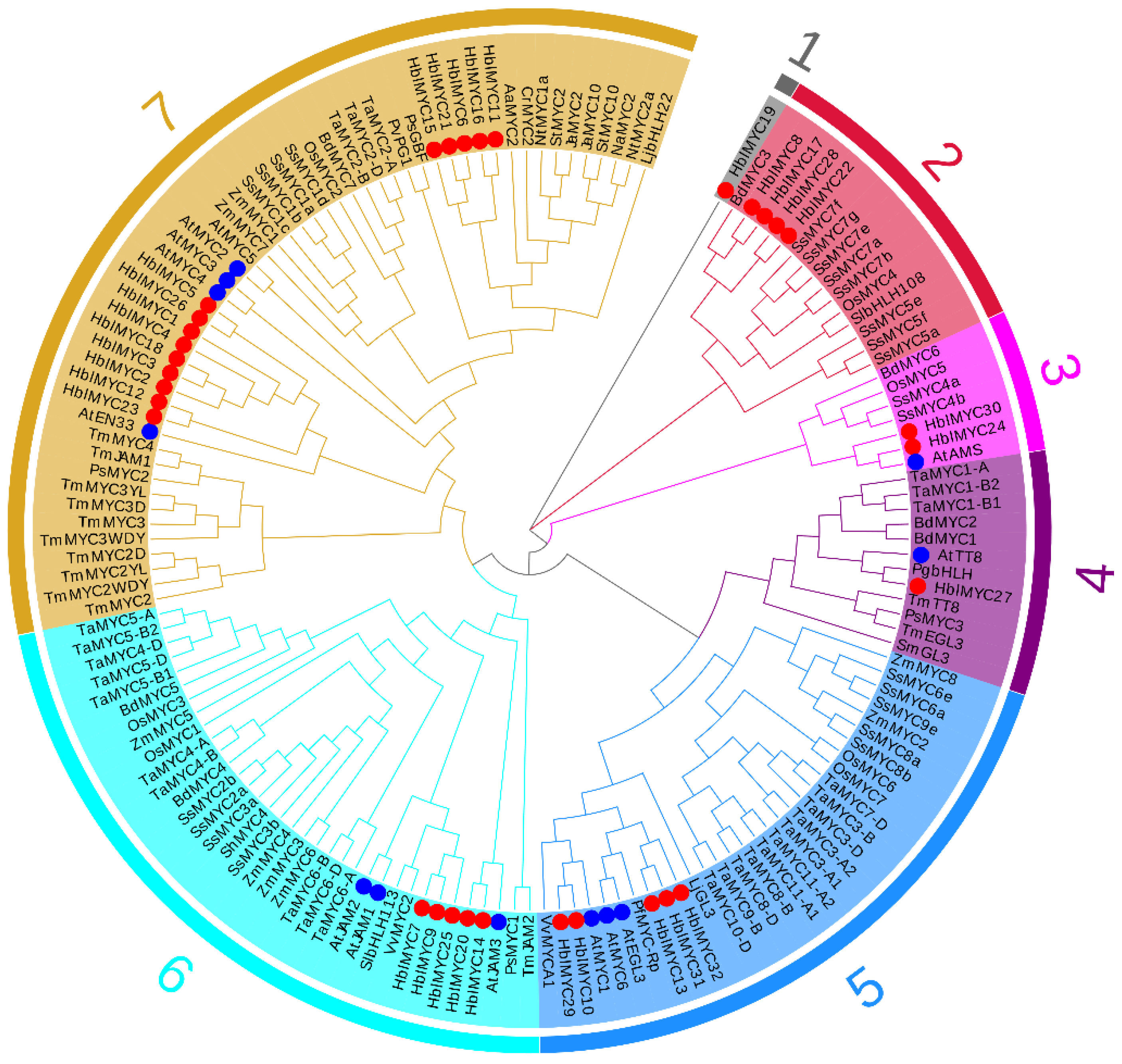
3.2. Phylogenetic Tree and Gene Structures of HblMYCs
3.3. Gene Structures and Conserved Motifs of HblMYCs
3.4. Promoter Analysis of the HblMYC Genes
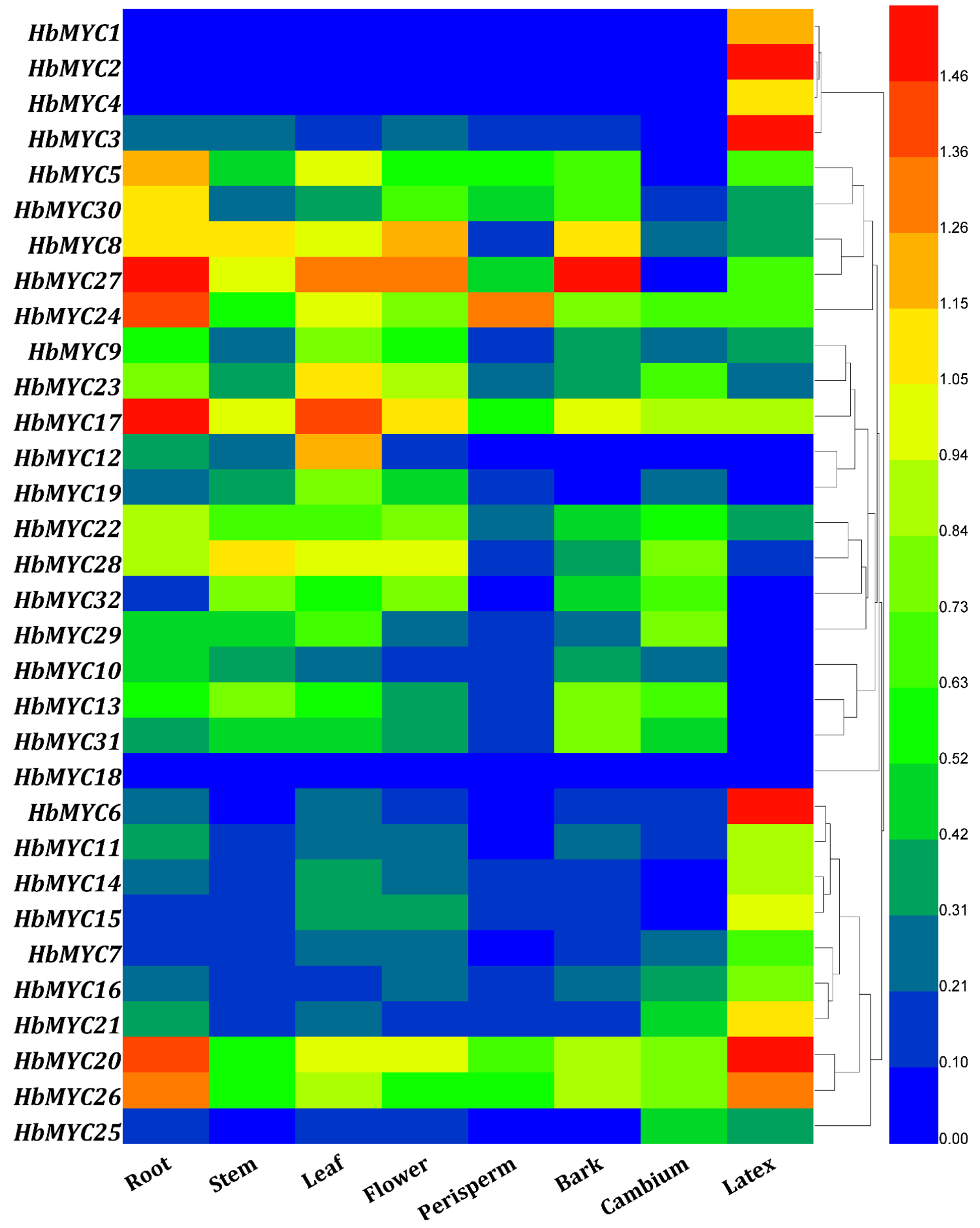
3.5. Tissue-Specific Expression Patterns of HblMYC Genes
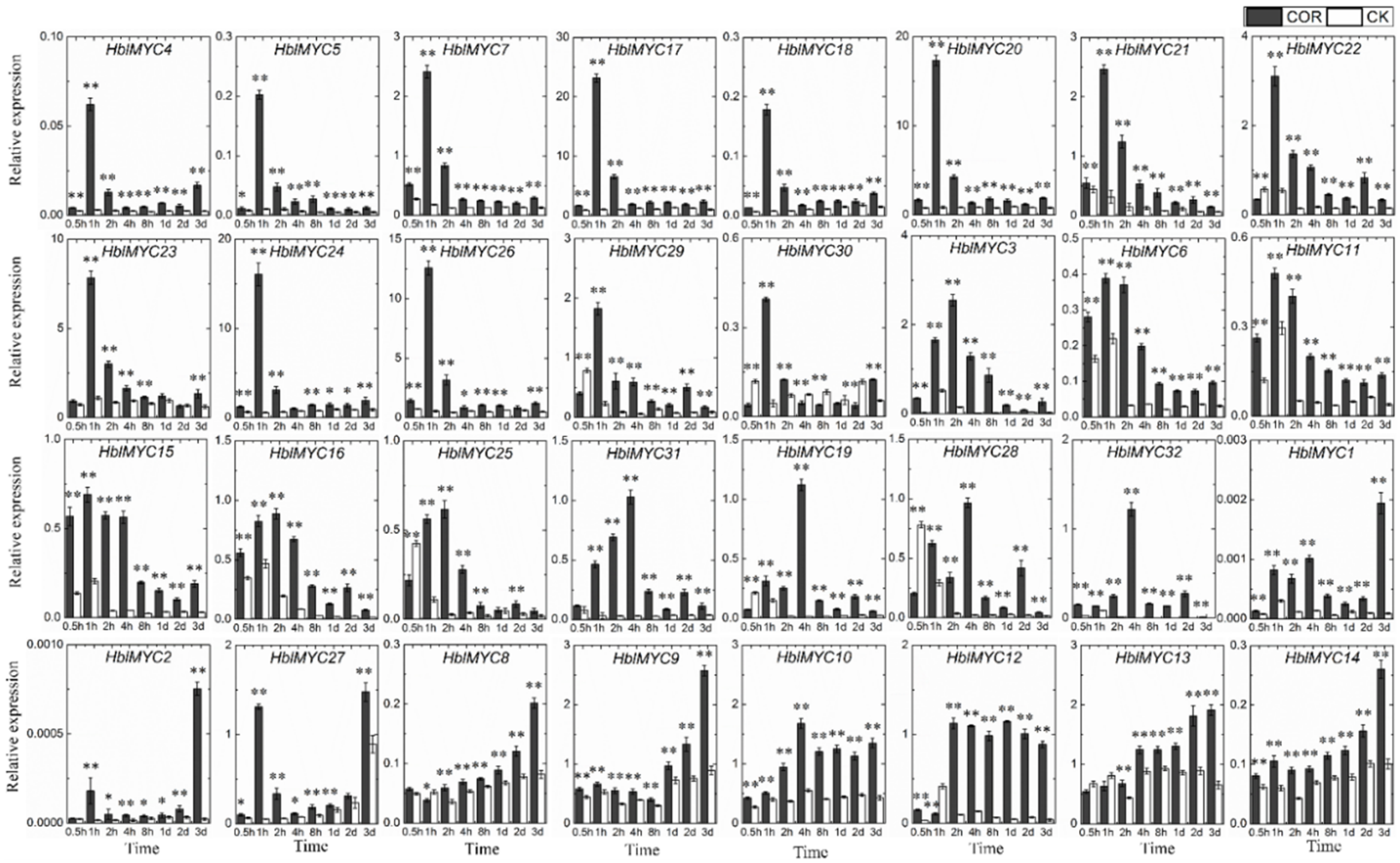
3.6. Expression Patterns of HblMYCs in Response to Coronatine (COR)
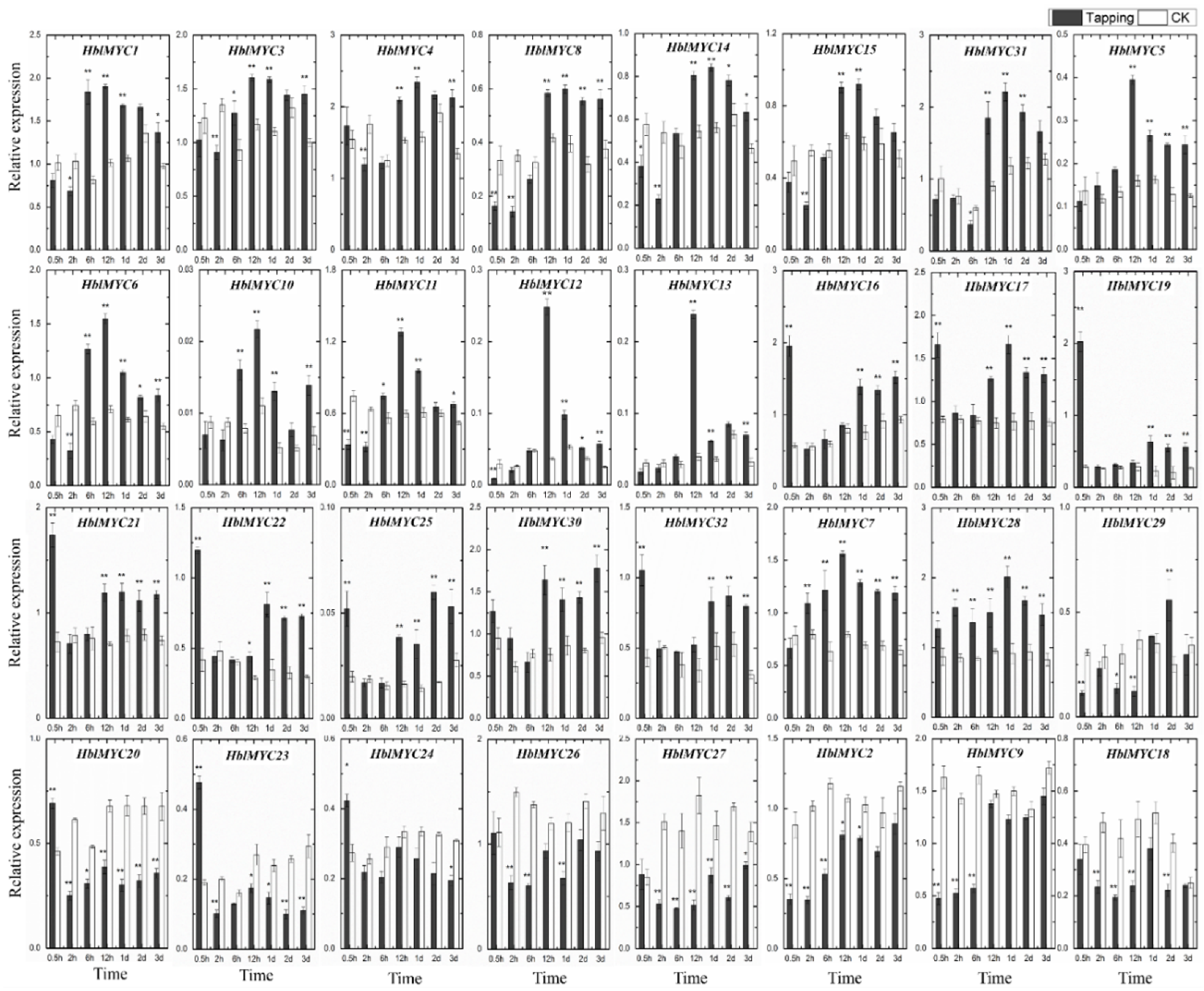
3.7. Expression Patterns of HblMYCs in Response to Tapping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F.; Chen, R.; Jiang, H.; Wang, H.; Chen, Q.; et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, E8930–E8939. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The Basic Helix-Loop-Helix Transcription Factor MYC2 Directly Represses PLETHORA Expression during Jasmonate-Mediated Modulation of the Root Stem Cell Niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Zhai, Q.; An, C.; Li, C. LEUNIG_HOMOLOG Mediates MYC2-Dependent Transcriptional Activation in Cooperation with the Coactivators HAC1 and MED25. Plant Cell 2019, 31, 2187–2205. [Google Scholar] [CrossRef]
- Hao, B.-Z.; Wu, J.-L. Laticifer Differentiation in Hevea brasiliensis: Induction by Exogenous Jasmonic Acid and Linolenic Acid. Ann. Bot. 2000, 85, 37–43. [Google Scholar] [CrossRef]
- Tian, W.; Shi, M.; Yu, F.; Wu, J.; Hao, B.; Cui, K. Localized Efects of Mechanical Wounding and Exogenous Jasmonic Acid on the Induction of Secondary Laticifer Diferentiation in Relation to the Distribution of Jasmonic Acid in Hevea brasiliensis. Acta Bot. Sin. 2003, 45, 1366–1372. [Google Scholar]
- Wu, J.-L.; Hao, B.-Z.; Tan, H.Y. Wound-induced differentiation in Hevea brasiliensis shoots mediated by jasmonic acid. J. Rubber Res. 2002, 5, 53–63. [Google Scholar]
- Zhang, S.; Wu, S.; Tian, W. The secondary laticifer differentiation in rubber tree is induced by trichostatin A, an inhibitor of histone acetylation. Front. Agric. Sci. Eng. 2016, 3, 357–362. [Google Scholar] [CrossRef][Green Version]
- Zhang, S.-X.; Wu, S.-H.; Chen, Y.-Y.; Tian, W.-M. Analysis of Differentially Expressed Genes Associated with Coronatine-Induced Laticifer Differentiation in the Rubber Tree by Subtractive Hybridization Suppression. PLoS ONE 2015, 10, e0132070. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, S.; Chao, J.; Deng, X.; Chen, Y.; Shi, M.; Tian, W.-M. Transcriptome Analysis of the Signalling Networks in Coronatine-Induced Secondary Laticifer Differentiation from Vascular Cambia in Rubber Trees. Sci. Rep. 2016, 6, 36384. [Google Scholar] [CrossRef] [PubMed]
- Ichihara, A.; Shiraishi, K.; Sato, H.; Sakamura, S.; Nishiyama, K.; Sakai, R.; Furusak, A.; Matsumoto, T. The structure of coronatine. J. Am. Chem. Soc. 1977, 99, 636–637. [Google Scholar] [CrossRef]
- Tian, W.-M.; Yang, S.-G.; Shi, M.-J.; Zhang, S.-X.; Wu, J.-L. Mechanical wounding-induced laticifer differentiation in rubber tree: An indicative role of dehydration, hydrogen peroxide, and jasmonates. J. Plant Physiol. 2015, 182, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Guo, N.; Yang, S.; Shi, M.; Chao, J.; Li, H.; Peng, S.; Tian, W.-M. Jasmonate signalling in the regulation of rubber biosynthesis in laticifer cells of rubber tree, Hevea brasiliensis. J. Exp. Bot. 2018, 69, 3559–3571. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Li, H.-L.; Wang, Y.; Zhu, J.-H.; Peng, S.-Q. A myelocytomatosis transcription factor from Hevea brasiliensis positively regulates the expression of the small rubber particle protein gene. Ind. Crops Prod. 2019, 133, 90–97. [Google Scholar] [CrossRef]
- Pirrello, J.; Leclercq, J.; Dessailly, F.; Rio, M.; Piyatrakul, P.; Kuswanhadi, K.; Tang, C.; Montoro, P. Transcriptional and post-transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol. 2014, 14, 341. [Google Scholar] [CrossRef]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Chao, J.; Chen, Y.; Wu, S.; Tian, W.-M. Comparative transcriptome analysis of latex from rubber tree clone CATAS8-79 and PR107 reveals new cues for the regulation of latex regeneration and duration of latex flow. BMC Plant Biol. 2015, 15, 104. [Google Scholar] [CrossRef]
- Bai, J.-F.; Wang, Y.-K.; Guo, L.-P.; Guo, X.-M.; Guo, H.-Y.; Yuan, S.-H.; Duan, W.-J.; Liu, Z.; Zhao, C.-P.; Zhang, F.-T.; et al. Genomic identification and characterization of MYC family genes in wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 1032. [Google Scholar] [CrossRef]
- Ren, Y.; Zou, W.; Feng, J.; Zhang, C.; Su, W.; Zhao, Z.; Wang, D.; Sun, T.; Wang, W.; Cen, G.; et al. Characterization of the sugarcane MYC gene family and the negative regulatory role of ShMYC4 in response to pathogen stress. Ind. Crops Prod. 2021, 176, 114292. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, L.-M.; Chen, Y.-Y.; Yang, S.-G.; Tian, W.-M. MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis Muell. Arg.). J. Plant Physiol. 2011, 168, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Yang, S.; Chen, Y.; Tian, W.-M. Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis of the Gene Expression in Laticifers on the Basis of Latex Flow in Rubber Tree (Hevea brasiliensis Muell. Arg.). Front. Plant Sci. 2016, 7, 1149. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Chakraborty, M.; Gangappa, S.; Gangappa, S.; Maurya, J.P.; Maurya, J.P.; Sethi, V.; Sethi, V.; Srivastava, A.K.; Srivastava, A.K.; et al. Functional interrelation of MYC 2 and HY 5 plays an important role in Arabidopsis seedling development. Plant J. 2019, 99, 1080–1097. [Google Scholar] [CrossRef]
- Jiu, S.; Guan, L.; Leng, X.; Zhang, K.; Haider, M.S.; Yu, X.; Zhu, X.; Zheng, T.; Ge, M.; Wang, C.; et al. The role of VvMYBA2r and VvMYBA2w alleles of the MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnol. J. 2021, 19, 1216–1239. [Google Scholar] [CrossRef]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-Triggered and Dark-Induced Leaf Senescence Require the bHLH Transcription Factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Xiong, R.; Liu, H.; Wu, M.; Chen, F.; Yan, H.; Xiang, Y. Basic helix-loop-helix gene family: Genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol. Biochem. 2018, 132, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Wei, K.; Chen, H. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 2018, 18, 309. [Google Scholar] [CrossRef]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef]
- Tian, Y.; Pu, X.; Yu, H.; Ji, A.; Gao, R.; Hu, Y.; Xu, Z.; Wang, H. Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Crocin Biosynthesis in Gardenia jasminoides Ellis (Rubiaceae). BioMed Res. Int. 2020, 2020, 2903861. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, Y.; Kim, S.-U.; Chen, Z.; Nie, G.; Cheng, S.; Ye, J.; Xu, F. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 2020, 10, 13723. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, H.; Luo, T.; Liu, Y.; Nie, X.; Li, H. Characteristics and Expression Pattern of MYC Genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon. Plants 2019, 8, 274. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Lu, X.; Yan, T.; Fu, X.; Lv, Z.; Zhang, F.; Pan, Q.; Wang, G.; Sun, X.; Tang, K. The jasmonate-responsive Aa MYC 2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 2016, 210, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Singh, S.K.; Patra, B.; Schluttenhofer, C.; Guo, W.; Pattanaik, S.; Yuan, L. Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. J. Exp. Bot. 2018, 69, 4267–4281. [Google Scholar] [CrossRef]
- Chen, R.; Huang, K.; Pan, S.; Xu, T.; Tan, J.; Hao, D. Jasmonate induced terpene-based defense in Pinus massoniana depresses Monochamus alternatus adult feeding. Pest Manag. Sci. 2021, 77, 731–740. [Google Scholar] [CrossRef]
- Lou, Y.; Zhou, H.; Han, Y.; Zeng, Q.; Zhu, J.; Yang, Z. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for another development in Arabidopsis thaliana. New Phytol. 2018, 217, 378–391. [Google Scholar] [CrossRef]
- Xiao, L.; Li, X.; Liu, F.; Zhao, Z.; Xu, L.; Chen, C.; Wang, Y.; Shang, G.; Du, D. Mutations in the CDS and promoter of BjuA07.CLV1 cause a multilocular trait in Brassica juncea. Sci. Rep. 2018, 8, 5339. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.-X.; Wu, S.-H.; Chao, J.-Q.; Yang, S.-G.; Bao, J.; Tian, W.-M. Genome-Wide Identification and Expression Analysis of MYC Transcription Factor Family Genes in Rubber Tree (Hevea brasiliensis Muell. Arg.). Forests 2022, 13, 531. https://doi.org/10.3390/f13040531
Zhang S-X, Wu S-H, Chao J-Q, Yang S-G, Bao J, Tian W-M. Genome-Wide Identification and Expression Analysis of MYC Transcription Factor Family Genes in Rubber Tree (Hevea brasiliensis Muell. Arg.). Forests. 2022; 13(4):531. https://doi.org/10.3390/f13040531
Chicago/Turabian StyleZhang, Shi-Xin, Shao-Hua Wu, Jin-Quan Chao, Shu-Guang Yang, Jie Bao, and Wei-Min Tian. 2022. "Genome-Wide Identification and Expression Analysis of MYC Transcription Factor Family Genes in Rubber Tree (Hevea brasiliensis Muell. Arg.)" Forests 13, no. 4: 531. https://doi.org/10.3390/f13040531
APA StyleZhang, S.-X., Wu, S.-H., Chao, J.-Q., Yang, S.-G., Bao, J., & Tian, W.-M. (2022). Genome-Wide Identification and Expression Analysis of MYC Transcription Factor Family Genes in Rubber Tree (Hevea brasiliensis Muell. Arg.). Forests, 13(4), 531. https://doi.org/10.3390/f13040531





