Abstract
Gene-editing methods, particularly CRISPR, provide extraordinary opportunities for scientific insights and applications in the life sciences. However, the prospects for near-term applications to commercial forestry appear limited. Loss-of-function phenotypes that can be imparted by mutation of one or a few conserved genes offer the best opportunities in the near term. For traits with complex inheritance, there is insufficient science to guide gene-editing efforts, and Genome-Wide Association Studies (GWASs), without strong validation, typically cannot provide high-confidence gene identification. Other obstacles include the difficulty of transformation in many important genotypes, difficulties of transient editing or complete editor removal, and complexity of use in breeding programs. Gene edits that cause loss-of-function traits will generally be recessive, and thus not be expressed among outbred progeny, so vegetative propagules (clones) will be required in most cases. There are also important societal constraints, such as strict regulations for field trials in most countries, and market certification systems that do not allow any kinds of recombinant DNA-modified trees, including those produced by gene-editing, in certified production forests. We conclude that gene-editing applications will be extremely limited for the foreseeable future (i.e., at least 10 years). Nevertheless, gene-editing is a very powerful scientific tool that will be widely used by molecular forest scientists and can lead to important applications in the longer term, if research advances are made on key fronts and regulatory and market obstacles greatly attenuated.
1. What Is Gene-Editing?
Gene-editing is the ability to make precisely targeted modifications to genes. It differs from its close relative “genetic engineering” (GE) (also widely called “genetic modification” or GM) in that the goal is not to impart new traits using inserted genes (transgenics), but to modify innate characteristics of native genomes. In practice, however, the distinction is not absolute as many forms of gene-editing also impart novel changes to genomes. Though earlier forms of gene-editing tools have been in use for several decades, their efficiency in plants has been so low that they received little attention in science or application. That changed with the development of CRISPR systems, which have extraordinary and near universal efficiency and precision, especially for loss-of-function mutations in plants (reviewed in [1,2]).
In most applications gene-editing causes targeted, double-strand breaks in DNA which are then usually repaired by cells imperfectly, causing frame shift or deletion mutations that give loss-of-function phenotypes. Because of its high efficiency (often 50-100%), using gene-editing it is also easy to obtain biallelic loss-of-function mutations, often termed knock-outs (KOs), so inbreeding is not required to cause phenotypic effects from recessive mutations (e.g., [3]). This is especially useful for forest trees, where strong inbreeding is typically avoided, heterozygosity is high, and the onset of reproduction is often delayed for years [4,5]. Thus, if clonal propagation is feasible, a gene edited KO that imparts a useful trait could potentially be deployed directly into field trials and commercial plantings (i.e., unless prevented by regulatory and/or societal barriers, as described below). In addition, high efficiency makes it feasible to mutate multiple gene-targets in a single transformation experiment, especially where the goal is to combine multiple gene KOs. The frequency of off-target mutations, a significant concern in animals, appears to be extremely low in plants, at least with the well-studied Cas9 nuclease [5], and of course far lower than the extensive unintended genetic diversity produced via comparable conventional breeding methods such as introgression. However, the somaclonal variation that can be introduced by genetic transformation and regeneration methods can still be significant and requires analysis during further breeding and field evaluations.
In addition to KOs, gene-editing can be used to produce targeted changes to genes and genomes of many kinds, a capability whose diversity is growing as new forms of gene-editing reagents continue to be developed. In these applications, a base-editor or DNA template is often used to direct the kinds of mutations that occur, such as to impart specific new amino acids to change the property of an enzyme, including, for example, susceptibility to herbicide toxicity. It can also swap-in a major change to a promoter, thus modifying gene expression in a minor or major way. Both of these types of modifications were needed for successful gene-editing to produce glyphosate-resistant cassava [6]. Recent innovations include the ability to make large deletions, such as to remove one or several entire genes and beyond (e.g., [7]). Furthermore, by using a deactivated nuclease fused to other proteins, such as transcription factors or enzymes that modify epigenetic states, gene-targeted and transient regulation of gene expression can be achieved [8].
2. Knowledge Base to Enable Gene-Editing in Tree Breeding
To implement gene-editing, it is essential to know the identities and sequences of genes that control phenotypes of interest. In contrast, conventional breeding requires no knowledge of genic control of traits, nor requires technology to insert DNA (or protein-RNA complexes) and regenerate non-chimeric organisms. Knowledge of the genic control of traits comes from two main sources: functional annotation of genomes, or gene discovery based on genetic variation in populations of the target trees.
Functional annotation is usually derived indirectly from tractable herbaceous plants, such as Arabidopsis, the main engine of gene discovery for all plants. However, enabled by rapid and low-cost sequencing technology, genome and transcriptome resources are now available for most commercially important tree species. This makes it feasible to target homologous genes in tree species that were first identified in Arabidopsis or other plant models and is the main method that has been used to modify flowering, wood chemistry, and other traits in gene-editing studies of trees and clonal crops (reviewed in [5]). This approach is particularly useful where the traits of interest can be simply obtained from a KO or, in some cases, by simple overexpression (such as to enhance transformation or regeneration rate). For example, herbicide resistance can be obtained by targeted modification of herbicide binding sites and/or increased expression of resistance alleles in highly conserved genes, as cited above for cassava. It can also be directly used to KO genes essential for fertility, such as to produce male-sterile trees to promote hybridization, enhance growth rate, or prevent male and/or female gene flow to aid regulatory compliance, public acceptance, or ecological mitigation (reviewed in [9]). Targeting of homologous genes is also feasible for commercially important wood quality traits such as lignin composition [10] and can help to produce disease resistance by targeting of well-known classes of susceptibility (S) genes [11].
However, most of the effort in forest tree breeding is to improve polygenic traits such as wood volume and quality, and adaptation to complex stresses. It is likely that variation in such traits is due to the combined effects of hundreds to thousands of genes, and the identity of these genes cannot be usefully inferred from model plants like Arabidopsis. A common approach for generating hypotheses about these genes is via phenotype-genotype association studies, which were initially limited to selected candidate genes [12,13]. This approach then evolved into Genome-Wide Association Studies (GWASs), which have been widely used to statistically implicate loci underlying complex phenotypic traits in other organisms, most notably humans [14,15]. GWAS approaches have also been applied to forest trees, though we contend that, thus far, such studies have largely failed to identify clear gene-targets for editing.
3. GWASs in Forest Trees—Where Are We?
The arrival of GWASs around 2014-2015 generated considerable excitement, particularly as new sequencing-based genotyping technologies [16] quickly blurred the distinction between model and non-model organisms for genomic studies. In theory, GWASs provide one of the most promising frameworks currently available for identifying functionally important genes in the absence of a priori knowledge. Biomedical studies clearly demonstrated that this potential is achievable [14,15], including the rapid identification of genetic risk factors for severe SARS-CoV-2/COVID-19 [17,18]. However, typical study population sizes in early forest tree GWASs (reviewed in [19,20,21]) were very small, likely limiting the reliability of the reported associations.
A survey of more recent GWASs in forest trees (i.e., since 2018, Table 1) shows a great variety of study populations, sample sizes, analysis methods, and numbers of identified candidate genes, but there are several clear patterns. First, sample sizes even in very recent forest tree GWASs remain low, typically less than 1000 (Table 1). Based on experience with human GWASs and theoretical expectations, this would be expected to yield only a handful of significant hits at best [14,15]. Thus, the high numbers of reported associations in many studies (column “Hits” in Table 1) are surprising. Unresolved statistical confounding caused by population structure and relatedness, a perennial caveat of GWASs [22,23,24], seems like the most likely explanation, though that does not necessarily apply to every single association reported. Second, the statistical methodology used in forest tree GWASs is evolving. Multi-SNP and multi-trait GWAS analyses are becoming more common and may partly mitigate the severe lack of statistical power [25,26,27,28]. Similarly, integrative “multi-omic” network analyses that combine methylome, transcriptome, and metabolome data sets are being used to rank candidate genes identified in GWASs (e.g., [25,26]), although we found no examples of directly leveraging gene expression (or data from other omic layers) into association tests for forest trees. Transcriptome-wide association study (TWAS) approaches, for example, offer distinct advantages over conventional GWASs [29], and will likely see increasing use in research with forest trees over the next few years. However, for complex traits, there are substantial challenges as to what tissues, time points, and environments to sample for such studies. Third, none of the methodological advances applied in recent forest tree GWASs addresses the key issue of confounding, although effective diagnostic methods do exist. For example, we did not find examples of using Linkage Disequilibrium (LD) Score Regression [24] in forest trees, even though it is now a well-established approach for statistically teasing apart confounding from high polygenicity in human GWASs. By its nature, the application of LD Score Regression is limited to systems with well-developed and integrated genomic toolboxes (i.e., genome assembly anchored to linkage maps and detailed knowledge of genome-wide LD), which are only available in a few species of forest trees such as Populus and Eucalyptus [30,31,32,33]. We expect to see this approach applied both retrospectively and in future GWASs to potentially increase confidence in the results from studies in which confounding was well accounted for. In the meantime, even less sophisticated means of visualizing or quantifying confounding, such as inclusion of quantile-quantile plots and/or reporting the genomic control inflation factor (λGC), can be used more consistently as they provide a reasonable indication of potential confounding issues [24]. The extensive population structure/admixture in most forest tree GWASs can also provide advantages, as strikingly illustrated by the recent examples of the importance of Neanderthal haplotypes as COVID-19 risk factors [34]. To turn this challenge into an opportunity, a methodological transition may be necessary from conventional mixed-linear model approaches (i.e., which were developed and tested in much more homogeneous populations) to trans-ancestry meta-analyses and explicit inference of chromosome segment ancestry [35,36,37,38]. Finally, examples of good practices, such as validation through independent GWAS [39] or direct confirmation of gene function [26,27]—for which gene editing is a very powerful tool—are starting to appear and will hopefully become more common. Given the legitimate concerns about false positive GWAS results, some sort of validation is critical.

Table 1.
Summary of recently published (i.e., since 2018) GWAS’s in forest trees.
The road ahead with GWASs in forest trees will be long and challenging. Sample size remains the main limiting factor, and a complete change of perspective may be necessary to make significant progress. For example, it may be possible to move beyond common garden plantations and data sets from breeding programs, which have dominated forest tree GWASs so far, to attempting larger-scale phenotyping and genotyping in natural or planted stands [40]. However, even if genotyping costs were not prohibitive, this approach would only be applicable to highly heritable traits that are also amenable to remote-sensed phenotyping (e.g., pest/pathogen resistance and phenology). Continuing the current trend of using broad, often range-wide, GWAS populations is also appealing, given the expected rapid pace of climate change. However, this must be balanced against the critical need to start building confidence in GWAS results by defining clear quality standards, including explicit quantitative measures of confounding (discussed above), and acceptable validation practices [41].
In the near term, the main value of GWASs is likely to be to inform models of biological processes (e.g., biosynthetic and signaling pathways) that control traits of interest, rather than directly identifying target genes for editing. Target genes for pathway inferences can be inferred by examination of combinations of interacting genes by physiological and computational inference. In silico models of plant growth and productivity that explicitly represent metabolic and regulatory pathways at the resolution of individual genes and regulatory elements could inform data integration and prediction [42]. These models might then enable researchers to target the edits that create the most leverage for altering phenotypes while also minimizing undesirable pleiotropic effects. Initial efforts have been effective at improving the power of GWAS by evaluating the effects of mutations on metabolic pathways [43]. However, practical implementation presents an extraordinary challenge given the biological and environmental complexity inherent to tree and forest productivity.
4. Factors Affecting the Use of Gene-Editing
The ability to apply gene-editing depends not just on biology, but also on a host of technical and social factors (Figure 1). Biological knowledge that is required includes the identities of at least some of the genes whose structure and expression affects the target traits, but also how the expression of those genes at the RNA and protein level interacts with that of other genes in molecular networks. This knowledge would allow predictions about how changes in specific genes are likely to affect both the target traits and non-target traits (such as basic stress tolerance) that we must be careful not to adversely impact while seeking to modify traits for production purposes. Forest trees are likely to require more retention of their basic adaptive qualities compared to food crops, as their environments are generally less controlled and less accessible. Because the basic structure of transcriptional and metabolic networks is often highly conserved among species [61], high-quality inferences in trees can generally be made from the core networks in model organisms, especially Arabidopsis. However, making refinements to networks, for example, to precisely modify wood structure and chemistry, will require detailed information from forest trees growing in relevant environments, and such information is lacking even for the most advanced and sophisticated network models, such as for lignin biosynthesis in Populus [62].
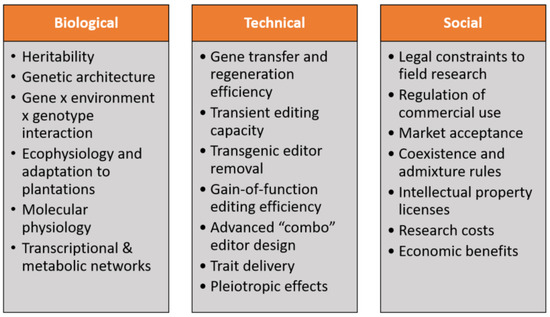
Figure 1.
Biological, technical, and social factors that affect the use of gene-editing in production forests. Biological characteristics that affect the value of gene-editing include the extent of genetic vs. environmental control of traits, complexity of genetic architecture, and how much traits can be modified, while maintaining adaptation to the environment. Technical capacity for gene-editing includes how well edits can usefully affect traits in diverse populations and the ability to edit and regenerate healthy trees in commercial genotypes. Social constraints include the extent of economic value created from edits vs. required research and commercial cost to create usefully edited trees, and the costs and political obstacles to obtaining legal and commercial license to plant edited trees.
Traits that can be affected by one or a few genes and are highly heritable (i.e., are not strongly affected by environment) will be much simpler to modify successfully by gene-editing than traits that need to be controlled by large numbers of genes and whose expression strongly depends on growth environment. Although most traits show polygenic inheritance in natural populations, there are multiple examples of useful trait modification by one or a few genes in agriculture or forestry. Well-known examples include cell wall and terpenoid chemical composition, resistance to biotrophic pathogens, flowering characteristics, and plant stature (semi-dwarfism or apical dominance) [63,64]. For example, the time of flowering onset tends to vary quantitatively and is affected by many genes [65], however, overexpression of a single gene can dramatically accelerate or delay flowering, including in forest trees (e.g., [66,67]). Similarly, modifying the expression of loci that explains less than 1% of the phenotypic variance in a GWAS population can result in relative phenotypic changes of 43–66% in RNAi transformants [26], at least in laboratory or greenhouse environments. For this reason, effect sizes observed in GWASs, and even detection of a gene itself through GWAS, should not be the only means of prioritizing gene-targets for editing. This is in part because GWAS detection and estimated effect sizes are strongly dependent on allele frequency; they are therefore limited by the standing range of natural variation, itself a product of a long, complex, and stochastic evolutionary processes. A much wider genotypic and phenotypic space could be available in managed plantation environments where trees may be more protected from biotic and abiotic stresses through density and weed control, and in some cases by fertilization or pesticide use. Plantation trees are also typically grown for much shorter time periods compared to their natural longevity, in theory allowing more rapid growth and a reduced investment in stress mitigation and pest protection.
The technical capacity for gene-editing is a limiting factor for most trees and crops, which is a direct result of the common recalcitrance to gene insertion and regeneration in most genotypes [68]. In cases where the final product needs to be free of all transgenic DNA for regulatory or market acceptance—which is most commonly done by segregation of the editing agents away from the edited gene-targets—this presents another major challenge due to the delayed reproduction and high heterozygosity of forest trees. Efforts are underway to use transient, often protein based or viral editing, to avoid this problem; however, there is little indication that this can as yet be done at scale in any forest trees or other perennial and clonal crops [5].
The target population for gene-editing is a critical consideration. The deployment of gene edited trees thorough sexual propagules (vs. vegetative propagation) presents serious constraints in forest trees as the large majority of edits produced to date are KOs, and thus likely to produce recessive gene action. If one or a small number of gene edited trees are outcrossed to other trees with functional alleles at the target locus, the trait is unlikely to be expressed in progeny. Thus, dominant gene edits, such as those that upregulate or downregulate expression in trans, are likely to be highly desirable for such situations. However, these are also likely to be regulated as GMOs, which may be problematic for acceptance. Knock-in modifications, where genes or promoter elements are inserted, could mitigate this problem, but a rule of thumb is that such targeted modifications—which generally require some type of controlled recombination—are substantially more difficult than KOs in plants, though improvements to CRISPR technology seem to occur continuously [69]. However, such edits would also likely be regulated as GMOs. In general, when transformation and regeneration efficiency is an obstacle, as is the rule in commercially important forest trees, “clean” gene edited plants lacking the CRISPR machinery will be very difficult to produce at scale.
Where clonal deployment is practiced, gene edited propagules are much more readily employed. The advent of clonal systems has had major impacts on intensive forestry in many countries [70]—particularly for Eucalyptus, Pinus, and Populus—and gene-editing may further amplify its impacts. In these systems, additive gene action does not need to be the dominant form; non-additive gene action in all its forms, including the use of recessive allelic configurations, can be readily deployed [71]. In addition, the edits can be customized to specific genotypes, which can be very important for target traits like modified wood quality and pest resistance (because there is extensive genetic variability that will interact with the edits imparted).
Societal considerations are major factors affecting the feasibility of gene-editing. First, they affect the willingness of governments and companies to invest in science and technology. After initial zeal for recombinant biotechnology shown by a number of major forestry companies worldwide in the 1980s and 1990s, nearly all of that effort disappeared over the ensuing decade, presumably due to the increasingly strict regulation, market barriers, and globally publicized negative events that characterized agricultural and forest biotechnology generally [72,73,74]. It is also possible that the transgenic traits offered, such as herbicide and pest tolerance, were of insufficient economic importance in forestry systems (compared to their benefits for annual crop agriculture), to provide a strong enough economic incentive for their use, especially as technical obstacles remained high (e.g., transformation efficiency). Second, regulations in much of the world use the recombinant DNA method as the trigger for regulation; in effect, this trigger means a crop is presumptively harmful and cannot be grown unless proven “safe,” which is normally possible only after extensive and costly multi-year studies. During the period of research and breeding prior to a decision to commercialize, every pollen grain, seed, and vegetative propagule must be contained and killed, something that is nearly impossible to do with the large size, delayed reproduction, and extensive potential for gene dispersal from pollen, and sometimes also seed, in forest trees during breeding trials. Such releases and admixtures in agricultural crops have in the past led to billions of dollars in legal penalties and fines, so is a very serious risk. Exploring the benefit of a gene edited tree in a normal breeding program, unless it is fully exempt from regulation, is effectively impossible, unless drastic isolation measures are taken. There are regulatory exemptions for gene-editing in some countries, but these only apply to “clean” and simple edits, particularly those that could have been produced by natural breeding or hybridization [75]. In reality, it will be difficult to fully remove all transgenic elements in forest trees, as discussed above, and complex gene edits or those that use a co-inserted DNA template (that acts as an editing guide) will not be exempt in the USA or other countries that we are aware of. In addition, given the costs and difficulty of transformation and regeneration, we expect that products will often have a combination of valuable transgenic traits such as herbicide, stress, or pest resistance, as well as edits, to make the investment in transformation and regulatory compliance economically worthwhile.
Another barrier is the restrictions imposed by forestry certification programs. Like organically certified food, all of the major certification programs for forestry forbid any use of recombinant DNA methods in certified forests—no matter how limited or what the ecological benefits might be [76]. Extensive areas of forest plantations in the world have one kind of green certification or another (i.e., 426 million hectares in 2019, which is about 11% of the global forest area and 30% of roundwood production [77,78]), so the impacts on forestry operations are globally significant. The no-GMO policy has remained in place for nearly 30 years despite longstanding protests from biotechnology scientists [79,80]. This may have been the most important reason for the rapid cessation of investment in transgenic biotechnology, as markets increasingly and rapidly demanded certified (and thus GMO-free) products, while there was initially no allowance even for field research [79]. However, as discussed above, it is also not clear if the benefits from transgenic technology were large enough to spur further development in forestry, though some very promising results for yield and wood improvement were documented in field trials in poplar [81,82,83,84,85]. A possibly encouraging sign is the creation of a “learning process” for FSC certified companies to experiment with genetically engineered or edited trees outside of certified areas, and where the trees are not used in products, certified or not [86]. However, the program is highly controversial within FSC, thus its very existence and the parameters of its operation—and therefore its ultimate impact on the use of GE trees in certified forests—is completely unknown.
5. Future Prospects and Research Priorities
The first priority to enable application of gene-editing in trees is to improve the identification of useful target genes. As described above, GWASs are currently limited in their ability to identify loci that would be impactful and reliable targets. This is partly due to the very low statistical power of most tree GWASs and the very limited extent of adequate validation of candidates. Community standards are needed to build confidence in GWAS results, particularly with respect to statistical power and control of confounding factors (discussed above). However, for most traits targeted by breeding programs, it is unclear whether more powerful and reliable GWAS would in fact lead to identification of candidates for which gene-editing would make an important contribution beyond what can be achieved with conventional breeding or genomic selection [87]. Moreover, because of the large and complex multigene families of forest trees (that show various degrees of functional redundancy), gene-editing studies with GWAS candidates are unlikely to provide simple results that lead directly to useful applications. Instead, initial studies are likely to require further investigation, often including multiple gene-targeting—thus are a first rather than a final step in technology development. It is clear that, in the near term, the main value of gene-editing is likely to be as a research tool for exploring candidate genes and processes.
Second, it is essential to alleviate the bottleneck for functional testing by improving genome editing approaches. Much progress has been made in developing knock-ins and base editing, though efficiency remains low in plants. The age of epigenome editing—where expression-modifying non-sequence modifications such as methylation are made—is also beginning to dawn. This will provide another suite of tools that can be used to simultaneously activate or repress large numbers of genes, at least on a temporary basis (e.g., that could help overcome the transformation-regeneration bottleneck) [88]. Multiplex editing using multiple concatenated guide RNAs in a single construct can also enhance efficiency dramatically, thereby opening the possibility of pathway engineering [89]. Transient viral editing methods have been developed for several plant species and may even find use in the field for helping high-value trees respond to stressful environments or pathogen outbreaks. Delivery is typically done using Agrobacterium, but it can also use packaged RNA particles [90] or nanotechnology [91]. However, no such functional systems appear to have been reported for forest trees. Another important application of nanotechnology might be if it could provide efficient gene-editing in pollen without stable insertion, as this should have both the lowest regulatory barriers and easiest incorporation into breeding programs. However, as haploid KOs, the edits would not be expressed in heterozygous condition in progeny; expression would require pollination of previously edited females. The possibilities continue to expand, but progress will be stymied until the efficiency of transformation, or possibly transient editing methods, can be greatly improved and regulatory barriers reduced.
Third, early flowering is an important trait for the success of editing in tree breeding programs. This is because early flowering would allow (1) removal of gene-editing constructs from the edited germplasm via Mendelian segregation to ease regulatory barriers; (2) accelerate genomic selection— if it could be imparted to many genotypes (perhaps by systemic, transient gene expression methods as cited below); (3) aid in the introgression of edited loci into other genetic backgrounds; and (4) enhance the diversity of gene-edited plantations, which should improve prospects for maintaining performance over the long-term. A viral-based transient flowering system would be ideal given the recalcitrance of most forest trees to stable transformation; such a system has been demonstrated for apple [92], but we are not aware of similar progress for any forest trees.
Fourth, there is also a critical need for enhancing the efficiency of vegetative propagation systems in forest trees. Many commercially important trees cannot be readily propagated asexually, and this limits the rate at which gene edited clones can be scaled up for commercial production. Bypassing sexual reproduction during propagation enables non-additive genetic gains to be maintained in species for which inbreeding is poorly tolerated and inbred lines are not an option. Vegetative propagation also enables the use of sterile genotypes, which may improve system biosafety and reduce regulatory and public perception issues.
Finally, assuming that gene-editing field research is able to move forward in at least some amenable genotypes in the next few years despite social constraints, it will still require many additional years to assess phenotypic effects of the gene-edit modifications and ready trees for commercial use—including to obtain licenses for use of needed intellectual property in the complex and rapidly changing area of CRISPR technologies [93]. The initial phenotypic assessment will require several years, possibly a decade or more, for assessments of traits such as mature wood quality, flowering/sterility, and many kinds of pest and stress resistance. Then, if results are promising, the modifications would need to be re-tested in additional genotypes and geographies for a similar period. Finally, if the modification continues to be successful, those modified genotypes would need to be mass-propagated for commercial deployment as clones, and approvals obtained from regulatory, market, and intellectual property institutions. If instead sexual propagules are used, it is likely to take several more years to create flowering seed orchards or make controlled crosses at scale, especially where recessive KOs are being employed and specific genotypes must therefore be crossed for trait expression in progeny. Thus, optimistically, we will need a decade at minimum, and likely considerably longer, for gene-edited trees to make a significant impact in commercial forestry. Of course, were conditions to occur where gene-edited trees could solve an immense and existential challenge that other approaches cannot (such as to provide resistance to a new and fast spreading disease, or a marked improvement in heat or drought tolerance to help cope with climate change), this timeline would be advanced.
6. Conclusions
The scientific excitement surrounding gene-editing is extraordinary. It is one of the few methods with a reach across nearly all of biology, enabling much more precise science and many new therapeutic and agricultural applications. However, although others have expressed considerable enthusiasm for applications of gene editing in forestry (e.g., [1,2]), we believe that a combination of biological, technical, and societal constraints make the prospect for near-term, large-scale applications in commercial forestry remote. The best places are in clonally propagated, short rotation plantations, where well-known, conserved genes such as for flowering, herbicide tolerance, and wood chemistry can be targeted. For a larger and broader impact, we await much more science on gene-trait associations and improvements to genetic transformation/regeneration and editing systems. We also require innovations in regulatory and certification systems that recognize we are in a climate emergency world, and breeding progress needs to accelerate, not be slowed. Unfortunately, because of the powerful biological and social constraints limiting progress, we believe that significant commercial use of gene-editing will not begin for a least a decade—and perhaps much longer.
Author Contributions
Conceptualization, S.H.S., G.T.S. and S.P.D.; writing—review and editing, S.H.S., G.T.S. and S.P.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank Michael Nagle, Glenn Howe, the Editor, and three anonymous reviewers for their insightful comments on earlier versions of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cao, H.X.; Vu, G.T.; Gailing, O. From Genome Sequencing to CRISPR-Based Genome Editing for Climate-Resilient Forest Trees. Int. J. Mol. Sci. 2022, 23, 966. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, S.; Ballesta, P.; Ali, M.; Mora-Poblete, F. Achievements and Challenges of Genomics-Assisted Breeding in Forest Trees: From Marker-Assisted Selection to Genome Editing. Int. J. Mol. Sci. 2021, 22, 10583. [Google Scholar] [CrossRef] [PubMed]
- Elorriaga, E.; Klocko, A.L.; Ma, C.; du Plessis, M.; An, X.; Myburg, A.A.; Strauss, S.H. Genetic Containment in Vegetatively Propagated Forest Trees: CRISPR Disruption of LEAFY Function in Eucalyptus Gives Sterile Indeterminate Inflorescences and Normal Juvenile Development. Plant Biotechnol. J. 2021, 19, 1743–1755. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors Influencing Levels of Genetic Diversity in Woody Plant Species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Goralogia, G.S.; Redick, T.P.; Strauss, S.H. Gene Editing in Tree and Clonal Crops: Progress and Challenges. Vitro Cell. Dev. Biol. Plant 2021, 57, 683–699. [Google Scholar] [CrossRef]
- Hummel, A.W.; Chauhan, R.D.; Cermak, T.; Mutka, A.M.; Vijayaraghavan, A.; Boyher, A.; Starker, C.G.; Bart, R.; Voytas, D.F.; Taylor, N.J. Allele Exchange at the EPSPS Locus Confers Glyphosate Tolerance in Cassava. Plant Biotechnol. J. 2018, 16, 1275–1282. [Google Scholar] [CrossRef]
- Osakabe, K.; Wada, N.; Miyaji, T.; Murakami, E.; Marui, K.; Ueta, R.; Hashimoto, R.; Abe-Hara, C.; Kong, B.; Yano, K.; et al. Genome Editing in Plants Using CRISPR Type I-D Nuclease. Commun. Biol. 2020, 3, 648. [Google Scholar] [CrossRef]
- Gardiner, J.; Ghoshal, B.; Wang, M.; Jacobsen, S.E. CRISPR–Cas-Mediated Transcriptional Control and Epi-Mutagenesis. Plant Physiol. 2022, 188, 1811–1824. [Google Scholar] [CrossRef]
- Strauss, S.H.; Jones, K.N.; Lu, H.; Petit, J.D.; Klocko, A.L.; Betts, M.G.; Brosi, B.J.; Fletcher Jr, R.J.; Needham, M.D. Reproductive Modification in Forest Plantations: Impacts on Biodiversity and Society. New Phytol. 2017, 213, 1000–1021. [Google Scholar] [CrossRef]
- Tsai, C.-J.; Xu, P.; Xue, L.-J.; Hu, H.; Nyamdari, B.; Naran, R.; Zhou, X.; Goeminne, G.; Gao, R.; Gjersing, E.; et al. Compensatory Guaiacyl Lignin Biosynthesis at the Expense of Syringyl Lignin in 4CL1-Knockout Poplar. Plant Physiol. 2020, 183, 123–136. [Google Scholar] [CrossRef]
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-Edited Powdery Mildew Resistance in Wheat without Growth Penalties. Nature 2022, 602, 455–460. [Google Scholar] [CrossRef]
- Neale, D.B.; Savolainen, O. Association Genetics of Complex Traits in Conifers. Trends Plant Sci. 2004, 9, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Neale, D.B.; Kremer, A. Forest Tree Genomics: Growing Resources and Applications. Nat. Rev. Genet. 2011, 12, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five Years of GWAS Discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-Wide Genetic Marker Discovery and Genotyping Using next-Generation Sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Huffman, J.E.; Butler-Laporte, G.; Khan, A.; Pairo-Castineira, E.; Drivas, T.G.; Peloso, G.M.; Nakanishi, T.; Ganna, A.; Verma, A.; Baillie, J.K.; et al. Multi-Ancestry Fine Mapping Implicates OAS1 Splicing in Risk of Severe COVID-19. Nat. Genet. 2022, 54, 125–127. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Hall, D.; Hallingbäck, H.R.; Wu, H.X. Estimation of Number and Size of QTL Effects in Forest Tree Traits. Tree Genet. Genomes 2016, 12, 110. [Google Scholar] [CrossRef]
- Du, Q.; Lu, W.; Quan, M.; Xiao, L.; Song, F.; Li, P.; Zhou, D.; Xie, J.; Wang, L.; Zhang, D. Genome-Wide Association Studies to Improve Wood Properties: Challenges and Prospects. Front. Plant Sci. 2018, 9, 1912. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Silva-Junior, O.B.; Resende, R.T.; Cappa, E.P.; Müller, B.S.F.; Tan, B.; Isik, F.; Ratcliffe, B.; El-Kassaby, Y.A. Quantitative Genetics and Genomics Converge to Accelerate Forest Tree Breeding. Front. Plant Sci. 2018, 9, 1693. [Google Scholar] [CrossRef] [PubMed]
- Balding, D.J. A Tutorial on Statistical Methods for Population Association Studies. Nat. Rev. Genet. 2006, 7, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. New Approaches to Population Stratification in Genome-Wide Association Studies. Nat. Rev. Genet. 2010, 11, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.K.; Loh, P.-R.; Finucane, H.K.; Ripke, S.; Yang, J.; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M.; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score Regression Distinguishes Confounding from Polygenicity in Genome-Wide Association Studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- Chhetri, H.B.; Furches, A.; Macaya-Sanz, D.; Walker, A.R.; Kainer, D.; Jones, P.; Harman-Ware, A.E.; Tschaplinski, T.J.; Jacobson, D.; Tuskan, G.A.; et al. Genome-Wide Association Study of Wood Anatomical and Morphological Traits in Populus trichocarpa. Front. Plant Sci. 2020, 11, 545748. [Google Scholar] [CrossRef]
- Chhetri, H.B.; Macaya-Sanz, D.; Kainer, D.; Biswal, A.K.; Evans, L.M.; Chen, J.-G.; Collins, C.; Hunt, K.; Mohanty, S.S.; Rosenstiel, T.; et al. Multitrait Genome-Wide Association Analysis of Populus trichocarpa Identifies Key Polymorphisms Controlling Morphological and Physiological Traits. New Phytol. 2019, 223, 293–309. [Google Scholar] [CrossRef]
- Kainer, D.; Padovan, A.; Degenhardt, J.; Krause, S.; Mondal, P.; Foley, W.J.; Külheim, C. High Marker Density GWAS Provides Novel Insights into the Genomic Architecture of Terpene Oil Yield in Eucalyptus. New Phytol. 2019, 223, 1489–1504. [Google Scholar] [CrossRef]
- Nagle, M.F.; Yuan, J.; Kaur, D.; Ma, C.; Peremyslova, E.; Jiang, Y.; Willig, C.J.; Goralogia, G.S.; de Rivera, A.N.; McEldowney, M.; et al. GWAS Identifies Candidate Regulators of In Planta Regeneration in Populus trichocarpa. bioRxiv 2022, 495082. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.; Jansen, R.; de Geus, E.J.C.; Boomsma, D.I.; Wright, F.A.; et al. Integrative Approaches for Large-Scale Transcriptome-Wide Association Studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef]
- Abeyratne, C.R.; Macaya-Sanz, D.; Zhou, R.; Barry, K.W.; Daum, C.; Haiby, K.; Lipzen, A.; Stanton, B.; Yoshinaga, Y.; Zane, M.; et al. High Resolution Mapping Reveals Hotspots and Sex-Biased Recombination in Populus trichocarpa. bioRxiv 2022, 491397. [Google Scholar] [CrossRef]
- Slavov, G.T.; DiFazio, S.P.; Martin, J.; Schackwitz, W.; Muchero, W.; Rodgers-Melnick, E.; Lipphardt, M.F.; Pennacchio, C.P.; Hellsten, U.; Pennacchio, L.A.; et al. Genome Resequencing Reveals Multiscale Geographic Structure and Extensive Linkage Disequilibrium in the Forest Tree Populus trichocarpa. New Phytol. 2012, 196, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.B.; Freeman, J.S.; Potts, B.M.; Vaillancourt, R.E.; Kahrood, H.V.; Ades, P.K.; Rigault, P.; Tibbits, J.F.G. Patterns of Genomic Diversity and Linkage Disequilibrium across the Disjunct Range of the Australian Forest Tree Eucalyptus globulus. Tree Genet. Genomes 2022, 18, 28. [Google Scholar] [CrossRef]
- Silva-Junior, O.B.; Grattapaglia, D. Genome-Wide Patterns of Recombination, Linkage Disequilibrium and Nucleotide Diversity from Pooled Resequencing and Single Nucleotide Polymorphism Genotyping Unlock the Evolutionary History of Eucalyptus grandis. New Phytol. 2015, 208, 830–845. [Google Scholar] [CrossRef] [PubMed]
- Zeberg, H.; Pääbo, S. The Major Genetic Risk Factor for Severe COVID-19 Is Inherited from Neanderthals. Nature 2020, 587, 610–612. [Google Scholar] [CrossRef] [PubMed]
- Seldin, M.F.; Pasaniuc, B.; Price, A.L. New Approaches to Disease Mapping in Admixed Populations. Nat. Rev. Genet. 2011, 12, 523–528. [Google Scholar] [CrossRef]
- Lawson, D.J.; Davies, N.M.; Haworth, S.; Ashraf, B.; Howe, L.; Crawford, A.; Hemani, G.; Davey Smith, G.; Timpson, N.J. Is Population Structure in the Genetic Biobank Era Irrelevant, a Challenge, or an Opportunity? Hum. Genet. 2020, 139, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, E.G.; Maihofer, A.X.; Kanai, M.; Martin, A.R.; Karczewski, K.J.; Santoro, M.L.; Ulirsch, J.C.; Kamatani, Y.; Okada, Y.; Finucane, H.K.; et al. Tractor Uses Local Ancestry to Enable the Inclusion of Admixed Individuals in GWAS and to Boost Power. Nat. Genet. 2021, 53, 195–204. [Google Scholar] [CrossRef]
- DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Mahajan, A.; Go, M.J.; Zhang, W.; Below, J.E.; Gaulton, K.J.; Ferreira, T.; et al. Genome-Wide Trans-Ancestry Meta-Analysis Provides Insight into the Genetic Architecture of Type 2 Diabetes Susceptibility. Nat. Genet. 2014, 46, 234–244. [Google Scholar] [CrossRef]
- Chen, Z.-Q.; Zan, Y.; Milesi, P.; Zhou, L.; Chen, J.; Li, L.; Cui, B.; Niu, S.; Westin, J.; Karlsson, B.; et al. Leveraging Breeding Programs and Genomic Data in Norway Spruce (Picea abies L. Karst) for GWAS Analysis. Genome Biol. 2021, 22, 179. [Google Scholar] [CrossRef]
- Dungey, H.S.; Dash, J.P.; Pont, D.; Clinton, P.W.; Watt, M.S.; Telfer, E.J. Phenotyping Whole Forests Will Help to Track Genetic Performance. Trends Plant Sci. 2018, 23, 854–864. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-Wide Association Studies. Nat. Rev. Methods Primer 2021, 1, 59. [Google Scholar] [CrossRef]
- Clark, T.J.; Guo, L.; Morgan, J.; Schwender, J. Modeling Plant Metabolism: From Network Reconstruction to Mechanistic Models. Annu. Rev. Plant Biol. 2020, 71, 303–326. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Maranas, C.D. SNPeffect: Identifying Functional Roles of SNPs Using Metabolic Networks. Plant J. 2020, 103, 512–531. [Google Scholar] [CrossRef] [PubMed]
- Mukrimin, M.; Kovalchuk, A.; Neves, L.G.; Jaber, E.H.A.; Haapanen, M.; Kirst, M.; Asiegbu, F.O. Genome-Wide Exon-Capture Approach Identifies Genetic Variants of Norway Spruce Genes Associated With Susceptibility to Heterobasidion parviporum Infection. Front. Plant Sci. 2018, 9, 793. [Google Scholar] [CrossRef] [PubMed]
- Baison, J.; Vidalis, A.; Zhou, L.; Chen, Z.-Q.; Li, Z.; Sillanpää, M.J.; Bernhardsson, C.; Scofield, D.; Forsberg, N.; Grahn, T.; et al. Genome-Wide Association Study Identified Novel Candidate Loci Affecting Wood Formation in Norway Spruce. Plant J. 2019, 100, 83–100. [Google Scholar] [CrossRef]
- Milesi, P.; Berlin, M.; Chen, J.; Orsucci, M.; Li, L.; Jansson, G.; Karlsson, B.; Lascoux, M. Assessing the Potential for Assisted Gene Flow Using Past Introduction of Norway Spruce in Southern Sweden: Local Adaptation and Genetic Basis of Quantitative Traits in Trees. Evol. Appl. 2019, 12, 1946–1959. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Puiu, D.; Crepeau, M.W.; Stevens, K.; Salzberg, S.L.; Langley, C.H.; Neale, D.B. Genomic Architecture of Complex Traits in Loblolly Pine. New Phytol. 2019, 221, 1789–1801. [Google Scholar] [CrossRef]
- Bai, Q.; Cai, Y.; He, B.; Liu, W.; Pan, Q.; Zhang, Q. Core Set Construction and Association Analysis of Pinus massoniana from Guangdong Province in Southern China Using SLAF-Seq. Sci. Rep. 2019, 9, 13157. [Google Scholar] [CrossRef]
- Ding, X.; Diao, S.; Luan, Q.; Wu, H.X.; Zhang, Y.; Jiang, J. A Transcriptome-Based Association Study of Growth, Wood Quality, and Oleoresin Traits in a Slash Pine Breeding Population. PLoS Genet. 2022, 18, e1010017. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Wilhite, B.; Puiu, D.; St. Clair, J.B.; Crepeau, M.W.; Salzberg, S.L.; Langley, C.H.; Allen, B.; Neale, D.B. Dissecting the Polygenic Basis of Cold Adaptation Using Genome-Wide Association of Traits and Environmental Data in Douglas-fir. Genes 2021, 12, 110. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Sekhwal, M.K.; Puiu, D.; Salzberg, S.L.; Scott, A.D.; Allen, B.; Neale, D.B.; Chin, A.R.O.; Buckley, T.N. Genome-Wide Association Identifies Candidate Genes for Drought Tolerance in Coast Redwood and Giant Sequoia. Plant J. 2022, 109, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, C.E.; Ballesta, P.; Ahmar, S.; Fiaz, S.; Heidari, P.; Maldonado, C.; Mora-Poblete, F. Haplotype- and SNP-Based GWAS for Growth and Wood Quality Traits in Eucalyptus cladocalyx Trees under Arid Conditions. Plants 2021, 10, 148. [Google Scholar] [CrossRef]
- Müller, B.S.F.; de Almeida Filho, J.E.; Lima, B.M.; Garcia, C.C.; Missiaggia, A.; Aguiar, A.M.; Takahashi, E.; Kirst, M.; Gezan, S.A.; Silva-Junior, O.B.; et al. Independent and Joint-GWAS for Growth Traits in Eucalyptus by Assembling Genome-Wide Data for 3373 Individuals across Four Breeding Populations. New Phytol 2019, 221, 818–833. [Google Scholar] [CrossRef]
- Yong, W.T.L.; Ades, P.K.; Runa, F.A.; Bossinger, G.; Sandhu, K.S.; Potts, B.M.; Tibbits, J.F.G. Genome-Wide Association Study of Myrtle Rust (Austropuccinia psidii) Resistance in Eucalyptus obliqua (Subgenus Eucalyptus). Tree Genet. Genomes 2021, 17, 31. [Google Scholar] [CrossRef]
- Muchero, W.; Sondreli, K.L.; Chen, J.-G.; Urbanowicz, B.R.; Zhang, J.; Singan, V.; Yang, Y.; Brueggeman, R.S.; Franco-Coronado, J.; Abraham, N.; et al. Association Mapping, Transcriptomics, and Transient Expression Identify Candidate Genes Mediating Plant–Pathogen Interactions in a Tree. Proc. Natl. Acad. Sci. USA 2018, 115, 11573. [Google Scholar] [CrossRef] [PubMed]
- McKown, A.D.; Klápště, J.; Guy, R.D.; Corea, O.R.A.; Fritsche, S.; Ehlting, J.; El-Kassaby, Y.A.; Mansfield, S.D. A Role for SPEECHLESS in the Integration of Leaf Stomatal Patterning with the Growth vs Disease Trade-off in Poplar. New Phytol. 2019, 223, 1888–1903. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.P.; Suren, H.; Holliday, J.; Richards, J.H.; Fiehn, O.; Famula, R.; Stanton, B.J.; Shuren, R.; Sykes, R.; Davis, M.F.; et al. Exome Resequencing and GWAS for Growth, Ecophysiology, and Chemical and Metabolomic Composition of Wood of Populus trichocarpa. BMC Genomics 2019, 20, 875. [Google Scholar] [CrossRef]
- Bdeir, R.; Muchero, W.; Yordanov, Y.; Tuskan, G.A.; Busov, V.; Gailing, O. Genome-Wide Association Studies of Bark Texture in Populus trichocarpa. Tree Genet. Genomes 2019, 15, 14. [Google Scholar] [CrossRef]
- Wang, J.; Ding, J.; Tan, B.; Robinson, K.M.; Michelson, I.H.; Johansson, A.; Nystedt, B.; Scofield, D.G.; Nilsson, O.; Jansson, S.; et al. A Major Locus Controls Local Adaptation and Adaptive Life History Variation in a Perennial Plant. Genome Biol. 2018, 19, 72. [Google Scholar] [CrossRef]
- Mähler, N.; Schiffthaler, B.; Robinson, K.M.; Terebieniec, B.K.; Vučak, M.; Mannapperuma, C.; Bailey, M.E.S.; Jansson, S.; Hvidsten, T.R.; Street, N.R. Leaf Shape in Populus tremula Is a Complex, Omnigenic Trait. Ecol. Evol. 2020, 10, 11922–11940. [Google Scholar] [CrossRef]
- Li, Y.; Pearl, S.A.; Jackson, S.A. Gene Networks in Plant Biology: Approaches in Reconstruction and Analysis. Trends Plant Sci. 2015, 20, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.; Tunlaya-Anukit, S.; Chen, H.-C.; Li, Q.; Liu, J.; Lin, C.-Y.; et al. Improving Wood Properties for Wood Utilization through Multi-Omics Integration in Lignin Biosynthesis. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Busov, V.B.; Brunner, A.M.; Strauss, S.H. Genes for Control of Plant Stature and Form. New Phytol. 2008, 177, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Mahon, E.L.; MacKay, H.A.; Rottmann, W.H.; Strauss, S.H.; Pijut, P.M.; Powell, W.A.; Coffey, V.; Lu, H.; Mansfield, S.D.; et al. Genetic Engineering of Trees: Progress and New Horizons. Vitro Cell. Dev. Biol. - Plant 2018, 54, 341–376. [Google Scholar] [CrossRef]
- Buckler, E.S.; Holland, J.B.; Bradbury, P.J.; Acharya, C.B.; Brown, P.J.; Browne, C.; Ersoz, E.; Flint-Garcia, S.; Garcia, A.; Glaubitz, J.C.; et al. The Genetic Architecture of Maize Flowering Time. Science 2009, 325, 714–718. [Google Scholar] [CrossRef]
- Goralogia, G.S.; Howe, G.T.; Brunner, A.M.; Helliwell, E.; Nagle, M.F.; Ma, C.; Lu, H.; Goddard, A.L.; Magnuson, A.C.; Klocko, A.L.; et al. Overexpression of SHORT VEGETATIVE PHASE-LIKE (SVL) in Populus Delays Onset and Reduces Abundance of Flowering in Field-Grown Trees. Hortic. Res. 2021, 8, 167. [Google Scholar] [CrossRef]
- Klocko, A.L.; Ma, C.; Robertson, S.; Esfandiari, E.; Nilsson, O.; Strauss, S.H. FT Overexpression Induces Precocious Flowering and Normal Reproductive Development in Eucalyptus. Plant Biotechnol. J. 2016, 14, 808–819. [Google Scholar] [CrossRef]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef]
- Duan, K.; Cheng, Y.; Ji, J.; Wang, C.; Wei, Y.; Wang, Y. Large Chromosomal Segment Deletions by CRISPR/LbCpf1-Mediated Multiplex Gene Editing in Soybean. J. Integr. Plant Biol. 2021, 63, 1620–1631. [Google Scholar] [CrossRef]
- Zobel, B. Vegetative Propagation in Production Forestry. J. For. 1992, 90, 29–33. [Google Scholar] [CrossRef]
- Libby, W.J.; Rauter, R.M. Advantages of Clonal Forestry. For. Chron. 1984, 60, 145–149. [Google Scholar] [CrossRef]
- Gressel, J. Genetic Glass Ceilings: Transgenics for Crop Biodiversity, 1st ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2008; p. 488. [Google Scholar]
- Miller, H.I.; Conko, G.P. The Frankenfood Myth: How Protest and Politics Threaten the Biotech Revolution; Greenwood Publishing Group: Westport, CT, USA, 2004; ISBN 978-0-275-97879-2. [Google Scholar]
- McWilliams, J.E. Just Food: Where Locavores Get It Wrong and How We Can Truly Eat Responsibly; Back Bay Books: New York, NY, USA, 2010; ISBN 978-0-316-05263-4. [Google Scholar]
- Hoffman, N.E. Revisions to USDA Biotechnology Regulations: The SECURE Rule. Proc. Natl. Acad. Sci. USA 2021, 118, e2004841118. [Google Scholar] [CrossRef] [PubMed]
- Strauss, S.H.; Costanza, A.; Séguin, A. Genetically Engineered Trees: Paralysis from Good Intentions. Science 2015, 349, 794–795. [Google Scholar] [CrossRef]
- FAO Forest Products Annual Market Review 2014–2015. Available online: https://unece.org/fileadmin/DAM/timber/publications/2015-FPAMR-E.pdf (accessed on 24 October 2022).
- FAO Global Forest Resources Assessment 2020. Available online: https://www.fao.org/documents/card/en/c/ca9825en (accessed on 24 October 2022).
- Strauss, S.H.; Campbell, M.M.; Pryor, S.N.; Coventry, P.; Burley, J. Plantation Certification and Genetic Engineering: FSC’s Ban on Research Is Counterproductive. J. For. 2001, 99, 4–7. [Google Scholar] [CrossRef]
- Strauss, S.H.; Boerjan, W.; Chiang, V.; Costanza, A.; Coleman, H.; Davis, J.M.; Lu, M.-Z.; Mansfield, S.D.; Merkle, S.; Myburg, A.; et al. Certification for Gene-Edited Forests. Science 2019, 365, 767–768. [Google Scholar] [CrossRef]
- Ault, K.; Viswanath, V.; Jayawickrama, J.; Ma, C.; Eaton, J.; Meilan, R.; Beauchamp, G.; Hohenschuh, W.; Murthy, G.; Strauss, S.H. Improved Growth and Weed Control of Glyphosate-Tolerant Poplars. New For. 2016, 47, 653–667. [Google Scholar] [CrossRef]
- Klocko, A.L.; Meilan, R.; James, R.R.; Viswanath, V.; Ma, C.; Payne, P.; Miller, L.; Skinner, J.S.; Oppert, B.; Cardineau, G.A.; et al. Bt-Cry3Aa Transgene Expression Reduces Insect Damage and Improves Growth in Field-Grown Hybrid Poplar. Can. J. For. Res. 2014, 44, 28–35. [Google Scholar] [CrossRef]
- Lu, H.; Viswanath, V.; Ma, C.; Etherington, E.; Dharmawardhana, P.; Shevchenko, O.; Strauss, S.H.; Pearce, D.W.; Rood, S.B.; Busov, V. Recombinant DNA Modification of Gibberellin Metabolism Alters Growth Rate and Biomass Allocation in Populus. Tree Genet. Genomes 2015, 11, 127. [Google Scholar] [CrossRef]
- Pilate, G.; Guiney, E.; Holt, K.; Petit-Conil, M.; Lapierre, C.; Leplé, J.-C.; Pollet, B.; Mila, I.; Webster, E.A.; Marstorp, H.G.; et al. Field and Pulping Performances of Transgenic Trees with Altered Lignification. Nat Biotechnol 2002, 20, 607–612. [Google Scholar] [CrossRef]
- Cho, J.-S.; Jeon, H.-W.; Kim, M.-H.; Vo, T.K.; Kim, J.; Park, E.-J.; Choi, Y.-I.; Lee, H.; Han, K.-H.; Ko, J.-H. Wood Forming Tissue-Specific Bicistronic Expression of PdGA20ox1 and PtrMYB221 Improves Both the Quality and Quantity of Woody Biomass Production in a Hybrid Poplar. Plant Biotechnol. J. 2019, 17, 1048–1057. [Google Scholar] [CrossRef]
- FSC Genetic Engineering Learning Process. Available online: https://fsc.org/en/newscentre/fsc-to-begin-learning-process-on-genetic-engineering-in-forestry-outside-of-fsc (accessed on 24 October 2022).
- Grattapaglia, D. Twelve Years into Genomic Selection in Forest Trees: Climbing the Slope of Enlightenment of Marker Assisted Tree Breeding. Forests 2022, 13, 1554. [Google Scholar] [CrossRef]
- Ghoshal, B.; Picard, C.L.; Vong, B.; Feng, S.; Jacobsen, S.E. CRISPR-Based Targeting of DNA Methylation in Arabidopsis thaliana by a Bacterial CG-Specific DNA Methyltransferase. Proc. Natl. Acad. Sci. 2021, 118, e2125016118. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for Highly Efficient Multiplexed Gene Activation in Plants. Nat. Plants 2021, 7, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Torti, S.; Schlesier, R.; Thümmler, A.; Bartels, D.; Römer, P.; Koch, B.; Werner, S.; Panwar, V.; Kanyuka, K.; von Wirén, N.; et al. Transient Reprogramming of Crop Plants for Agronomic Performance. Nat. Plants 2021, 7, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Demirer, G.S.; Zhang, H.; Goh, N.S.; Pinals, R.L.; Chang, R.; Landry, M.P. Carbon Nanocarriers Deliver SiRNA to Intact Plant Cells for Efficient Gene Knockdown. Sci. Adv. 2020, 6, eaaz0495. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, N.; Sasaki, S.; Yamagata, K.; Komori, S.; Nagase, M.; Wada, M.; Yamamoto, T.; Yoshikawa, N. Promotion of Flowering and Reduction of a Generation Time in Apple Seedlings by Ectopical Expression of the Arabidopsis thaliana FT Gene Using the Apple Latent Spherical Virus Vector. Plant Mol. Biol. 2011, 75, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Feeney, O.; Cockbain, J.; Morrison, M.; Diependaele, L.; Van Assche, K.; Sterckx, S. Patenting Foundational Technologies: Lessons From CRISPR and Other Core Biotechnologies. Am. J. Bioeth. 2018, 18, 36–48. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).